

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147011.8 - phase: 0
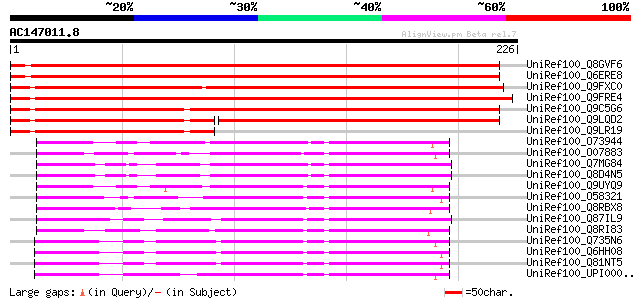
(226 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8GVF6 Pyrrolidone carboxyl peptidase-like protein [Or... 337 1e-91
UniRef100_Q6ERE8 Pyrrolidone carboxyl peptidase-like protein [Or... 333 3e-90
UniRef100_Q9FXC0 F25P12.86 protein [Arabidopsis thaliana] 321 8e-87
UniRef100_Q9FRE4 Putative pyrrolidone carboxyl peptidase [Oryza ... 309 4e-83
UniRef100_Q9C5G6 Hypothetical protein At1g23430 [Arabidopsis tha... 294 1e-78
UniRef100_Q9LQD2 F28C11.8 [Arabidopsis thaliana] 182 4e-45
UniRef100_Q9LR19 F26F24.31 [Arabidopsis thaliana] 106 4e-22
UniRef100_O73944 Pyrrolidone-carboxylate peptidase [Pyrococcus f... 85 1e-15
UniRef100_O07883 Pyrrolidone-carboxylate peptidase [Thermococcus... 79 1e-13
UniRef100_Q7MG84 Pyrrolidone-carboxylate peptidase [Vibrio vulni... 74 3e-12
UniRef100_Q8D4N5 Pyrrolidone-carboxylate peptidase [Vibrio vulni... 74 3e-12
UniRef100_Q9UYQ9 Pyrrolidone-carboxylate peptidase [Pyrococcus a... 73 5e-12
UniRef100_O58321 Pyrrolidone-carboxylate peptidase [Pyrococcus h... 70 5e-11
UniRef100_Q8RBX8 Pyrrolidone-carboxylate peptidase 1 [Thermoanae... 69 9e-11
UniRef100_Q87IL9 Pyrrolidone-carboxylate peptidase [Vibrio parah... 69 1e-10
UniRef100_Q8RI83 Pyrrolidone-carboxylate peptidase [Fusobacteriu... 67 5e-10
UniRef100_Q735N6 Pyrrolidone-carboxylate peptidase [Bacillus cer... 67 5e-10
UniRef100_Q6HH08 Pyrrolidone-carboxylate peptidase [Bacillus thu... 66 6e-10
UniRef100_Q81NT5 Pyrrolidone-carboxylate peptidase [Bacillus ant... 66 6e-10
UniRef100_UPI00003CB1FB UPI00003CB1FB UniRef100 entry 66 8e-10
>UniRef100_Q8GVF6 Pyrrolidone carboxyl peptidase-like protein [Oryza sativa]
Length = 219
Score = 337 bits (864), Expect = 1e-91
Identities = 163/218 (74%), Positives = 187/218 (85%), Gaps = 2/218 (0%)
Query: 1 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 60
MGSEGPS V TV++TGFKKFHGV+ENPTE IV NL +V+KKGLPK L +GSC++L+TA
Sbjct: 1 MGSEGPS--VVTVHVTGFKKFHGVAENPTEKIVGNLKSFVEKKGLPKNLVLGSCTVLETA 58
Query: 61 GQGALVPLYQTLQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDE 120
GQGAL LY+ L+SAI +E+ SS+ ++IW+HFGVNSGATRFA+E QAVNEATFRCPDE
Sbjct: 59 GQGALGTLYKVLESAIAERENGSSAQGQVIWIHFGVNSGATRFALENQAVNEATFRCPDE 118
Query: 121 MGWKPQKVPIVPSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSL 180
+GWKPQ+VPIVPSDG ISR RETTLPV E+TK+L GYDVM SDDAGRFVCNYVYYHSL
Sbjct: 119 LGWKPQRVPIVPSDGAISRTRETTLPVNELTKSLRKTGYDVMPSDDAGRFVCNYVYYHSL 178
Query: 181 RFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLA 218
RFAEQ+G KSLFVHVPLF TI+EE QM F ASLLE LA
Sbjct: 179 RFAEQHGIKSLFVHVPLFLTIDEEVQMHFVASLLEALA 216
>UniRef100_Q6ERE8 Pyrrolidone carboxyl peptidase-like protein [Oryza sativa]
Length = 216
Score = 333 bits (853), Expect = 3e-90
