

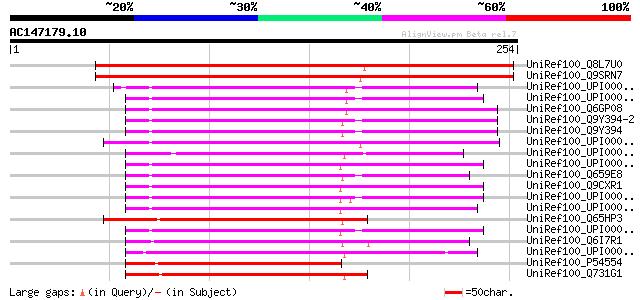
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147179.10 - phase: 1 /pseudo
(254 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8L7U0 AT3g03330/T21P5_25 [Arabidopsis thaliana] 314 1e-84
UniRef100_Q9SRN7 T21P5.25 protein [Arabidopsis thaliana] 307 2e-82
UniRef100_UPI00003AE075 UPI00003AE075 UniRef100 entry 127 3e-28
UniRef100_UPI00003AE074 UPI00003AE074 UniRef100 entry 126 6e-28
UniRef100_Q6GP08 MGC80755 protein [Xenopus laevis] 125 8e-28
UniRef100_Q9Y394-2 Splice isoform 2 of Q9Y394 [Homo sapiens] 125 1e-27
UniRef100_Q9Y394 Dehydrogenase/reductase SDR family member 7 pre... 125 1e-27
UniRef100_UPI00002E4D3C UPI00002E4D3C UniRef100 entry 119 6e-26
UniRef100_UPI0000432199 UPI0000432199 UniRef100 entry 118 1e-25
UniRef100_UPI0000247A31 UPI0000247A31 UniRef100 entry 117 2e-25
UniRef100_Q659E8 Hypothetical protein DKFZp564H1664 [Homo sapiens] 115 1e-24
UniRef100_Q9CXR1 Dehydrogenase/reductase SDR family member 7 pre... 112 9e-24
UniRef100_UPI000036210E UPI000036210E UniRef100 entry 111 2e-23
UniRef100_UPI00001CF9D7 UPI00001CF9D7 UniRef100 entry 110 3e-23
UniRef100_Q65HP3 YqjQ [Bacillus licheniformis] 107 3e-22
UniRef100_UPI00002BB840 UPI00002BB840 UniRef100 entry 104 2e-21
UniRef100_Q6I7R1 Down-regulated in nephrectomized rat kidney #3 ... 103 4e-21
UniRef100_UPI000042928A UPI000042928A UniRef100 entry 102 9e-21
UniRef100_P54554 Hypothetical oxidoreductase yqjQ [Bacillus subt... 96 7e-19
UniRef100_Q731G1 Oxidoreductase, short-chain dehydrogenase/reduc... 87 5e-16
>UniRef100_Q8L7U0 AT3g03330/T21P5_25 [Arabidopsis thaliana]
Length = 328
Score = 314 bits (805), Expect = 1e-84
Identities = 151/214 (70%), Positives = 181/214 (84%), Gaps = 5/214 (2%)
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
+ VV++A SLFP +GVDY++HNAA+ERPKS D +EE+LK TFDVNVFGTI+LT+L+ P
Sbjct: 111 KHVVEQAVSLFPGAGVDYLVHNAAFERPKSKASDASEETLKTTFDVNVFGTISLTKLVAP 170
Query: 104 FMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGP 163
ML++G GHFVV+SSAAGK P+PGQA+YSASK+AL+GYFHSLRSE CQKGI+VTVVCPGP
Sbjct: 171 HMLKQGGGHFVVISSAAGKVPSPGQAIYSASKHALHGYFHSLRSEFCQKGIKVTVVCPGP 230
Query: 164 IETANNSGSQVPS-----EKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMP 218
IET N +G+ EKRVS+E+C ELTIIAA+H LKEAWISYQPVL VMYLVQYMP
Sbjct: 231 IETWNGTGTSTSEDSKSPEKRVSSERCAELTIIAASHNLKEAWISYQPVLLVMYLVQYMP 290
Query: 219 TIGYWLMDKVGKNRVEAAKEKGNAYSLSLLFGKK 252
++G+WLMDKVG RVE A++KGN YS LLFGKK
Sbjct: 291 SLGFWLMDKVGGKRVEVAEKKGNTYSWDLLFGKK 324
>UniRef100_Q9SRN7 T21P5.25 protein [Arabidopsis thaliana]
Length = 345
Score = 307 bits (786), Expect = 2e-82
Identities = 150/231 (64%), Positives = 182/231 (77%), Gaps = 22/231 (9%)
