

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.4 - phase: 0
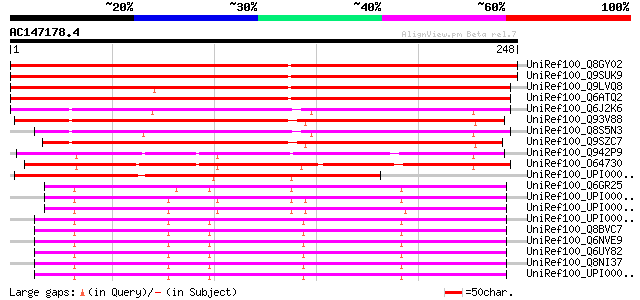
(248 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8GY02 Hypothetical protein At4g16580/dl4315c [Arabido... 357 2e-97
UniRef100_Q9SUK9 Hypothetical protein AT4g16580 [Arabidopsis tha... 357 2e-97
UniRef100_Q9LVQ8 Similarity to unknown protein [Arabidopsis thal... 342 5e-93
UniRef100_Q6ATQ2 Expressed protein [Oryza sativa] 337 2e-91
UniRef100_Q6J2K6 BTH-induced protein phosphatase 1 [Oryza sativa] 187 3e-46
UniRef100_Q93V88 Hypothetical protein At4g33500 [Arabidopsis tha... 177 2e-43
UniRef100_Q8S5N3 Hypothetical protein OJ1003C07.6 [Oryza sativa] 174 2e-42
UniRef100_Q9SZC7 Hypothetical protein F17M5.260 [Arabidopsis tha... 170 3e-41
UniRef100_Q942P9 Putative senescence-associated protein [Oryza s... 155 9e-37
UniRef100_O64730 Expressed protein [Arabidopsis thaliana] 154 2e-36
UniRef100_UPI00002B0976 UPI00002B0976 UniRef100 entry 149 8e-35
UniRef100_Q6GR25 MGC81279 protein [Xenopus laevis] 144 3e-33
UniRef100_UPI0000360007 UPI0000360007 UniRef100 entry 142 8e-33
UniRef100_UPI000043985D UPI000043985D UniRef100 entry 142 1e-32
UniRef100_UPI00001D0928 UPI00001D0928 UniRef100 entry 142 1e-32
UniRef100_Q8BVC7 Mus musculus adult male cecum cDNA, RIKEN full-... 142 1e-32
UniRef100_Q6NVE9 T-cell activation protein phosphatase 2C [Mus m... 142 1e-32
UniRef100_Q6UY82 T-cell activation protein phosphatase 2C-like p... 142 1e-32
UniRef100_Q8NI37 T-cell activation protein phosphatase 2C [Homo ... 142 1e-32
UniRef100_UPI000045667C UPI000045667C UniRef100 entry 140 2e-32
>UniRef100_Q8GY02 Hypothetical protein At4g16580/dl4315c [Arabidopsis thaliana]
Length = 300
Score = 357 bits (915), Expect = 2e-97
Identities = 170/248 (68%), Positives = 209/248 (83%), Gaps = 1/248 (0%)
Query: 1 MLSGSCYLPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSAR 60
++SGSCYLPHPDK ATGGEDAHFIC +EQA+GVADGVGGWA++G++AG Y++EL++NS
Sbjct: 51 LVSGSCYLPHPDKEATGGEDAHFICAEEQALGVADGVGGWAELGIDAGYYSRELMSNSVN 110
Query: 61 AIREEPKGSFNPVRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVI 120
AI++EPKGS +P RVLEKAH+ TK+ GSST CIIAL ++ L+AINLGDSGF+V+R+G +
Sbjct: 111 AIQDEPKGSIDPARVLEKAHTCTKSQGSSTACIIALTNQGLHAINLGDSGFMVVREGHTV 170
Query: 121 FKSPVQQRGFNFPYQLARSGTEGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVG 180
F+SPVQQ FNF YQL SG GDLPSSG+VFTV VAPGD+I+AGTDGLFDN+YNN+I
Sbjct: 171 FRSPVQQHDFNFTYQL-ESGRNGDLPSSGQVFTVAVAPGDVIIAGTDGLFDNLYNNEITA 229
Query: 181 VVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVVVS 240
+VV A RA + PQ TAQKIAALARQRA D RQ+PFS AA + G+R+ GGKLDD+TVVVS
Sbjct: 230 IVVHAVRANIDPQVTAQKIAALARQRAQDKNRQTPFSTAAQDAGFRYYGGKLDDITVVVS 289
Query: 241 YISNSVNE 248
Y++ S E
Sbjct: 290 YVAASKEE 297
>UniRef100_Q9SUK9 Hypothetical protein AT4g16580 [Arabidopsis thaliana]
Length = 335
Score = 357 bits (915), Expect = 2e-97
Identities = 170/248 (68%), Positives = 209/248 (83%), Gaps = 1/248 (0%)