Identities = 160/218 (73%), Positives = 186/218 (84%), Gaps = 2/218 (0%)
Query: 1 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 60
MGSEGPS V TV++TGFKKFHGV+ENPTE IV NL +V+KKGLPK L +GSC++L+TA
Sbjct: 1 MGSEGPS--VVTVHVTGFKKFHGVAENPTEKIVTNLKSFVEKKGLPKNLVLGSCTVLETA 58
Query: 61 GQGALVPLYQTLQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDE 120
GQGAL LY+ L+S+I +E+ SS+ ++IW+HFGVNSGATRFA+E QAVNEATFRCPDE
Sbjct: 59 GQGALGTLYKVLESSIAERENGSSAQGQVIWIHFGVNSGATRFALENQAVNEATFRCPDE 118
Query: 121 MGWKPQKVPIVPSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSL 180
+GWKPQ+ PIVPSDG ISR RETTLPV E+TK+L GYDVM SDDAGRFVCNYVYYHSL
Sbjct: 119 LGWKPQRAPIVPSDGGISRTRETTLPVNELTKSLRKTGYDVMPSDDAGRFVCNYVYYHSL 178
Query: 181 RFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLA 218
RFAEQ+G KSLFVHVPLF TI+EE QM F ASLLE L+
Sbjct: 179 RFAEQHGIKSLFVHVPLFLTIDEEVQMHFVASLLEALS 216
>UniRef100_Q9FXC0 F25P12.86 protein [Arabidopsis thaliana]
Length = 219
Score = 321 bits (823), Expect = 8e-87
Identities = 155/220 (70%), Positives = 185/220 (83%), Gaps = 3/220 (1%)
Query: 1 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 60
MGSEGP+ T++ITGFKKFHGV+ENPTE + NNL EY+ K + K + +GSC++L+TA
Sbjct: 1 MGSEGPTGV--TIHITGFKKFHGVAENPTEKMANNLKEYLAKNCVSKDVNLGSCTVLETA 58
Query: 61 GQGALVPLYQTLQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDE 120
GQGAL LYQ LQSA+ KESES + K IW+HFGVNSGAT+FAIE+QAVNEATFRCPDE
Sbjct: 59 GQGALASLYQLLQSAVNTKESESLTG-KTIWVHFGVNSGATKFAIEQQAVNEATFRCPDE 117
Query: 121 MGWKPQKVPIVPSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSL 180
+GWKPQ +PIVPSDGPIS +R+T LPVEEITK L G++V+TSDDAGRFVCNYVYYHSL
Sbjct: 118 LGWKPQNLPIVPSDGPISTVRKTNLPVEEITKALEKNGFEVITSDDAGRFVCNYVYYHSL 177
Query: 181 RFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLANV 220
RFAEQN +SLFVHVPLF ++EETQM+F SLLEVLA++
Sbjct: 178 RFAEQNKTRSLFVHVPLFVAVDEETQMRFTVSLLEVLASI 217
>UniRef100_Q9FRE4 Putative pyrrolidone carboxyl peptidase [Oryza sativa]
Length = 222
Score = 309 bits (791), Expect = 4e-83
Identities = 148/224 (66%), Positives = 179/224 (79%), Gaps = 2/224 (0%)
Query: 1 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 60
MGSEGPS TV++TGFKKFHGV+ENPTE IV NL +++K+GLPKGL +GSC++L+TA
Sbjct: 1 MGSEGPSGV--TVHVTGFKKFHGVAENPTEKIVRNLESFMEKRGLPKGLTLGSCTVLETA 58
Query: 61 GQGALVPLYQTLQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDE 120
GQG L PLY+ +SAI+ KE + ++I LHFGVNSG TRFA+E QA+NEATFRCPDE
Sbjct: 59 GQGGLGPLYEVFESAIVDKEYGLNDQGQVILLHFGVNSGTTRFALENQAINEATFRCPDE 118
Query: 121 MGWKPQKVPIVPSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSL 180
+GWKPQ+ PIV SDG IS +R+TT+PV E+ K+L G+DV SDDAGRFVCNYVYY SL
Sbjct: 119 LGWKPQRAPIVSSDGSISNLRKTTVPVNEVNKSLQQMGFDVAPSDDAGRFVCNYVYYQSL 178
Query: 181 RFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLANVSNTQ 224
RFAEQ G KSLFVH PLF TI+EE QM F A+LLEVLA+ + Q
