

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.6 + phase: 0
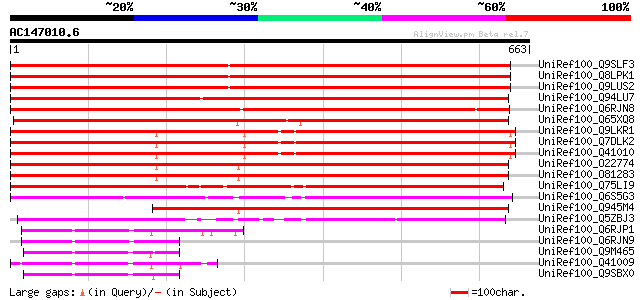
(663 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SLF3 Putative chloroplast outer membrane protein [Ar... 1039 0.0
UniRef100_Q8LPK1 Putative chloroplast outer membrane protein [Ar... 1039 0.0
UniRef100_Q9LUS2 Chloroplast outer envelope protein-like [Arabid... 1024 0.0
UniRef100_Q94LU7 Putative outer envelope protein [Oryza sativa] 909 0.0
UniRef100_Q6RJN8 Chloroplast Toc125 [Physcomitrella patens] 817 0.0
UniRef100_Q65XQ8 Putative chloroplast outer envelope 86-like pro... 697 0.0
UniRef100_Q9LKR1 Chloroplast protein import component Toc159 [Pi... 660 0.0
UniRef100_Q7DLK2 Chloroplast outer envelope protein 86 [Pisum sa... 660 0.0
UniRef100_Q41010 GTP-binding protein [Pisum sativum] 654 0.0
UniRef100_O22774 Putative chloroplast outer envelope 86-like pro... 646 0.0
UniRef100_O81283 T14P8.24 protein [Arabidopsis thaliana] 646 0.0
UniRef100_Q75LI9 Putative GTP-binding protein, having alternativ... 545 e-153
UniRef100_Q6S5G3 Chloroplast import receptor Toc90 [Arabidopsis ... 486 e-136
UniRef100_Q945M4 AT4g02510/T10P11_19 [Arabidopsis thaliana] 446 e-123
UniRef100_Q5ZBJ3 Putative OEP86=outer envelope protein [Oryza sa... 368 e-100
UniRef100_Q6RJP1 Chloroplast Toc34-1 [Physcomitrella patens] 145 5e-33
UniRef100_Q6RJN9 Chloroplast Toc34-3 [Physcomitrella patens] 144 6e-33
UniRef100_Q9M465 Toc34-2 protein [Zea mays] 136 2e-30
UniRef100_Q41009 Translocase of chloroplast 34 [Pisum sativum] 135 5e-30
UniRef100_Q9SBX0 Toc34-1 protein [Zea mays] 133 1e-29
>UniRef100_Q9SLF3 Putative chloroplast outer membrane protein [Arabidopsis thaliana]
Length = 1206
Score = 1039 bits (2686), Expect = 0.0
Identities = 511/640 (79%), Positives = 577/640 (89%), Gaps = 1/640 (0%)
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
MAEQLE+AGQ+PLDFSCTIMVLGKSGVGKS+TINSIFDEVKF TDAF MGTK+VQDV G+
Sbjct: 559 MAEQLEAAGQDPLDFSCTIMVLGKSGVGKSATINSIFDEVKFCTDAFQMGTKRVQDVEGL 618
Query: 61 VQGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMP 120
VQGIKVRVIDTPGLLPSWSDQ NEKIL+SVK FIKK PPDIVLYLDRLDMQSRD DMP
Sbjct: 619 VQGIKVRVIDTPGLLPSWSDQAKNEKILNSVKAFIKKNPPDIVLYLDRLDMQSRDSGDMP 678
Query: 121 LLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGD 180
LLRTI+D+FGP IWFNAIV LTHAAS PPDGPNGT SSYDMFVTQRSHV+QQAIRQAAGD
Sbjct: 679 LLRTISDVFGPSIWFNAIVGLTHAASVPPDGPNGTASSYDMFVTQRSHVIQQAIRQAAGD 738
Query: 181 MRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPR 240
MRLMNPVSLVENHSACRTN AGQRVLPNGQVWKP LLLLSFASKILAEANALLKLQDN
Sbjct: 739 MRLMNPVSLVENHSACRTNRAGQRVLPNGQVWKPHLLLLSFASKILAEANALLKLQDNIP 798
Query: 241 EKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLP 300
+P+ AR++APPLPFLLSSLLQSRPQ KLPE Q+ DE+ DDL+E SDS +E++ D LP
Sbjct: 799 GRPFAARSKAPPLPFLLSSLLQSRPQPKLPEQQYGDEED-EDDLEESSDSDEESEYDQLP 857
Query: 301 PFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSD 360