Query: 1 MLSGSCYLPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSAR 60
++SGSCYLPHPDK ATGGEDAHFIC +EQA+GVADGVGGWA++G++AG Y++EL++NS
Sbjct: 86 LVSGSCYLPHPDKEATGGEDAHFICAEEQALGVADGVGGWAELGIDAGYYSRELMSNSVN 145
Query: 61 AIREEPKGSFNPVRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVI 120
AI++EPKGS +P RVLEKAH+ TK+ GSST CIIAL ++ L+AINLGDSGF+V+R+G +
Sbjct: 146 AIQDEPKGSIDPARVLEKAHTCTKSQGSSTACIIALTNQGLHAINLGDSGFMVVREGHTV 205
Query: 121 FKSPVQQRGFNFPYQLARSGTEGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVG 180
F+SPVQQ FNF YQL SG GDLPSSG+VFTV VAPGD+I+AGTDGLFDN+YNN+I
Sbjct: 206 FRSPVQQHDFNFTYQL-ESGRNGDLPSSGQVFTVAVAPGDVIIAGTDGLFDNLYNNEITA 264
Query: 181 VVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVVVS 240
+VV A RA + PQ TAQKIAALARQRA D RQ+PFS AA + G+R+ GGKLDD+TVVVS
Sbjct: 265 IVVHAVRANIDPQVTAQKIAALARQRAQDKNRQTPFSTAAQDAGFRYYGGKLDDITVVVS 324
Query: 241 YISNSVNE 248
Y++ S E
Sbjct: 325 YVAASKEE 332
>UniRef100_Q9LVQ8 Similarity to unknown protein [Arabidopsis thaliana]
Length = 414
Score = 342 bits (877), Expect = 5e-93
Identities = 167/246 (67%), Positives = 207/246 (83%), Gaps = 2/246 (0%)
Query: 1 MLSGSCYLPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSAR 60
++SGSCYLPHP+K ATGGEDAHFIC++EQAIGVADGVGGWA+VGVNAGL+++EL++ S
Sbjct: 170 LVSGSCYLPHPEKEATGGEDAHFICDEEQAIGVADGVGGWAEVGVNAGLFSRELMSYSVS 229
Query: 61 AIREEPKGS-FNPVRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSV 119
AI+E+ KGS +P+ VLEKAHS+TKA GSST CII L D+ L+AINLGDSGF V+R+G+
Sbjct: 230 AIQEQHKGSSIDPLVVLEKAHSQTKAKGSSTACIIVLKDKGLHAINLGDSGFTVVREGTT 289
Query: 120 IFKSPVQQRGFNFPYQLARSGTEGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIV 179
+F+SPVQQ GFNF YQL SG D+PSSG+VFT+ V GD+IVAGTDG++DN+YN +I
Sbjct: 290 VFQSPVQQHGFNFTYQL-ESGNSADVPSSGQVFTIDVQSGDVIVAGTDGVYDNLYNEEIT 348
Query: 180 GVVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVVV 239
GVVV + RA L P+ TAQKIA LARQRA+D KRQSPF+ AA E GYR+ GGKLDD+T VV
Sbjct: 349 GVVVSSVRAGLDPKGTAQKIAELARQRAVDKKRQSPFATAAQEAGYRYYGGKLDDITAVV 408
Query: 240 SYISNS 245
SY+++S
Sbjct: 409 SYVTSS 414
>UniRef100_Q6ATQ2 Expressed protein [Oryza sativa]
Length = 479
Score = 337 bits (863), Expect = 2e-91
Identities = 161/245 (65%), Positives = 204/245 (82%), Gaps = 1/245 (0%)
Query: 1 MLSGSCYLPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSAR 60
++SGSCYLPHP K ATGGED HFIC DEQAIGVADGVGGWAD GV+AGLYA+EL++NS
Sbjct: 233 LVSGSCYLPHPAKEATGGEDGHFICVDEQAIGVADGVGGWADHGVDAGLYAKELMSNSMS 292
Query: 61 AIREEPKGSFNPVRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVI 120
AI++EP+G+ +P RVLEKA++ TKA GSST CI+AL ++ ++A+NLGDSGFI++RDG +
Sbjct: 293 AIKDEPQGTIDPSRVLEKAYTCTKARGSSTACIVALKEQGIHAVNLGDSGFIIVRDGRTV 352
Query: 121 FKSPVQQRGFNFPYQLARSGTEGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVG 180
+SPVQQ FNF YQL SG DLPSS + F PVAPGD+I+AGTDGLFDN+Y+N+I
Sbjct: 353 LRSPVQQHDFNFTYQL-ESGGGSDLPSSAQTFHFPVAPGDVIIAGTDGLFDNLYSNEISA 411
Query: 181 VVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVVVS 240