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
+ VV++A SLFP +GVDY++HNAA+ERPKS D +EE+LK TFDVNVFGTI+LT+L+ P
Sbjct: 111 KHVVEQAVSLFPGAGVDYLVHNAAFERPKSKASDASEETLKTTFDVNVFGTISLTKLVAP 170
Query: 104 FMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGP 163
ML++G GHFVV+SSAAGK P+PGQA+YSASK+AL+GYFHSLRSE CQKGI+VTVVCPGP
Sbjct: 171 HMLKQGGGHFVVISSAAGKVPSPGQAIYSASKHALHGYFHSLRSEFCQKGIKVTVVCPGP 230
Query: 164 IETANNSGSQV----------------------PSEKRVSAEKCVELTIIAATHGLKEAW 201
IET N +G+ ++KRVS+E+C ELTIIAA+H LKEAW
Sbjct: 231 IETWNGTGTSTSEDSKSPELLAQLDFSQASILFATQKRVSSERCAELTIIAASHNLKEAW 290
Query: 202 ISYQPVLAVMYLVQYMPTIGYWLMDKVGKNRVEAAKEKGNAYSLSLLFGKK 252
ISYQPVL VMYLVQYMP++G+WLMDKVG RVE A++KGN YS LLFGKK
Sbjct: 291 ISYQPVLLVMYLVQYMPSLGFWLMDKVGGKRVEVAEKKGNTYSWDLLFGKK 341
>UniRef100_UPI00003AE075 UPI00003AE075 UniRef100 entry
Length = 304
Score = 127 bits (319), Expect = 3e-28
Identities = 72/197 (36%), Positives = 113/197 (56%), Gaps = 21/197 (10%)
Query: 53 LFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGH 112
+FP+ +D +++N + +S +D + A ++N GTI+LT+ + M+ R +G
Sbjct: 114 IFPE--IDVLVNNGGRSQ-RSLFVDTNLDVFSAIMELNYLGTISLTKHVLNHMIERRRGK 170
Query: 113 FVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPIETA---- 167
V +SS G AP + Y ASK+AL G+F+SLR+EL I + +CPGP+++
Sbjct: 171 IVTVSSVMGIMGAPLASGYCASKHALQGFFNSLRTELTDYPEISIINICPGPVQSKIIQN 230
Query: 168 ----------NNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYM 217
NSG Q ++ E+CV LT+++ + LKEAWIS P LAV YL QY
Sbjct: 231 VFTEKLDKSIENSGDQ---SHKMPTERCVRLTLVSTANDLKEAWISDHPYLAVCYLWQYA 287
Query: 218 PTIGYWLMDKVGKNRVE 234
PT +WLM+++GK R++
Sbjct: 288 PTWAWWLMNRMGKRRIQ 304
>UniRef100_UPI00003AE074 UPI00003AE074 UniRef100 entry
Length = 319
Score = 126 bits (316), Expect = 6e-28
Identities = 71/194 (36%), Positives = 110/194 (56%), Gaps = 19/194 (9%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++N + +S +D + A ++N GTI+LT+ + M+ R +G V +SS
Sbjct: 127 IDVLVNNGGRSQ-RSLFVDTNLDVFSAIMELNYLGTISLTKHVLNHMIERRRGKIVTVSS 185
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPIETA---------- 167
G AP + Y ASK+AL G+F+SLR+EL I + +CPGP+++
Sbjct: 186 VMGIMGAPLASGYCASKHALQGFFNSLRTELTDYPEISIINICPGPVQSKIIQNVFTEKL 245
Query: 168 ----NNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYW 223
NSG Q ++ E+CV LT+++ + LKEAWIS P LAV YL QY PT +W
Sbjct: 246 DKSIENSGDQ---SHKMPTERCVRLTLVSTANDLKEAWISDHPYLAVCYLWQYAPTWAWW 302
Query: 224 LMDKVGKNRVEAAK 237
LM+++GK R++ K
Sbjct: 303 LMNRMGKRRIQNFK 316
>UniRef100_Q6GP08 MGC80755 protein [Xenopus laevis]
Length = 336
Score = 125 bits (315), Expect = 8e-28
Identities = 66/198 (33%), Positives = 115/198 (57%), Gaps = 13/198 (6%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++NA + +S ++ + +A ++N GTI++T+ + P M++R +G + +SS