PFK LTKAQ+ LS++QKK YLDE+EYREKL MKKQ+K E+K+RKM K+ A +KDLP
Sbjct: 858 PFKSLTKAQMATLSKSQKKQYLDEMEYREKLLMKKQMKEERKRRKMFKKFAAEIKDLPDG 917
Query: 361 YVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWDHD 420
Y ENVEEESGG ASVPVPMPD+SLPASFDSD PTHRYR+LDSSNQWLVRPVLETHGWDHD
Sbjct: 918 YSENVEEESGGPASVPVPMPDLSLPASFDSDNPTHRYRYLDSSNQWLVRPVLETHGWDHD 977
Query: 421 VGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQTV 480
+GYEG+N ERLFV+K+KIP+S SGQVTKDKKDANVQ+EM SSVK+GEGK+TSLGFDMQTV
Sbjct: 978 IGYEGVNAERLFVVKEKIPISVSGQVTKDKKDANVQLEMASSVKHGEGKSTSLGFDMQTV 1037
Query: 481 GKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGAM 540
GK+LAYTLRSET+F NF RNKA AGLS T LGD++SAG+KVEDK IA+K F++V++GGAM
Sbjct: 1038 GKELAYTLRSETRFNNFRRNKAAAGLSVTHLGDSVSAGLKVEDKFIASKWFRIVMSGGAM 1097
Query: 541 TGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLV 600
T R D AYGG+LEAQLRDK+YPLGR L+TLGLSVMDWHGDLA+G N+QSQ+PIGR +NL+
Sbjct: 1098 TSRGDFAYGGTLEAQLRDKDYPLGRFLTTLGLSVMDWHGDLAIGGNIQSQVPIGRSSNLI 1157
Query: 601 ARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVIGY 640
ARANLNNRGAGQ+S+R+NSSEQLQ+A++ ++PL KK++ Y
Sbjct: 1158 ARANLNNRGAGQVSVRVNSSEQLQLAMVAIVPLFKKLLSY 1197
>UniRef100_Q8LPK1 Putative chloroplast outer membrane protein [Arabidopsis thaliana]
Length = 1202
Score = 1039 bits (2686), Expect = 0.0
Identities = 511/640 (79%), Positives = 577/640 (89%), Gaps = 1/640 (0%)
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
MAEQLE+AGQ+PLDFSCTIMVLGKSGVGKS+TINSIFDEVKF TDAF MGTK+VQDV G+
Sbjct: 559 MAEQLEAAGQDPLDFSCTIMVLGKSGVGKSATINSIFDEVKFCTDAFQMGTKRVQDVEGL 618
Query: 61 VQGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMP 120
VQGIKVRVIDTPGLLPSWSDQ NEKIL+SVK FIKK PPDIVLYLDRLDMQSRD DMP
Sbjct: 619 VQGIKVRVIDTPGLLPSWSDQAKNEKILNSVKAFIKKNPPDIVLYLDRLDMQSRDSGDMP 678
Query: 121 LLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGD 180
LLRTI+D+FGP IWFNAIV LTHAAS PPDGPNGT SSYDMFVTQRSHV+QQAIRQAAGD
Sbjct: 679 LLRTISDVFGPSIWFNAIVGLTHAASVPPDGPNGTASSYDMFVTQRSHVIQQAIRQAAGD 738
Query: 181 MRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPR 240
MRLMNPVSLVENHSACRTN AGQRVLPNGQVWKP LLLLSFASKILAEANALLKLQDN
Sbjct: 739 MRLMNPVSLVENHSACRTNRAGQRVLPNGQVWKPHLLLLSFASKILAEANALLKLQDNIP 798
Query: 241 EKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLP 300
+P+ AR++APPLPFLLSSLLQSRPQ KLPE Q+ DE+ DDL+E SDS +E++ D LP
Sbjct: 799 GRPFAARSKAPPLPFLLSSLLQSRPQPKLPEQQYGDEED-EDDLEESSDSDEESEYDQLP 857
Query: 301 PFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSD 360
PFK LTKAQ+ LS++QKK YLDE+EYREKL MKKQ+K E+K+RKM K+ A +KDLP
Sbjct: 858 PFKSLTKAQMATLSKSQKKQYLDEMEYREKLLMKKQMKEERKRRKMFKKFAAEIKDLPDG 917
Query: 361 YVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWDHD 420
Y ENVEEESGG ASVPVPMPD+SLPASFDSD PTHRYR+LDSSNQWLVRPVLETHGWDHD
Sbjct: 918 YSENVEEESGGPASVPVPMPDLSLPASFDSDNPTHRYRYLDSSNQWLVRPVLETHGWDHD 977