+VV A R L P+ATA+KIAALA+Q+A+D RQSPF+AAA E GYR+ GGKLDD+TV+VS
Sbjct: 412 IVVEALRTGLEPEATAKKIAALAQQKAMDRNRQSPFAAAAQEAGYRYFGGKLDDITVIVS 471
Query: 241 YISNS 245
Y++++
Sbjct: 472 YVTSA 476
>UniRef100_Q6J2K6 BTH-induced protein phosphatase 1 [Oryza sativa]
Length = 569
Score = 187 bits (474), Expect = 3e-46
Identities = 110/248 (44%), Positives = 147/248 (58%), Gaps = 8/248 (3%)
Query: 1 MLSGSCYLPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSAR 60
+ SG+ LPHP KV TGGEDA+FI D GVADGVG W+ G+NAGLYA+EL+ +
Sbjct: 325 LASGAAMLPHPSKVLTGGEDAYFIACDGW-FGVADGVGQWSFEGINAGLYARELMDGCKK 383
Query: 61 AIREEPKG-SFNPVRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSV 119
A+ E VL KA + ++ GSSTV + + L+A N+GDSGF+VIR+G +
Sbjct: 384 AVMESQGAPEMRTEEVLAKAADEARSPGSSTVLVAHFDGQVLHACNIGDSGFLVIRNGEI 443
Query: 120 IFKSPVQQRGFNFPYQLARSGTEGDLP-SSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDI 178
KS GFNFP Q+ + GD P + +T+ + GD IV TDGLFDN+Y +I
Sbjct: 444 YQKSKPMTYGFNFPLQIEK----GDDPFKLVQKYTIDLQEGDAIVTATDGLFDNVYEEEI 499
Query: 179 VGVVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGY-RFDGGKLDDLTV 237
V+ + A L P A+ + A A++ +SPFS AAL GY + GGKLDD+TV
Sbjct: 500 AAVISKSLEAGLKPSEIAEFLVARAKEVGRSATCRSPFSDAALAVGYLGYSGGKLDDVTV 559
Query: 238 VVSYISNS 245
VVS + S
Sbjct: 560 VVSVVRKS 567
>UniRef100_Q93V88 Hypothetical protein At4g33500 [Arabidopsis thaliana]
Length = 724
Score = 177 bits (450), Expect = 2e-43
Identities = 101/242 (41%), Positives = 147/242 (60%), Gaps = 7/242 (2%)
Query: 3 SGSCYLPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSARAI 62
SG L P K G EDA+FI IG+ADGV W+ G+N G+YAQEL++N + I
Sbjct: 483 SGFASLQSPFKALAGREDAYFISHHNW-IGIADGVSQWSFEGINKGMYAQELMSNCEKII 541
Query: 63 REEPKGSFNPVRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVIFK 122
E +PV+VL ++ ++TK+ GSST I L + L+ N+GDSGF+VIRDG+V+
Sbjct: 542 SNETAKISDPVQVLHRSVNETKSSGSSTALIAHLDNNELHIANIGDSGFMVIRDGTVLQN 601
Query: 123 SPVQQRGFNFPYQLARSGTEG-DLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVGV 181
S F FP + T+G D+ EV+ V + GD+++A TDGLFDN+Y +IV +
Sbjct: 602 SSPMFHHFCFPLHI----TQGCDVLKLAEVYHVNLEEGDVVIAATDGLFDNLYEKEIVSI 657
Query: 182 VVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGYR-FDGGKLDDLTVVVS 240
V G+ + L PQ A+ +AA A++ ++PF+ AA E GY GGKLD +TV++S
Sbjct: 658 VCGSLKQSLEPQKIAELVAAKAQEVGRSKTERTPFADAAKEEGYNGHKGGKLDAVTVIIS 717
Query: 241 YI 242
++
Sbjct: 718 FV 719
>UniRef100_Q8S5N3 Hypothetical protein OJ1003C07.6 [Oryza sativa]
Length = 496
Score = 174 bits (440), Expect = 2e-42
Identities = 105/236 (44%), Positives = 141/236 (59%), Gaps = 8/236 (3%)
Query: 13 KVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSARAIRE-EPKGSFN 71
K ATGGEDA+FI D GVADGVG W+ G+NAGLYA+EL+ + I E +
Sbjct: 264 KAATGGEDAYFIACDGW-FGVADGVGQWSFEGINAGLYARELMDGCKKFIMENQGAADIK 322
Query: 72 PVRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVIFKSPVQQRGFN 131
P +VL KA + + GSSTV + + LNA N+GDSGF+VIR+G V KS GFN
Sbjct: 323 PEQVLSKAADEAHSPGSSTVLVAHFDGQFLNASNIGDSGFLVIRNGEVYQKSKPMVYGFN 382