Sbjct: 179 RFAEQRGIKSLFVHFPLFTTISEEVQMNFVATLLEVLASQNYAQ 222
>UniRef100_Q9C5G6 Hypothetical protein At1g23430 [Arabidopsis thaliana]
Length = 217
Score = 294 bits (752), Expect = 1e-78
Identities = 140/218 (64%), Positives = 176/218 (80%), Gaps = 4/218 (1%)
Query: 1 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 60
MGSEGP A T+++TGFKKF GVSENPTE I N L YV+K+GLP GL +GSCS+LDTA
Sbjct: 1 MGSEGPKAI--TIHVTGFKKFLGVSENPTEKIANGLKSYVEKRGLPSGLCLGSCSVLDTA 58
Query: 61 GQGALVPLYQTLQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDE 120
G+GA LY+ L+S++++ + +++ ++WLH GVNSGAT+FAIERQAVNEA FRCPDE
Sbjct: 59 GEGAKSKLYEVLESSVVS--GDKNNNGTVVWLHLGVNSGATKFAIERQAVNEAHFRCPDE 116
Query: 121 MGWKPQKVPIVPSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSL 180
+GW+PQ++PIV DG IS+ +ET+ E I + L KG++V+ SDDAGRFVCNYVYYHSL
Sbjct: 117 LGWQPQRLPIVVEDGGISKAKETSCSTESIFQLLKKKGFEVVQSDDAGRFVCNYVYYHSL 176
Query: 181 RFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLA 218
RFAEQ G+KSLFVHVPLF I+E+TQMQF ASLLE +A
Sbjct: 177 RFAEQKGHKSLFVHVPLFSKIDEDTQMQFVASLLEAIA 214
>UniRef100_Q9LQD2 F28C11.8 [Arabidopsis thaliana]
Length = 368
Score = 182 bits (463), Expect = 4e-45
Identities = 87/125 (69%), Positives = 103/125 (81%)
Query: 94 FGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIVPSDGPISRIRETTLPVEEITKT 153
FG GAT+FAIERQAVNEA FRCPDE+GW+PQ++PIV DG IS+ +ET+ E I +
Sbjct: 241 FGGQCGATKFAIERQAVNEAHFRCPDELGWQPQRLPIVVEDGGISKAKETSCSTESIFQL 300
Query: 154 LATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNGNKSLFVHVPLFFTINEETQMQFAASL 213
L KG++V+ SDDAGRFVCNYVYYHSLRFAEQ G+KSLFVHVPLF I+E+TQMQF ASL
Sbjct: 301 LKKKGFEVVQSDDAGRFVCNYVYYHSLRFAEQKGHKSLFVHVPLFSKIDEDTQMQFVASL 360
Query: 214 LEVLA 218
LE +A
Sbjct: 361 LEAIA 365
Score = 106 bits (265), Expect = 4e-22
Identities = 49/91 (53%), Positives = 69/91 (74%), Gaps = 4/91 (4%)
Query: 1 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 60
MGSEGP A T+++TGFKKF GVSENPTE I N L YV+K+GLP GL +GSCS+LDTA
Sbjct: 1 MGSEGPKAI--TIHVTGFKKFLGVSENPTEKIANGLKSYVEKRGLPSGLCLGSCSVLDTA 58
Query: 61 GQGALVPLYQTLQSAIIAKESESSSSNKIIW 91
G+GA LY+ L+S++++ + +++ ++W
Sbjct: 59 GEGAKSKLYEVLESSVVS--GDKNNNGTVVW 87
>UniRef100_Q9LR19 F26F24.31 [Arabidopsis thaliana]
Length = 157
Score = 106 bits (265), Expect = 4e-22
Identities = 49/91 (53%), Positives = 69/91 (74%), Gaps = 4/91 (4%)
Query: 1 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 60
MGSEGP A T+++TGFKKF GVSENPTE I N L YV+K+GLP GL +GSCS+LDTA
Sbjct: 1 MGSEGPKAI--TIHVTGFKKFLGVSENPTEKIANGLKSYVEKRGLPSGLCLGSCSVLDTA 58
Query: 61 GQGALVPLYQTLQSAIIAKESESSSSNKIIW 91
G+GA LY+ L+S++++ + +++ ++W
Sbjct: 59 GEGAKSKLYEVLESSVVS--GDKNNNGTVVW 87
>UniRef100_O73944 Pyrrolidone-carboxylate peptidase [Pyrococcus furiosus]
Length = 208
Score = 85.1 bits (209), Expect = 1e-15
Identities = 62/186 (33%), Positives = 89/186 (47%), Gaps = 22/186 (11%)