Sbjct: 132 IDILVNNAGRSQ-RSLYVETNLDVFRALIELNYLGTISITKHVLPHMIQRKRGRIINISS 190
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPIETA---------- 167
AG AP Y ASK+AL G+F+SLR+EL I ++ +CPGP+++
Sbjct: 191 VAGLIGAPLSTGYCASKHALQGFFNSLRTELTTYPDIIISNICPGPVQSKIVENAITEEC 250
Query: 168 -NNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYWLMD 226
S + +++ +C +L +I A + LKE WIS QP+L + YL QY+PT+ +W+
Sbjct: 251 DKVSSIKTDQSHKMATSRCAQLILITAANNLKETWISAQPILIIYYLWQYIPTLAWWITA 310
Query: 227 KVGKNRVEAAKEKGNAYS 244
KVG+ R++ K +A S
Sbjct: 311 KVGERRIKNFKSGVDADS 328
>UniRef100_Q9Y394-2 Splice isoform 2 of Q9Y394 [Homo sapiens]
Length = 289
Score = 125 bits (314), Expect = 1e-27
Identities = 70/201 (34%), Positives = 112/201 (54%), Gaps = 19/201 (9%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++N + +S +D + + + ++N GT++LT+ + P M+ R +G V ++S
Sbjct: 82 IDILVNNGGMSQ-RSLCMDTSLDVYRKLIELNYLGTVSLTKCVLPHMIERKQGKIVTVNS 140
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPIE------------ 165
G P Y ASK+AL G+F+ LR+EL GI V+ +CPGP++
Sbjct: 141 ILGIISVPLSIGYCASKHALRGFFNGLRTELATYPGIIVSNICPGPVQSNIVENSLAGEV 200
Query: 166 --TANNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYW 223
T N+G Q +++ +CV L +I+ + LKE WIS QP L V YL QYMPT +W
Sbjct: 201 TKTIGNNGDQ---SHKMTTSRCVRLMLISMANDLKEVWISEQPFLLVTYLWQYMPTWAWW 257
Query: 224 LMDKVGKNRVEAAKEKGNAYS 244
+ +K+GK R+E K +A S
Sbjct: 258 ITNKMGKKRIENFKSGVDADS 278
>UniRef100_Q9Y394 Dehydrogenase/reductase SDR family member 7 precursor [Homo
sapiens]
Length = 339
Score = 125 bits (314), Expect = 1e-27
Identities = 70/201 (34%), Positives = 112/201 (54%), Gaps = 19/201 (9%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++N + +S +D + + + ++N GT++LT+ + P M+ R +G V ++S
Sbjct: 132 IDILVNNGGMSQ-RSLCMDTSLDVYRKLIELNYLGTVSLTKCVLPHMIERKQGKIVTVNS 190
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPIE------------ 165
G P Y ASK+AL G+F+ LR+EL GI V+ +CPGP++
Sbjct: 191 ILGIISVPLSIGYCASKHALRGFFNGLRTELATYPGIIVSNICPGPVQSNIVENSLAGEV 250
Query: 166 --TANNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYW 223
T N+G Q +++ +CV L +I+ + LKE WIS QP L V YL QYMPT +W
Sbjct: 251 TKTIGNNGDQ---SHKMTTSRCVRLMLISMANDLKEVWISEQPFLLVTYLWQYMPTWAWW 307
Query: 224 LMDKVGKNRVEAAKEKGNAYS 244
+ +K+GK R+E K +A S
Sbjct: 308 ITNKMGKKRIENFKSGVDADS 328
>UniRef100_UPI00002E4D3C UPI00002E4D3C UniRef100 entry
Length = 285
Score = 119 bits (299), Expect = 6e-26
Identities = 73/230 (31%), Positives = 114/230 (48%), Gaps = 33/230 (14%)
Query: 48 DEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLR 107
D A+ D GVDY+ H A + ++ D +E + F++N I LT+ L P ML