Query: 132 FPYQLARSGTEGDLP-SSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVGVVVGATRARL 190
FP Q+ + GD P + +T+ + GD+IV +DGLFDN+Y ++ +V + +A L
Sbjct: 383 FPLQIEK----GDNPLKLVQNYTIELEDGDVIVTASDGLFDNVYEQEVATMVSKSLQADL 438
Query: 191 GPQATAQKIAALARQRALDTKRQSPFSAAALEYGY-RFDGGKLDDLTVVVSYISNS 245
P A+ +AA A++ +PFS AAL GY F GGKLDD+ VVVS + S
Sbjct: 439 KPTEIAEHLAAKAQEVGRSAAGSTPFSDAALAVGYLGFSGGKLDDIAVVVSIVRKS 494
>UniRef100_Q9SZC7 Hypothetical protein F17M5.260 [Arabidopsis thaliana]
Length = 1066
Score = 170 bits (431), Expect = 3e-41
Identities = 96/227 (42%), Positives = 141/227 (61%), Gaps = 7/227 (3%)
Query: 17 GGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSARAIREEPKGSFNPVRVL 76
G EDA+FI IG+ADGV W+ G+N G+YAQEL++N + I E +PV+VL
Sbjct: 480 GREDAYFISHHNW-IGIADGVSQWSFEGINKGMYAQELMSNCEKIISNETAKISDPVQVL 538
Query: 77 EKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVIFKSPVQQRGFNFPYQL 136
++ ++TK+ GSST I L + L+ N+GDSGF+VIRDG+V+ S F FP +
Sbjct: 539 HRSVNETKSSGSSTALIAHLDNNELHIANIGDSGFMVIRDGTVLQNSSPMFHHFCFPLHI 598
Query: 137 ARSGTEG-DLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVGVVVGATRARLGPQAT 195
T+G D+ EV+ V + GD+++A TDGLFDN+Y +IV +V G+ + L PQ
Sbjct: 599 ----TQGCDVLKLAEVYHVNLEEGDVVIAATDGLFDNLYEKEIVSIVCGSLKQSLEPQKI 654
Query: 196 AQKIAALARQRALDTKRQSPFSAAALEYGYR-FDGGKLDDLTVVVSY 241
A+ +AA A++ ++PF+ AA E GY GGKLD +TV++S+
Sbjct: 655 AELVAAKAQEVGRSKTERTPFADAAKEEGYNGHKGGKLDAVTVIISF 701
>UniRef100_Q942P9 Putative senescence-associated protein [Oryza sativa]
Length = 331
Score = 155 bits (392), Expect = 9e-37
Identities = 95/253 (37%), Positives = 153/253 (59%), Gaps = 21/253 (8%)
Query: 4 GSCYLPHPDKVATGGEDAHFICEDEQAI-GVADGVGGWADVGVNAGLYAQELVANSARAI 62
G+ +PHP K TGGEDA F+ D+ + VADGV GWA+ VN L+++EL+A+++ +
Sbjct: 47 GTHLIPHPRKAETGGEDAFFVNGDDGGVFAVADGVSGWAEKDVNPALFSRELMAHTSTFL 106
Query: 63 REEPKGSFNPVRVLEKAHSKTKAMGSSTVCIIALIDEA--LNAINLGDSGFIVIRDGSVI 120
++E + + +P +L KAH+ T ++GS+TV IIA++++ L ++GD G VIR G V+
Sbjct: 107 KDE-EVNHDPQLLLMKAHAATTSVGSATV-IIAMLEKTGILKIASVGDCGLKVIRKGQVM 164
Query: 121 FKSPVQQRGFNFPYQLARSGTEGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVG 180
F + Q+ F+ PYQL+ S G V TV + GD+IV+G+DG FDN+++ +IV
Sbjct: 165 FSTCPQEHYFDCPYQLS-SEAIGQTYLDALVCTVNLMEGDMIVSGSDGFFDNIFDQEIVS 223
Query: 181 VVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGY-----------RFDG 229
V+ + G A+ +A LAR+ ++D SP+S A G+ + G
Sbjct: 224 VISESP----GVDEAAKALAELARKHSVDVTFDSPYSMEARSRGFDVPSWKKFIGGKLIG 279
Query: 230 GKLDDLTVVVSYI 242
GK+DD+TV+V+ +
Sbjct: 280 GKMDDITVIVAQV 292
>UniRef100_O64730 Expressed protein [Arabidopsis thaliana]
Length = 298
Score = 154 bits (390), Expect = 2e-36
Identities = 91/253 (35%), Positives = 157/253 (61%), Gaps = 23/253 (9%)
Query: 8 LPHPDKVATGGEDAHFICEDEQAI-GVADGVGGWADVGVNAGLYAQELVANSARAIREEP 66
+PHPDKV GGEDA F+ + VADGV GWA+ V+ L+++EL+AN++R + ++
Sbjct: 54 IPHPDKVEKGGEDAFFVSSYRGGVMAVADGVSGWAEQDVDPSLFSKELMANASRLV-DDQ 112
Query: 67 KGSFNPVRVLEKAHSKTKAMGSSTVCIIALIDEA--LNAINLGDSGFIVIRDGSVIFKSP 124