Query: 421 VGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQTV 480
+GYEG+N ERLFV+K+KIP+S SGQVTKDKKDANVQ+EM SSVK+GEGK+TSLGFDMQTV
Sbjct: 978 IGYEGVNAERLFVVKEKIPISVSGQVTKDKKDANVQLEMASSVKHGEGKSTSLGFDMQTV 1037
Query: 481 GKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGAM 540
GK+LAYTLRSET+F NF RNKA AGLS T LGD++SAG+KVEDK IA+K F++V++GGAM
Sbjct: 1038 GKELAYTLRSETRFNNFRRNKAAAGLSVTHLGDSVSAGLKVEDKFIASKWFRIVMSGGAM 1097
Query: 541 TGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLV 600
T R D AYGG+LEAQLRDK+YPLGR L+TLGLSVMDWHGDLA+G N+QSQ+PIGR +NL+
Sbjct: 1098 TSRGDFAYGGTLEAQLRDKDYPLGRFLTTLGLSVMDWHGDLAIGGNIQSQVPIGRSSNLI 1157
Query: 601 ARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVIGY 640
ARANLNNRGAGQ+S+R+NSSEQLQ+A++ ++PL KK++ Y
Sbjct: 1158 ARANLNNRGAGQVSVRVNSSEQLQLAMVAIVPLFKKLLSY 1197
>UniRef100_Q9LUS2 Chloroplast outer envelope protein-like [Arabidopsis thaliana]
Length = 1089
Score = 1024 bits (2647), Expect = 0.0
Identities = 502/640 (78%), Positives = 574/640 (89%), Gaps = 1/640 (0%)
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
MAEQLE+A Q+PLDFSCTIMVLGKSGVGKS+TINSIFDE+K +TDAF +GTKKVQD+ G
Sbjct: 441 MAEQLEAAAQDPLDFSCTIMVLGKSGVGKSATINSIFDELKISTDAFQVGTKKVQDIEGF 500
Query: 61 VQGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMP 120
VQGIKVRVIDTPGLLPSWSDQ NEKIL SV+ FIKK+PPDIVLYLDRLDMQSRD DMP
Sbjct: 501 VQGIKVRVIDTPGLLPSWSDQHKNEKILKSVRAFIKKSPPDIVLYLDRLDMQSRDSGDMP 560
Query: 121 LLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGD 180
LLRTITD+FGP IWFNAIV LTHAASAPPDGPNGT SSYDMFVTQRSHV+QQAIRQAAGD
Sbjct: 561 LLRTITDVFGPSIWFNAIVGLTHAASAPPDGPNGTASSYDMFVTQRSHVIQQAIRQAAGD 620
Query: 181 MRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPR 240
MRLMNPVSLVENHSACRTN AGQRVLPNGQVWKP LLLLSFASKILAEANALLKLQDN
Sbjct: 621 MRLMNPVSLVENHSACRTNRAGQRVLPNGQVWKPHLLLLSFASKILAEANALLKLQDNIP 680
Query: 241 EKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLP 300
+ R++APPLP LLSSLLQSRPQ KLPE Q+ DED DDLDE SDS +E++ D+LP
Sbjct: 681 GGQFATRSKAPPLPLLLSSLLQSRPQAKLPEQQYDDEDD-EDDLDESSDSEEESEYDELP 739
Query: 301 PFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSD 360
PFK LTKA++ LS++QKK YLDE+EYREKLFMK+Q+K E+K+RK++K+ A +KD+P+
Sbjct: 740 PFKRLTKAEMTKLSKSQKKEYLDEMEYREKLFMKRQMKEERKRRKLLKKFAAEIKDMPNG 799
Query: 361 YVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWDHD 420
Y ENVEEE ASVPVPMPD+SLPASFDSD PTHRYR+LD+SNQWLVRPVLETHGWDHD
Sbjct: 800 YSENVEEERSEPASVPVPMPDLSLPASFDSDNPTHRYRYLDTSNQWLVRPVLETHGWDHD 859
Query: 421 VGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQTV 480
+GYEG+N ERLFV+KDKIPVSFSGQVTKDKKDA+VQ+E+ SSVK+GEG++TSLGFDMQ
Sbjct: 860 IGYEGVNAERLFVVKDKIPVSFSGQVTKDKKDAHVQLELASSVKHGEGRSTSLGFDMQNA 919
Query: 481 GKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGAM 540
GK+LAYT+RSET+F F +NKA AGLS TLLGD++SAG+KVEDKLIANKRF++V++GGAM
Sbjct: 920 GKELAYTIRSETRFNKFRKNKAAAGLSVTLLGDSVSAGLKVEDKLIANKRFRMVMSGGAM 979