Sbjct: 56 DGADGTDADIGVDYLFHVAGGSQ-HAAAEDTADEVDRDMFELNTMSAIALTKALLPSMLA 114
Query: 108 RGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETA 167
R KG+ V + S A K PAPGQA Y+A+K AL+ + HSLR E+ +G+ VTV PGPI T
Sbjct: 115 RRKGNIVAIGSMAVKCPAPGQATYAATKAALSAFCHSLRGEVADRGVSVTVAHPGPIATG 174
Query: 168 NNSGSQV--------------------------------PSEKRVSAEKCVELTIIAATH 195
++V E+R++AE + A
Sbjct: 175 LGGQTRVIFGATLAKSTCDTAADAAAAAAADPTRGAKVGKKERRLNAEWVAKRIAAGAAL 234
Query: 196 GLKEAWISYQPVLAVMYLVQYMPTIGYWLMDKVGKNRVEAAKEKGNAYSL 245
G E ++ QP++ + +Q++P++G+ +++KVG R AA+ + Y L
Sbjct: 235 GFDEVVLATQPIMLLRNFMQFVPSVGFRVLNKVGPKRTRAARLGVSMYEL 284
>UniRef100_UPI0000432199 UPI0000432199 UniRef100 entry
Length = 172
Score = 118 bits (296), Expect = 1e-25
Identities = 67/175 (38%), Positives = 101/175 (57%), Gaps = 9/175 (5%)
Query: 59 VDYMIHNAAY-ERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMS 117
+D +++NA +R K ++++ + K FD+NVF TI L+RL+ + + +GHFV+ S
Sbjct: 1 LDILVNNAGRSQRAKWENIELSVD--KELFDLNVFSTIALSRLVAKYFFQMNEGHFVINS 58
Query: 118 SAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET-----ANNSGS 172
S AG T P A Y ASK +L+ YF SL E K I +T+VCPGP+ET + S
Sbjct: 59 SIAGVTAVPFSATYCASKSSLHAYFESLSIEKINKNISITIVCPGPVETNFLAESFTEKS 118
Query: 173 QVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYWLMDK 227
+ P+ R++AE+C L IA + L WI +L ++YL Y P +G W + K
Sbjct: 119 EKPTH-RMTAERCATLIGIAIANKLSVVWICKSIILQMVYLRIYYPNVGTWYVGK 172
>UniRef100_UPI0000247A31 UPI0000247A31 UniRef100 entry
Length = 330
Score = 117 bits (294), Expect = 2e-25
Identities = 64/191 (33%), Positives = 107/191 (55%), Gaps = 13/191 (6%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +I+N + ++ +D + +A ++N GT+++T+ + P M+RRG G +SS
Sbjct: 129 IDVLINNGGRSQ-RALCVDADVDVYQALMELNYLGTVSITKQVLPHMIRRGTGIIATVSS 187
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPI-----------ET 166
AG AP Y+ASK+AL G+F+SLR+EL I ++ +CPGP+ E
Sbjct: 188 VAGFVGAPLATGYAASKHALQGFFNSLRTELSDCPNILISNICPGPVISSIVQNAFTEEL 247
Query: 167 ANNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYWLMD 226
+ ++S E+CV LT++ + +KE WI+ QP L Y+ QY PT+ ++L +
Sbjct: 248 GKPVATAGDQTHKMSTERCVHLTLVGLANRVKEMWIAEQPFLLFCYVWQYTPTLAWYLTN 307
Query: 227 KVGKNRVEAAK 237
+GK RV+ K
Sbjct: 308 VLGKKRVQNFK 318
>UniRef100_Q659E8 Hypothetical protein DKFZp564H1664 [Homo sapiens]
Length = 375
Score = 115 bits (288), Expect = 1e-24
Identities = 63/186 (33%), Positives = 103/186 (54%), Gaps = 18/186 (9%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++N + +S +D + + + ++N GT++LT+ + P M+ R +G V ++S
Sbjct: 132 IDILVNNGGMSQ-RSLCMDTSLDVYRKLIELNYLGTVSLTKCVLPHMIERKQGKIVTVNS 190
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPIE------------ 165
G P Y ASK+AL G+F+ LR+EL GI V+ +CPGP++