+ ++P +++KAH+ T + GS+T+ I+A+++E L N+GD G ++R+G +IF +
Sbjct: 113 EVRYDPGFLIDKAHTATTSRGSATI-ILAMLEEVGILKIGNVGDCGLKLLREGQIIFATA 171
Query: 125 VQQRGFNFPYQLARSGT-EGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVGVVV 183
Q+ F+ PYQL+ G+ + L +S + V V GD+IV G+DGLFDN+++++IV +V
Sbjct: 172 PQEHYFDCPYQLSSEGSAQTYLDASFSI--VEVQKGDVIVMGSDGLFDNVFDHEIVSIVT 229
Query: 184 GATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGY-----------RFDGGKL 232
T +++ +A +A + DT+ +SP++ A G+ + GGKL
Sbjct: 230 KHTDV----AESSRLLAEVASSHSRDTEFESPYALEARAKGFDVPLWKKVLGKKLTGGKL 285
Query: 233 DDLTVVVSYISNS 245
DD+TV+V+ + +S
Sbjct: 286 DDVTVIVAKVVSS 298
>UniRef100_UPI00002B0976 UPI00002B0976 UniRef100 entry
Length = 236
Score = 149 bits (375), Expect = 8e-35
Identities = 76/182 (41%), Positives = 115/182 (62%), Gaps = 6/182 (3%)
Query: 3 SGSCYLPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSARAI 62
+G+ +PHPDK GGEDA F+ + + A GV DGVGGWA+ GV+ Y+++ SA+++
Sbjct: 58 AGAILVPHPDKADKGGEDACFVLKRKGAFGVFDGVGGWAEEGVDPAEYSEQFAERSAQSV 117
Query: 63 REEPKGSFNPVRVLEKAHSKTKAMGSSTVCIIALID-EALNAINLGDSGFIVIRDGSVIF 121
G +PV+V+++AH KT+ +GS T CI L D L+ NLGD+G ++ R G IF
Sbjct: 118 L---NGERDPVKVMKEAHEKTQVIGSCTACITVLKDGNVLDIANLGDAGVMMARKGKTIF 174
Query: 122 KSPVQQRGFNFPYQL--ARSGTEGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIV 179
++ QQ FN PYQL + EGD PS + + + PGD IV G+DGL+DN+ ++++
Sbjct: 175 RTESQQHEFNLPYQLGWVKVYPEGDKPSCADRTEIEMKPGDAIVLGSDGLWDNVPHDEVA 234
Query: 180 GV 181
+
Sbjct: 235 AL 236
>UniRef100_Q6GR25 MGC81279 protein [Xenopus laevis]
Length = 297
Score = 144 bits (362), Expect = 3e-33
Identities = 91/242 (37%), Positives = 128/242 (52%), Gaps = 16/242 (6%)
Query: 18 GEDAHFICEDEQA--IGVADGVGGWADVGVNAGLYAQELVANSARAIREEPKGSFNPVRV 75
G+DA FI A +GVADGVGGW D GV+ +++ L+ R ++E NPV +
Sbjct: 52 GDDACFIARHRTADVLGVADGVGGWRDYGVDPSQFSETLMRTCERLVKEGRFVPTNPVGI 111
Query: 76 LEKAH-----SKTKAMGSSTVCIIAL--IDEALNAINLGDSGFIVIRDGSVIFKSPVQQR 128
L ++ +K +GSST C++ L L+ NLGDSGF+V+R G V+ +S QQ
Sbjct: 112 LTSSYRELLQNKVPLLGSSTACLVVLDRTSHRLHTANLGDSGFLVVRAGEVVHRSDEQQH 171
Query: 129 GFNFPYQL------ARSGTEGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVGVV 182
FN P+QL A D P + + + V GDII+ TDGLFDNM + I+ +
Sbjct: 172 YFNTPFQLSIAPPEAEGAVLSDSPDAADSNSFDVQLGDIILTATDGLFDNMPDYMILQEL 231
Query: 183 VGATRARL-GPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVVVSY 241
Q TA+ IA A A D SPF+ A +YG GGK DD+TV++S
Sbjct: 232 KKLKNTNYESIQQTARSIAEQAHDLAYDPNYMSPFAQFACDYGLNVRGGKPDDITVLLSI 291
Query: 242 IS 243
++
Sbjct: 292 VA 293
>UniRef100_UPI0000360007 UPI0000360007 UniRef100 entry
Length = 297
Score = 142 bits (358), Expect = 8e-33
Identities = 96/242 (39%), Positives = 130/242 (53%), Gaps = 16/242 (6%)
Query: 18 GEDAHFICEDEQA--IGVADGVGGWADVGVNAGLYAQELVANSARAIREEPKGSFNPVRV 75
G+DA FI A +GVADGVGGW D GV+ ++ L+ R ++E +PV V
Sbjct: 52 GDDACFIARHRSADVLGVADGVGGWRDYGVDPSQFSSTLMKTCERLVKEGRFVPSSPVGV 111
Query: 76 L-----EKAHSKTKAMGSSTVCIIALIDEA--LNAINLGDSGFIVIRDGSVIFKSPVQQR 128