Query: 13 VYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQTL 72
V +TGF+ F G NPTE I +L G+ IG + G ++P+
Sbjct: 3 VLVTGFEPFGGEKINPTERIAKDLD----------GIKIGDAQVF-----GRVLPVVFGK 47
Query: 73 QSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIVP 132
++ K E + I +H G+ G + +IER AVN R PD G K + PIVP
Sbjct: 48 AKEVLEKTLEEIKPD--IAIHVGLAPGRSAISIERIAVNAIDARIPDNEGKKIEDEPIVP 105
Query: 133 SDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNG--NKS 190
P + +TLP+++I K L +G S+ AG ++CNYV Y SL + G S
Sbjct: 106 G-APTAYF--STLPIKKIMKKLHERGIPAYISNSAGLYLCNYVMYLSLHHSATKGYPKMS 162
Query: 191 LFVHVP 196
F+HVP
Sbjct: 163 GFIHVP 168
>UniRef100_O07883 Pyrrolidone-carboxylate peptidase [Thermococcus litoralis]
Length = 220
Score = 78.6 bits (192), Expect = 1e-13
Identities = 62/186 (33%), Positives = 92/186 (49%), Gaps = 22/186 (11%)
Query: 13 VYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQTL 72
V ITGF+ F G S+NPTE I +Y +K + + G +L + + A + L + L
Sbjct: 4 VLITGFEPFGGDSKNPTEQIA----KYFDRKQIGNAMVYGR--VLPVSVKRATIELKRYL 57
Query: 73 QSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIVP 132
+ I E I ++ G+ + +ER AVN R PD G++P I
Sbjct: 58 EE--IKPE---------IVINLGLAPTYSNITVERIAVNIIDARIPDNDGYQPIDEKI-E 105
Query: 133 SDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNGN--KS 190
D P++ + TLPV ITKTL G S AG ++CNYV + +L F++ G K+
Sbjct: 106 EDAPLAYM--ATLPVRAITKTLRDNGIPATISYSAGTYLCNYVMFKTLHFSKIEGYPLKA 163
Query: 191 LFVHVP 196
F+HVP
Sbjct: 164 GFIHVP 169
>UniRef100_Q7MG84 Pyrrolidone-carboxylate peptidase [Vibrio vulnificus]
Length = 212
Score = 73.9 bits (180), Expect = 3e-12
Identities = 57/185 (30%), Positives = 85/185 (45%), Gaps = 20/185 (10%)
Query: 13 VYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQTL 72
V +TGF+ F G S NP+ +V + + LP+ IG C + V YQ +
Sbjct: 4 VLLTGFEPFGGESINPSLELVKQMAS----RALPQVEIIG-CEVP--------VVRYQAI 50
Query: 73 QSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIVP 132
++ + A E+ + L G SG ER A+N +R D G +P PI+
Sbjct: 51 ETVLQAVETHQPD----LVLMIGQASGRCAITPERVAINLDDYRIEDNAGHQPVDEPIIA 106
Query: 133 SDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNGNKSLF 192
+ GP + +TLPV+ IT L G S AG FVCN+++Y +S F
Sbjct: 107 T-GPAAYF--STLPVKAITHALQQAGIPCQISHSAGTFVCNHLFYGVQHHLHTRAIRSGF 163
Query: 193 VHVPL 197
+H+PL
Sbjct: 164 IHIPL 168
>UniRef100_Q8D4N5 Pyrrolidone-carboxylate peptidase [Vibrio vulnificus]
Length = 212
Score = 73.9 bits (180), Expect = 3e-12
Identities = 57/185 (30%), Positives = 85/185 (45%), Gaps = 20/185 (10%)
Query: 13 VYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQTL 72
V +TGF+ F G S NP+ +V + + LP+ IG C + V YQ +
Sbjct: 4 VLLTGFEPFGGESINPSLELVKQMAS----RALPQVEIIG-CEVP--------VVRYQAI 50
Query: 73 QSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIVP 132
++ + A E+ + L G SG ER A+N +R D G +P PI+
Sbjct: 51 ETVLQAVETHQPD----LVLMIGQASGRCAITPERVAINLDDYRIEDNAGHQPVDEPIIA 106