Sbjct: 191 ILGIISVPLSIGYCASKHALRGFFNGLRTELATYPGIIVSNICPGPVQSNIVENSLAEVT 250
Query: 166 -TANNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYWL 224
T N+G Q +++ +CV L +I+ + LKE WIS QP L V YL QYMPT +W+
Sbjct: 251 KTIGNNGDQ---SHKMTTSRCVRLMLISMANDLKEVWISEQPFLLVTYLWQYMPTWAWWI 307
Query: 225 MDKVGK 230
+ G+
Sbjct: 308 TTRWGR 313
>UniRef100_Q9CXR1 Dehydrogenase/reductase SDR family member 7 precursor [Mus
musculus]
Length = 338
Score = 112 bits (280), Expect = 9e-24
Identities = 65/191 (34%), Positives = 102/191 (53%), Gaps = 13/191 (6%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++N + +S VL+ + K ++N GT++LT+ + P M+ R +G V ++S
Sbjct: 132 IDILVNNGGRSQ-RSLVLETNLDVFKELINLNYIGTVSLTKCVLPHMIERKQGKIVTVNS 190
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPI-----------ET 166
AG + Y ASK+AL G+F++L SEL Q GI V PGP+ E
Sbjct: 191 IAGIASVSLSSGYCASKHALRGFFNALHSELGQYPGITFCNVYPGPVQSDIVKNAFTEEV 250
Query: 167 ANNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYWLMD 226
+ + + ++ +CV L +I+ + LKE WIS PVL Y+ QYMPT WL
Sbjct: 251 TKSMRNNIDQSYKMPTSRCVRLMLISMANDLKEVWISDHPVLLGAYIWQYMPTWAAWLNC 310
Query: 227 KVGKNRVEAAK 237
K+GK R++ K
Sbjct: 311 KLGKERIQNFK 321
>UniRef100_UPI000036210E UPI000036210E UniRef100 entry
Length = 319
Score = 111 bits (278), Expect = 2e-23
Identities = 64/195 (32%), Positives = 108/195 (54%), Gaps = 20/195 (10%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +I+N + +S +D + + +A ++N GT+++T+ + P M +R G V +SS
Sbjct: 126 IDILINNGGRTQ-RSLCIDTSVDVYQALMELNFLGTVSITKQVLPHMTQRRAGSIVTVSS 184
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPI--ETANN------ 169
G AP Y+A+K+AL G+F+SLR+EL I ++ VCPGP+ + N
Sbjct: 185 VVGLAAAPLGTGYAATKHALQGFFNSLRTELTDFPNIHISTVCPGPVVSKIVQNAFTEEV 244
Query: 170 -------SGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGY 222
SGSQ E +++ +CV L ++ + +KE WI+ QP L YL QY PT+ +
Sbjct: 245 DKVMRFISGSQ---EHKMATSRCVRLMLVGIANNIKEMWIAQQPFLLFYYLWQYTPTLAW 301
Query: 223 WLMDKVGKNRVEAAK 237
+ + +G+ RV+ K
Sbjct: 302 CVTNILGRKRVQNFK 316
>UniRef100_UPI00001CF9D7 UPI00001CF9D7 UniRef100 entry
Length = 338
Score = 110 bits (276), Expect = 3e-23
Identities = 63/188 (33%), Positives = 101/188 (53%), Gaps = 13/188 (6%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++N + +S VL+ E K ++N GT++LT+ + P M+ R +G V ++S
Sbjct: 132 IDILVNNGGRSQ-RSLVLETNLEVFKELMNLNYLGTVSLTKCVLPHMVERKQGKIVTVNS 190
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPI-----------ET 166
AG + Y ASK+AL G+F++L SEL + GI + V PGP+ E
Sbjct: 191 LAGIASVSLSSGYCASKHALRGFFNALHSELGKYPGITLCNVYPGPVQSNVVKNALTEEL 250
Query: 167 ANNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYWLMD 226
+ ++ +CV L +I+ + LKE WIS QP+L Y+ QYMPT WL