L E +K +GSST CI+ L ++ L+ NLGDSGF+V+R G V+ +S QQ
Sbjct: 112 LTTSYYELLQNKVPLLGSSTACIVILDRQSHRLHTANLGDSGFLVVRGGEVVHRSDEQQH 171
Query: 129 GFNFPYQL--ARSGTEG----DLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVGVV 182
FN P+QL A G EG D P + + + V GDII+ TDGLFDNM + I+ +
Sbjct: 172 YFNTPFQLSIAPPGAEGAVLSDSPDAADSSSFDVQLGDIILTATDGLFDNMPDYMILQEL 231
Query: 183 VGATRARL-GPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVVVSY 241
Q TAQ IA A A D SPF+ A + G GGK DD+TV++S
Sbjct: 232 KKLKNTNYESIQQTAQSIAEQAHILAYDPNYMSPFAQFACDNGLNVRGGKPDDITVLLSI 291
Query: 242 IS 243
++
Sbjct: 292 VA 293
>UniRef100_UPI000043985D UPI000043985D UniRef100 entry
Length = 297
Score = 142 bits (357), Expect = 1e-32
Identities = 93/242 (38%), Positives = 131/242 (53%), Gaps = 16/242 (6%)
Query: 18 GEDAHFICEDEQA--IGVADGVGGWADVGVNAGLYAQELVANSARAIREEPKGSFNPVRV 75
G+DA FI + A +GVADGVGGW D GV+ ++ L+ R ++E +PV +
Sbjct: 52 GDDACFIARHKSADVLGVADGVGGWRDYGVDPSQFSATLMKTCERLVKEGRFTPSSPVGI 111
Query: 76 L-----EKAHSKTKAMGSSTVCIIALIDEA--LNAINLGDSGFIVIRDGSVIFKSPVQQR 128
L E +K +GSST CI+ L + ++ NLGDSGF+V+R G V+ +S QQ
Sbjct: 112 LTSGYYELLQNKVPLLGSSTACIVVLDRRSHRIHTCNLGDSGFLVVRGGEVVHRSDEQQH 171
Query: 129 GFNFPYQL--ARSGTEG----DLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVGVV 182
FN P+QL A G EG D P + + + V GDII+ TDGLFDNM + I+ +
Sbjct: 172 YFNTPFQLSIAPPGAEGVVLSDSPEAADSSSFDVQLGDIILTATDGLFDNMPDYMILQEL 231
Query: 183 VGATRARLGP-QATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVVVSY 241
Q TA+ IA A + A D SPF+ A + G GGK DD+TV++S
Sbjct: 232 KKLKNTNYDSIQQTARSIAEQAHELAYDPNYMSPFAQFACDNGLNVRGGKPDDITVLLSI 291
Query: 242 IS 243
++
Sbjct: 292 VA 293
>UniRef100_UPI00001D0928 UPI00001D0928 UniRef100 entry
Length = 307
Score = 142 bits (357), Expect = 1e-32
Identities = 93/247 (37%), Positives = 131/247 (52%), Gaps = 16/247 (6%)
Query: 13 KVATGGEDAHFICEDEQA--IGVADGVGGWADVGVNAGLYAQELVANSARAIREEPKGSF 70
K A G+DA F+ A +GVADGVGGW D GV+ ++ L+ R ++E
Sbjct: 57 KGACYGDDACFVARHRSADVLGVADGVGGWRDYGVDPSQFSGTLMRTCERLVKEGRFVPS 116
Query: 71 NPVRVL-----EKAHSKTKAMGSSTVCIIAL--IDEALNAINLGDSGFIVIRDGSVIFKS 123
NPV +L E +K +GSST CI+ L L+ NLGDSGF+V+R G V+ +S
Sbjct: 117 NPVGILTTSYCELLQNKVPLLGSSTACIVVLDRSSHRLHTANLGDSGFLVVRGGEVVHRS 176
Query: 124 PVQQRGFNFPYQLARSGTE------GDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNND 177
QQ FN P+QL+ + E D P + + + V GDII+ TDGLFDNM +
Sbjct: 177 DEQQHYFNTPFQLSIAPPEAEGVVLSDSPDAADSTSFDVQLGDIILTATDGLFDNMPDYM 236
Query: 178 IVGVVVGATRARL-GPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLT 236
I+ + + Q TA+ IA A + A D SPF+ A + G GGK DD+T
Sbjct: 237 ILQELKKLKNSNYESIQRTARSIAEQAHELAYDPNYMSPFAQFACDNGLNVRGGKPDDIT 296
Query: 237 VVVSYIS 243
V++S ++
Sbjct: 297 VLLSIVA 303
>UniRef100_Q8BVC7 Mus musculus adult male cecum cDNA, RIKEN full-length enriched
library, clone:9130017A15 product:weakly similar to
CG12091 PROTEIN [Mus musculus]
Length = 310
Score = 142 bits (357), Expect = 1e-32