Query: 133 SDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNGNKSLF 192
+ GP + +TLPV+ IT L G S AG FVCN+++Y +S F
Sbjct: 107 T-GPAAYF--STLPVKAITHALQQAGIPCQLSHSAGTFVCNHLFYGVQHHLHTRAIRSGF 163
Query: 193 VHVPL 197
+H+PL
Sbjct: 164 IHIPL 168
>UniRef100_Q9UYQ9 Pyrrolidone-carboxylate peptidase [Pyrococcus abyssi]
Length = 200
Score = 73.2 bits (178), Expect = 5e-12
Identities = 56/187 (29%), Positives = 88/187 (46%), Gaps = 24/187 (12%)
Query: 13 VYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPL-YQT 71
V +TGF+ F G +NPT IV L G IG ++ G ++P+ ++
Sbjct: 3 VLVTGFEPFGGDDKNPTMEIVKFLD----------GKEIGGAKVI-----GRVLPVSFKR 47
Query: 72 LQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIV 131
+ ++A E + ++ G+ G T ++ER AVN R PD G KP PIV
Sbjct: 48 ARKELVAILDEIKPD---VTINLGLAPGRTHISVERVAVNIIDARIPDNDGEKPIDEPIV 104
Query: 132 PSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNG--NK 189
+GP + T+P EI + + + S AG ++CN+V Y +L + G K
Sbjct: 105 -ENGPAAYF--ATIPTREIVEEMKRNNIPAVLSYTAGTYLCNFVMYLTLHHSATKGYPRK 161
Query: 190 SLFVHVP 196
+ F+HVP
Sbjct: 162 AGFIHVP 168
>UniRef100_O58321 Pyrrolidone-carboxylate peptidase [Pyrococcus horikoshii]
Length = 206
Score = 69.7 bits (169), Expect = 5e-11
Identities = 54/186 (29%), Positives = 84/186 (45%), Gaps = 25/186 (13%)
Query: 13 VYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQTL 72
+ +TGF+ F G +NPT IV L+E + + +G IL + + A L + L
Sbjct: 3 ILLTGFEPFGGDDKNPTMDIVEALSERIPE-------VVGE--ILPVSFKRAREKLLKVL 53
Query: 73 QSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIVP 132
I ++ G+ G T ++ER AVN R PD G +P+ PIV
Sbjct: 54 DDV-----------RPDITINLGLAPGRTHISVERVAVNMIDARIPDNDGEQPKDEPIV- 101
Query: 133 SDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNGNKSL- 191
GP + T+P EI + + G + S AG ++CN+ Y +L + G +
Sbjct: 102 EGGPAAYF--ATIPTREIVEEMKKNGIPAVLSYTAGTYLCNFAMYLTLHTSATKGYPKIA 159
Query: 192 -FVHVP 196
F+HVP
Sbjct: 160 GFIHVP 165
>UniRef100_Q8RBX8 Pyrrolidone-carboxylate peptidase 1 [Thermoanaerobacter
tengcongensis]
Length = 203
Score = 68.9 bits (167), Expect = 9e-11
Identities = 50/186 (26%), Positives = 87/186 (45%), Gaps = 23/186 (12%)
Query: 13 VYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQTL 72
+ +T F F G S NP+ ++ NL + ++ + K L + + Y ++
Sbjct: 3 ILVTAFDPFGGESVNPSYEVLKNLKDNIEGAEIIK-LQVPTA-------------FYVSV 48
Query: 73 QSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIVP 132
+ AI + N L G G ++ER A+N R PD MG +P +PI P
Sbjct: 49 EKAI----EKIKEVNPDAVLSIGQAGGRYDISVERVAINIDDARIPDNMGQQPIDIPIDP 104
Query: 133 SDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQN--GNKS 190
+GP + T+P++EI + + + S+ AG +VCN++ Y L + +N K+
Sbjct: 105 -EGPPAYF--ATIPIKEIVEAIKKENLPASVSNSAGTYVCNHLMYGILNYIHKNKLNIKA 161
Query: 191 LFVHVP 196
F+H+P
Sbjct: 162 GFIHIP 167
>UniRef100_Q87IL9 Pyrrolidone-carboxylate peptidase [Vibrio parahaemolyticus]