Sbjct: 251 TKPMRENIDQSYKMPTSRCVRLMLISMANDLKEVWISDQPILLGAYIWQYMPTWALWLTC 310
Query: 227 KVGKNRVE 234
K+G+ R++
Sbjct: 311 KLGEKRIQ 318
>UniRef100_Q65HP3 YqjQ [Bacillus licheniformis]
Length = 260
Score = 107 bits (267), Expect = 3e-22
Identities = 59/138 (42%), Positives = 89/138 (63%), Gaps = 7/138 (5%)
Query: 48 DEAESLFPDSG-VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFML 106
DE E+ F ++G VD +++NA + +S+ LD + E +KA F+VNVFG I T++ P M+
Sbjct: 70 DEVEAAFQEAGPVDILVNNAGFGIFESA-LDASLEDMKAMFEVNVFGLIACTKMALPHMI 128
Query: 107 RRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIE- 165
R+ KGH + ++S AGK P ++Y+A+K+A+ GY +SLR EL + G+ VT V PGPI+
Sbjct: 129 RQDKGHIINIASQAGKIATPKSSLYAATKHAVLGYSNSLRMELAETGVNVTTVNPGPIQT 188
Query: 166 ----TANNSGSQVPSEKR 179
TA+ G V S R
Sbjct: 189 DFFKTADKKGDYVKSVGR 206
>UniRef100_UPI00002BB840 UPI00002BB840 UniRef100 entry
Length = 337
Score = 104 bits (260), Expect = 2e-21
Identities = 62/195 (31%), Positives = 106/195 (53%), Gaps = 20/195 (10%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGK-GHFVVMS 117
+D +++N+ + +S D + E +A ++N GT+++T+ + M +R + G V +S
Sbjct: 131 IDILVNNSGRSQ-RSLFTDTSLEVYQALMELNFLGTVSITKQVLAHMTQRQRAGTIVTVS 189
Query: 118 SAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPI------------ 164
S G AP Y+A+K+AL G+F+SLR EL I ++ VCPGP+
Sbjct: 190 SIVGLAGAPLGTGYAATKHALQGFFNSLRPELTDYPNIHISTVCPGPVISNIVQNVFTEE 249
Query: 165 --ETANNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGY 222
+ + GSQ E ++ +CV L ++ + +KE WI+ QP L V YL QY PT+ +
Sbjct: 250 VNKPLTSPGSQ---EHKMPTSRCVRLMLVGIANNIKEMWIADQPFLLVFYLWQYTPTLAW 306
Query: 223 WLMDKVGKNRVEAAK 237
+ + +G+ RV+ K
Sbjct: 307 FATNILGRKRVQNFK 321
>UniRef100_Q6I7R1 Down-regulated in nephrectomized rat kidney #3 [Rattus norvegicus]
Length = 324
Score = 103 bits (257), Expect = 4e-21
Identities = 59/183 (32%), Positives = 101/183 (54%), Gaps = 12/183 (6%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++N S V + + K +VN GT++LT+ + P M+ R +G VVM S
Sbjct: 131 IDILVNNGGVAHA-SLVENTNMDIFKVLIEVNYLGTVSLTKCVLPHMMERNQGKIVVMKS 189
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPIE--------TANN 169
G P P + Y+ASK AL G+F LR+EL GI ++++CPGP+ T +
Sbjct: 190 LVGIVPRPLCSGYAASKLALRGFFDVLRTELFDYPGITLSMICPGPVHSNIFQNAFTGDF 249
Query: 170 SGSQVPSEK--RVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYWLMDK 227
+ +++P ++ +CV+L +++ + L++ WI+ QPVL Y+ QY+P + L +
Sbjct: 250 TETRLPKIPLFKMETSRCVQLILVSLANDLEDIWIANQPVLLRAYVWQYVPFRDWILQGR 309
Query: 228 VGK 230
GK
Sbjct: 310 YGK 312
>UniRef100_UPI000042928A UPI000042928A UniRef100 entry
Length = 272
Score = 102 bits (254), Expect = 9e-21