Query: 541 TGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLV 600
T R DVAYGG+LEAQ RDK+YPLGR LSTLGLSVMDWHGDLA+G N+QSQ+PIGR +NL+
Sbjct: 980 TSRGDVAYGGTLEAQFRDKDYPLGRFLSTLGLSVMDWHGDLAIGGNIQSQVPIGRSSNLI 1039
Query: 601 ARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVIGY 640
ARANLNNRGAGQ+SIR+NSSEQLQ+A++ L+PL KK++ Y
Sbjct: 1040 ARANLNNRGAGQVSIRVNSSEQLQLAVVALVPLFKKLLTY 1079
>UniRef100_Q94LU7 Putative outer envelope protein [Oryza sativa]
Length = 1008
Score = 909 bits (2350), Expect = 0.0
Identities = 446/638 (69%), Positives = 536/638 (83%), Gaps = 4/638 (0%)
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
MAE+LE+AG EPLDFSCTI+VLGK+GVGKS+TINSIFD+V+ T+AF T+KVQ+VVG
Sbjct: 368 MAERLEAAGNEPLDFSCTILVLGKTGVGKSATINSIFDDVRLETNAFDTSTRKVQEVVGA 427
Query: 61 VQGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMP 120
V+GIKV+VIDTPGL S SDQ HN+KIL+SVKR I + PPDIVLY DRLDMQ+RD+ D+P
Sbjct: 428 VEGIKVKVIDTPGLSCSSSDQHHNQKILNSVKRLISRNPPDIVLYFDRLDMQTRDYGDVP 487
Query: 121 LLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGD 180
LL+TIT +FG IWFNAIVVLTHAASAPPDG NG P SY+MFVTQRSHVVQQAIRQAAGD
Sbjct: 488 LLQTITRVFGASIWFNAIVVLTHAASAPPDGLNGIPLSYEMFVTQRSHVVQQAIRQAAGD 547
Query: 181 MRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPR 240
+RLMNPVSLVENHSACRTN AGQRVLPNG VWKPQLLLL FASK+LAEANALLKLQDNP
Sbjct: 548 VRLMNPVSLVENHSACRTNRAGQRVLPNGHVWKPQLLLLCFASKVLAEANALLKLQDNPA 607
Query: 241 EKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLP 300
KP R R PPLPFLLSSLLQSR LKLPE+QF D+D + DDL + SDS D +D DDLP
Sbjct: 608 GKP---RMRIPPLPFLLSSLLQSRAPLKLPEEQFGDDDDIEDDLADDSDSDDGSDYDDLP 664
Query: 301 PFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSD 360
PFK LTKAQ+ L+ AQ+KAYL+E++YREKLF KKQLK E+ +RK+MK+MA D
Sbjct: 665 PFKRLTKAQLAKLNHAQRKAYLEELDYREKLFYKKQLKEERMRRKIMKKMAAEASARTDD 724
Query: 361 YVE-NVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWDH 419
+ N+E++ +V VPMPDM LP+SFDSD P+HRYR LD+ ++WLVRPVLET GWDH
Sbjct: 725 FSNSNLEDDGSAPTNVAVPMPDMVLPSSFDSDHPSHRYRFLDTPSEWLVRPVLETQGWDH 784
Query: 420 DVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQT 479
DVGYEGLNVERLF +K K+P+S SGQ++KDKKD ++QME+ SS+K+GEGK TSLG D+Q+
Sbjct: 785 DVGYEGLNVERLFAVKGKVPLSVSGQLSKDKKDCSLQMEVASSLKHGEGKTTSLGLDLQS 844
Query: 480 VGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGA 539
VGKD+AYTLR E++F NF RN AG+S TLLGD++SAGVKVEDKL+ NK+ +++++GGA
Sbjct: 845 VGKDMAYTLRGESRFKNFRRNNTAAGISATLLGDSVSAGVKVEDKLVVNKQLRVLVSGGA 904
Query: 540 MTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNL 599
M+GR DVAYGG LEA L+DK+YP+GR LST+ LSV+DWHGDLAVGCN+QSQIP GR +NL
Sbjct: 905 MSGRGDVAYGGRLEATLKDKDYPIGRMLSTIALSVVDWHGDLAVGCNIQSQIPAGRASNL 964
Query: 600 VARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKV 637
+ ANL+N+G GQ+ IRLNSSE L+IALI L+P+ + +
Sbjct: 965 IGHANLSNKGTGQVGIRLNSSEHLEIALIALVPIYQNI 1002
>UniRef100_Q6RJN8 Chloroplast Toc125 [Physcomitrella patens]