Length = 212
Score = 68.6 bits (166), Expect = 1e-10
Identities = 54/185 (29%), Positives = 83/185 (44%), Gaps = 20/185 (10%)
Query: 13 VYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQTL 72
V ITGF+ F G + NP V L E G+ I +C + T ++++
Sbjct: 4 VLITGFEPFGGDAINPALEAVKRLEETSLDGGI-----IVTCQVPVTR--------FESI 50
Query: 73 QSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIVP 132
+ I A E+ + G +G ER A+N FR PD G +P PI+
Sbjct: 51 SAVIDAIEAYQPDCVITV----GQAAGRAAITPERVAINVDDFRIPDNGGNQPIDEPII- 105
Query: 133 SDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNGNKSLF 192
GP + ++LP++ I +TL G S+ AG FVCN+++Y + + F
Sbjct: 106 EQGPDAYF--SSLPIKRIAQTLHESGIPCQVSNSAGTFVCNHLFYGVQHYLRDKSIRHGF 163
Query: 193 VHVPL 197
VH+PL
Sbjct: 164 VHIPL 168
>UniRef100_Q8RI83 Pyrrolidone-carboxylate peptidase [Fusobacterium nucleatum]
Length = 214
Score = 66.6 bits (161), Expect = 5e-10
Identities = 58/186 (31%), Positives = 80/186 (42%), Gaps = 23/186 (12%)
Query: 13 VYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQTL 72
+ +TGF F G NP ++ K LPK + IL+ +P
Sbjct: 4 ILVTGFDPFGGEKINPALEVI---------KLLPKKIGENEIKILE-------IPTVYKK 47
Query: 73 QSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIVP 132
I KE ES + I L G G T +IER A+N FR D G +P I
Sbjct: 48 SIEKIDKEIESYDPDYI--LSIGQAGGRTDISIERIAINIDDFRIKDNEGNQPIDEKIY- 104
Query: 133 SDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQ--NGNKS 190
DG + +TLP++ I + G S+ AG FVCN+V+Y E+ G KS
Sbjct: 105 LDGDNAYF--STLPIKAIQSEITKNGIPASISNTAGTFVCNHVFYGVRYLIEKKYKGKKS 162
Query: 191 LFVHVP 196
F+H+P
Sbjct: 163 GFIHIP 168
>UniRef100_Q735N6 Pyrrolidone-carboxylate peptidase [Bacillus cereus]
Length = 215
Score = 66.6 bits (161), Expect = 5e-10
Identities = 57/187 (30%), Positives = 82/187 (43%), Gaps = 22/187 (11%)
Query: 12 TVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQT 71
TV +TGF F G + NP + NL E IG I+ VP
Sbjct: 3 TVLLTGFDPFGGENINPAWEVAKNLHEKT----------IGEYKIISKQ-----VPTVFH 47
Query: 72 LQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIV 131
+++ + E + II + G G IER A+N R D G +P VP+V
Sbjct: 48 KSISVLKEYIEELAPEFIICI--GQAGGRPDITIERVAINIDDARIADNEGNQPVDVPVV 105
Query: 132 PSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNGN--K 189
+GP + +TLP++ I K L +G S AG FVCN+++Y + E+ K
Sbjct: 106 -EEGPTAYW--STLPMKAIVKRLQEEGIPASVSQTAGTFVCNHLFYGLMHELEKRDKKIK 162
Query: 190 SLFVHVP 196
FVH+P
Sbjct: 163 GGFVHIP 169
>UniRef100_Q6HH08 Pyrrolidone-carboxylate peptidase [Bacillus thuringiensis]
Length = 215
Score = 66.2 bits (160), Expect = 6e-10
Identities = 56/187 (29%), Positives = 83/187 (43%), Gaps = 22/187 (11%)
Query: 12 TVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQT 71
TV +TGF F G S NP + +L E IG I+ VP
Sbjct: 3 TVLLTGFDPFGGESINPAWEVAKSLHEKT----------IGEYKIISKQ-----VPTVFH 47