Identities = 61/189 (32%), Positives = 105/189 (55%), Gaps = 15/189 (7%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++N E +D + K +VN GT++LT+ + P M+ R +G V+M+S
Sbjct: 79 IDILVNNGG-ETNYCFAVDANLDVFKVLMEVNYLGTVSLTKSVLPHMMERKQGKIVIMNS 137
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELC-QKGIQVTVVCPGPIET----------A 167
AG P+P Y ASK+AL G+ ++LR+EL GI ++ +CPGP+ + A
Sbjct: 138 LAGIVPSPLCCGYIASKHALRGFANALRTELLDHPGITLSTICPGPVHSNIYQNYLTREA 197
Query: 168 NNSGSQVPSE-KRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYWLMD 226
+ P E +++ +CV+L +++ + L+E WI+ QP L Y+ QY+P I W++
Sbjct: 198 IKARWPKPQEFPKMATSRCVQLILVSMANDLEEVWIANQPALFRTYVWQYVP-IRDWIIS 256
Query: 227 KV-GKNRVE 234
+ K R+E
Sbjct: 257 RTQWKKRIE 265
>UniRef100_P54554 Hypothetical oxidoreductase yqjQ [Bacillus subtilis]
Length = 259
Score = 96.3 bits (238), Expect = 7e-19
Identities = 51/108 (47%), Positives = 73/108 (67%), Gaps = 1/108 (0%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +I+NA + ++ VLD T + +KA FDVNVFG I T+ + P ML + KGH + ++S
Sbjct: 81 IDVLINNAGFGIFET-VLDSTLDDMKAMFDVNVFGLIACTKAVLPQMLEQKKGHIINIAS 139
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
AGK P ++YSA+K+A+ GY ++LR EL GI VT V PGPI+T
Sbjct: 140 QAGKIATPKSSLYSATKHAVLGYSNALRMELSGTGIYVTTVNPGPIQT 187
>UniRef100_Q731G1 Oxidoreductase, short-chain dehydrogenase/reductase family
[Bacillus cereus]
Length = 264
Score = 86.7 bits (213), Expect = 5e-16
Identities = 46/126 (36%), Positives = 77/126 (60%), Gaps = 6/126 (4%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++NA + K+ D + + +K F VNVFG + T+ + P+M++R +GH + ++S
Sbjct: 86 IDILVNNAGFGIFKTFE-DASMDEVKDMFQVNVFGLVACTKAVLPYMVKRNEGHIINVAS 144
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET-----ANNSGSQ 173
AGK P + Y+A+K+A+ G+ +SLR EL I VT + PGPI+T A+ SG+
Sbjct: 145 LAGKIATPKSSAYAATKHAVLGFTNSLRMELSSTNIFVTAINPGPIDTNFFEIADQSGTY 204
Query: 174 VPSEKR 179
V + R
Sbjct: 205 VKNMGR 210
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.134 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 406,240,979
Number of Sequences: 2790947
Number of extensions: 15200586
Number of successful extensions: 48715
Number of sequences better than 10.0: 7863
Number of HSP's better than 10.0 without gapping: 5708
Number of HSP's successfully gapped in prelim test: 2155
Number of HSP's that attempted gapping in prelim test: 41699
Number of HSP's gapped (non-prelim): 7979
length of query: 254
length of database: 848,049,833
effective HSP length: 124
effective length of query: 130
effective length of database: 501,972,405
effective search space: 65256412650
effective search space used: 65256412650
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 73 (32.7 bits)
Medicago: description of AC147179.10