Identities = 93/247 (37%), Positives = 131/247 (52%), Gaps = 16/247 (6%)
Query: 13 KVATGGEDAHFICEDEQA--IGVADGVGGWADVGVNAGLYAQELVANSARAIREEPKGSF 70
K A G+DA F+ A +GVADGVGGW D GV+ ++ L+ R ++E
Sbjct: 60 KGACYGDDACFVARHRSADVLGVADGVGGWRDYGVDPSQFSGTLMRTCERLVKEGRFVPS 119
Query: 71 NPVRVL-----EKAHSKTKAMGSSTVCIIAL--IDEALNAINLGDSGFIVIRDGSVIFKS 123
NPV +L E +K +GSST CI+ L L+ NLGDSGF+V+R G V+ +S
Sbjct: 120 NPVGILTTSYCELLQNKVPLLGSSTACIVVLDRSSHRLHTANLGDSGFLVVRGGEVVHRS 179
Query: 124 PVQQRGFNFPYQLARSGTE------GDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNND 177
QQ FN P+QL+ + E D P + + + V GDII+ TDGLFDNM +
Sbjct: 180 DEQQHYFNTPFQLSIAPPEAEGVVLSDSPDAADSTSFDVQLGDIILTATDGLFDNMPDYM 239
Query: 178 IVGVVVGATRARL-GPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLT 236
I+ + + Q TA+ IA A + A D SPF+ A + G GGK DD+T
Sbjct: 240 ILQELKKLKNSNYESIQRTARSIAEQAHELAYDPNYMSPFAQFACDNGLNVRGGKPDDIT 299
Query: 237 VVVSYIS 243
V++S ++
Sbjct: 300 VLLSIVA 306
>UniRef100_Q6NVE9 T-cell activation protein phosphatase 2C [Mus musculus]
Length = 310
Score = 142 bits (357), Expect = 1e-32
Identities = 93/247 (37%), Positives = 131/247 (52%), Gaps = 16/247 (6%)
Query: 13 KVATGGEDAHFICEDEQA--IGVADGVGGWADVGVNAGLYAQELVANSARAIREEPKGSF 70
K A G+DA F+ A +GVADGVGGW D GV+ ++ L+ R ++E
Sbjct: 60 KGACYGDDACFVARHRSADVLGVADGVGGWRDYGVDPSQFSGTLMRTCERLVKEGRFVPS 119
Query: 71 NPVRVL-----EKAHSKTKAMGSSTVCIIAL--IDEALNAINLGDSGFIVIRDGSVIFKS 123
NPV +L E +K +GSST CI+ L L+ NLGDSGF+V+R G V+ +S
Sbjct: 120 NPVGILTTSYCELLQNKVPLLGSSTACIVVLDRSSHRLHTANLGDSGFLVVRGGEVVHRS 179
Query: 124 PVQQRGFNFPYQLARSGTE------GDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNND 177
QQ FN P+QL+ + E D P + + + V GDII+ TDGLFDNM +
Sbjct: 180 DEQQHYFNTPFQLSIAPPEAEGVVLSDSPDAADSTSFDVQLGDIILTATDGLFDNMPDYM 239
Query: 178 IVGVVVGATRARL-GPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLT 236
I+ + + Q TA+ IA A + A D SPF+ A + G GGK DD+T
Sbjct: 240 ILQELKKLKNSNYESIQRTARSIAEQAHELAYDPNYMSPFAQFACDNGLNVRGGKPDDIT 299
Query: 237 VVVSYIS 243
V++S ++
Sbjct: 300 VLLSIVA 306
>UniRef100_Q6UY82 T-cell activation protein phosphatase 2C-like protein [Homo
sapiens]
Length = 303
Score = 142 bits (357), Expect = 1e-32
Identities = 92/247 (37%), Positives = 131/247 (52%), Gaps = 16/247 (6%)
Query: 13 KVATGGEDAHFICEDEQA--IGVADGVGGWADVGVNAGLYAQELVANSARAIREEPKGSF 70
K A G+DA F+ A +GVADGVGGW D GV+ ++ L+ R ++E
Sbjct: 53 KGACYGDDACFVARHRSADVLGVADGVGGWRDYGVDPSQFSGTLMRTCERLVKEGRFVPS 112
Query: 71 NPVRVL-----EKAHSKTKAMGSSTVCIIAL--IDEALNAINLGDSGFIVIRDGSVIFKS 123
NP+ +L E +K +GSST CI+ L L+ NLGDSGF+V+R G V+ +S
Sbjct: 113 NPIGILTTSYRELLQNKVPLLGSSTACIVVLDRTSHRLHTANLGDSGFLVVRGGEVVHRS 172
Query: 124 PVQQRGFNFPYQLARSGTE------GDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNND 177
QQ FN P+QL+ + E D P + + + V GDII+ TDGLFDNM +
Sbjct: 173 DEQQHYFNTPFQLSIAPPEAEGVVLSDSPDAADSTSFDVQLGDIILTATDGLFDNMPDYM 232
Query: 178 IVGVVVGATRARL-GPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLT 236
I+ + + Q TA+ IA A + A D SPF+ A + G GGK DD+T