Query: 72 LQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIV 131
+++ + E + II + G G IER A+N R D G +P VP+V
Sbjct: 48 KSISVLKEYIEELAPEFIICM--GQAGGRPDITIERVAINIDDARIADNEGNQPVDVPVV 105
Query: 132 PSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNGNKSL 191
+GP + +TLP++ I K L +G S AG FVCN+++Y + E++ K
Sbjct: 106 -EEGPAAYW--STLPMKAIVKKLQEEGIPASVSQTAGTFVCNHLFYGLMHELEKHDTKMK 162
Query: 192 --FVHVP 196
F+H+P
Sbjct: 163 GGFIHIP 169
>UniRef100_Q81NT5 Pyrrolidone-carboxylate peptidase [Bacillus anthracis]
Length = 215
Score = 66.2 bits (160), Expect = 6e-10
Identities = 56/187 (29%), Positives = 83/187 (43%), Gaps = 22/187 (11%)
Query: 12 TVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQT 71
TV +TGF F G S NP + +L E IG I+ VP
Sbjct: 3 TVLLTGFDPFGGESINPAWEVAKSLHEKT----------IGEYKIISKQ-----VPTVFH 47
Query: 72 LQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIV 131
+++ + E + II + G G IER A+N R D G +P VP+V
Sbjct: 48 KSISVLKEYIEELAPEFIICI--GQAGGRPDITIERVAINIDDARIADNEGNQPVDVPVV 105
Query: 132 PSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNGNKSL 191
+GP + +TLP++ I K L +G S AG FVCN+++Y + E++ K
Sbjct: 106 -EEGPAAYW--STLPMKAIVKKLQEEGIPASVSQTAGTFVCNHLFYGLMHELEKHDTKMK 162
Query: 192 --FVHVP 196
F+H+P
Sbjct: 163 GGFIHIP 169
>UniRef100_UPI00003CB1FB UPI00003CB1FB UniRef100 entry
Length = 215
Score = 65.9 bits (159), Expect = 8e-10
Identities = 57/187 (30%), Positives = 77/187 (40%), Gaps = 22/187 (11%)
Query: 12 TVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTAGQGALVPLYQT 71
TV +TGF F G S NP + +L E IG I+ T
Sbjct: 3 TVLLTGFDPFGGESINPAWEVAKSLHEKT----------IGEYKIISKQVPTVFHKSIST 52
Query: 72 LQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDEMGWKPQKVPIV 131
L+ I N I + G G IER A+N R D G +P VP+V
Sbjct: 53 LKEYI-------EQLNPEIIICIGQAGGRPDITIERVAINIDDARIADNEGNQPVDVPVV 105
Query: 132 PSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNGN--K 189
+GP + +TLP++ I K L +G S AG FVCN+++Y + E K
Sbjct: 106 -EEGPAAYW--STLPMKAIVKRLQAEGIPASVSQTAGTFVCNHLFYGLMHELENRDKKIK 162
Query: 190 SLFVHVP 196
FVH+P
Sbjct: 163 GGFVHIP 169
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.132 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 351,601,141
Number of Sequences: 2790947
Number of extensions: 13779127
Number of successful extensions: 37951
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 68
Number of HSP's successfully gapped in prelim test: 48
Number of HSP's that attempted gapping in prelim test: 37768
Number of HSP's gapped (non-prelim): 127
length of query: 226
length of database: 848,049,833
effective HSP length: 123
effective length of query: 103
effective length of database: 504,763,352
effective search space: 51990625256
effective search space used: 51990625256
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 72 (32.3 bits)
Medicago: description of AC147011.8