Sbjct: 233 ILQELKKLKNSNYESIQQTARSIAEQAHELAYDPNYMSPFAQFACDNGLNVRGGKPDDIT 292
Query: 237 VVVSYIS 243
V++S ++
Sbjct: 293 VLLSIVA 299
>UniRef100_Q8NI37 T-cell activation protein phosphatase 2C [Homo sapiens]
Length = 304
Score = 142 bits (357), Expect = 1e-32
Identities = 92/247 (37%), Positives = 131/247 (52%), Gaps = 16/247 (6%)
Query: 13 KVATGGEDAHFICEDEQA--IGVADGVGGWADVGVNAGLYAQELVANSARAIREEPKGSF 70
K A G+DA F+ A +GVADGVGGW D GV+ ++ L+ R ++E
Sbjct: 54 KGACYGDDACFVARHRSADVLGVADGVGGWRDYGVDPSQFSGTLMRTCERLVKEGRFVPS 113
Query: 71 NPVRVL-----EKAHSKTKAMGSSTVCIIAL--IDEALNAINLGDSGFIVIRDGSVIFKS 123
NP+ +L E +K +GSST CI+ L L+ NLGDSGF+V+R G V+ +S
Sbjct: 114 NPIGILTTSYCELLQNKVPLLGSSTACIVVLDRTSHRLHTANLGDSGFLVVRGGEVVHRS 173
Query: 124 PVQQRGFNFPYQLARSGTE------GDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNND 177
QQ FN P+QL+ + E D P + + + V GDII+ TDGLFDNM +
Sbjct: 174 DEQQHYFNTPFQLSIAPPEAEGVVLSDSPDAADSTSFDVQLGDIILTATDGLFDNMPDYM 233
Query: 178 IVGVVVGATRARL-GPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLT 236
I+ + + Q TA+ IA A + A D SPF+ A + G GGK DD+T
Sbjct: 234 ILQELKKLKNSNYESIQQTARSIAEQAHELAYDPNYMSPFAQFACDNGLNVRGGKPDDIT 293
Query: 237 VVVSYIS 243
V++S ++
Sbjct: 294 VLLSIVA 300
>UniRef100_UPI000045667C UPI000045667C UniRef100 entry
Length = 304
Score = 140 bits (354), Expect = 2e-32
Identities = 91/247 (36%), Positives = 131/247 (52%), Gaps = 16/247 (6%)
Query: 13 KVATGGEDAHFICEDEQA--IGVADGVGGWADVGVNAGLYAQELVANSARAIREEPKGSF 70
K A G+DA F+ A +GVADGVGGW D GV+ ++ L+ R ++E
Sbjct: 54 KGACYGDDACFVARHRSADVLGVADGVGGWRDYGVDPSQFSGTLMRTCERLVKEGRFVPS 113
Query: 71 NPVRVL-----EKAHSKTKAMGSSTVCIIAL--IDEALNAINLGDSGFIVIRDGSVIFKS 123
NP+ +L E +K +G+ST CI+ L L+ NLGDSGF+V+R G V+ +S
Sbjct: 114 NPIGILTTSYCELLQNKVPLLGNSTACIVVLDRTSHRLHTANLGDSGFLVVRGGEVVHRS 173
Query: 124 PVQQRGFNFPYQLARSGTE------GDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNND 177
QQ FN P+QL+ + E D P + + + V GDII+ TDGLFDNM +
Sbjct: 174 DEQQHYFNTPFQLSIAPPEAEGVVLSDSPDAADSTSFDVQLGDIILTATDGLFDNMPDYM 233
Query: 178 IVGVVVGATRARL-GPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLT 236
I+ + + Q TA+ IA A + A D SPF+ A + G GGK DD+T
Sbjct: 234 ILQELKKLKNSNYESIQQTARSIAEQAHELAYDPNYMSPFAQFACDNGLNVRGGKPDDIT 293
Query: 237 VVVSYIS 243
V++S ++
Sbjct: 294 VLLSIVA 300
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 412,725,788
Number of Sequences: 2790947
Number of extensions: 16954311
Number of successful extensions: 37621
Number of sequences better than 10.0: 153
Number of HSP's better than 10.0 without gapping: 88
Number of HSP's successfully gapped in prelim test: 65
Number of HSP's that attempted gapping in prelim test: 37264
Number of HSP's gapped (non-prelim): 183
length of query: 248
length of database: 848,049,833
effective HSP length: 124
effective length of query: 124
effective length of database: 501,972,405
effective search space: 62244578220
effective search space used: 62244578220
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC147178.4