Length = 1141
Score = 817 bits (2111), Expect = 0.0
Identities = 406/641 (63%), Positives = 508/641 (78%), Gaps = 6/641 (0%)
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
+AE+ E+A E LDF+CTI+VLGK+GVGKS+TINSIFDE K T A++ T KV +V G
Sbjct: 492 LAEEQEAAKYEDLDFACTILVLGKTGVGKSATINSIFDECKTVTSAYYPSTTKVHEVSGT 551
Query: 61 VQGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMP 120
V G+KVR IDTPGLLPS +DQ HN+ I+ VK++IKK PDIVLY DR+DMQ+RD D+P
Sbjct: 552 VLGVKVRFIDTPGLLPSTADQRHNKNIMRQVKKYIKKVSPDIVLYFDRMDMQTRDSGDVP 611
Query: 121 LLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGD 180
LLRTITD+FG +WFNA VVLTHA+ APPDG NGTP SYD FV QRSH VQQ IRQAAGD
Sbjct: 612 LLRTITDVFGAAVWFNATVVLTHASKAPPDGSNGTPMSYDYFVAQRSHFVQQTIRQAAGD 671
Query: 181 MRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQD-NP 239
RL NPVSLVENH ACR N +GQRVLPNGQ WK QLLLL FASKILAEAN LLKLQ+ +
Sbjct: 672 ARLQNPVSLVENHPACRINRSGQRVLPNGQPWKQQLLLLCFASKILAEANTLLKLQEAST 731
Query: 240 REKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDS-GDETDPDD 298
KP+ R+R PPLP+LLSSLLQSR QLK+P++Q + + +DD DE + GDE D D
Sbjct: 732 PGKPFGQRSRVPPLPYLLSSLLQSRAQLKMPDEQHGESEDSDDDSDEEDEEEGDEYD--D 789
Query: 299 LPPFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESV-KDL 357
LPPF+PL+K ++ +LS+ Q++ Y +E+ RE+LF KKQ + + ++R+ K+ A + K+
Sbjct: 790 LPPFRPLSKQELEDLSKEQRQEYAEELADRERLFQKKQYREQIRRRRERKKQASVMSKEG 849
Query: 358 PSDYVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGW 417
PS + E+ESG A+V VPMPDM+LP SFDSD PTHRYR+L+++NQWLVRPVLETHGW
Sbjct: 850 PSIPGDGAEDESGQPATVAVPMPDMALPPSFDSDNPTHRYRYLETANQWLVRPVLETHGW 909
Query: 418 DHDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDM 477
DHD GY+G NVE++FV+K+KIP S SGQVTKDKK+A V E +S+++GEGK T GFD+
Sbjct: 910 DHDAGYDGFNVEKMFVVKEKIPASVSGQVTKDKKEAQVNFEAAASLRHGEGKVTLTGFDV 969
Query: 478 QTVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAG 537
QT+GKDLAYT+R+ET+F NF RNK TAG++ T L D ++AGVK+ED+++ KR KLV+ G
Sbjct: 970 QTIGKDLAYTVRAETRFNNFKRNKTTAGVTATYLNDTIAAGVKLEDRVLIGKRVKLVVNG 1029
Query: 538 GAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYT 597
G +TG+ D AYGGSLEA LR K YPL R+LSTLGLSVMDWHGDLA+G NLQSQ +G+ T
Sbjct: 1030 GVLTGKGDKAYGGSLEATLRGKEYPLSRTLSTLGLSVMDWHGDLAIGGNLQSQFMVGK-T 1088
Query: 598 NLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVI 638
+V RANLNNRG+GQ+SIR +SSEQLQ+ LIG++P+L+ +I
Sbjct: 1089 MMVGRANLNNRGSGQVSIRASSSEQLQMVLIGIVPILRSLI 1129
>UniRef100_Q65XQ8 Putative chloroplast outer envelope 86-like protein [Oryza sativa]
Length = 1118
Score = 697 bits (1798), Expect = 0.0
Identities = 355/639 (55%), Positives = 463/639 (71%), Gaps = 8/639 (1%)
Query: 5 LESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGI 64
LE+ G+E L+FSC I+VLGK GVGKS+TINSIF E K TDAF T V+++VG V G+
Sbjct: 469 LEAEGKEELNFSCNILVLGKIGVGKSATINSIFGEEKSKTDAFSSATNSVREIVGNVDGV 528
Query: 65 KVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRT 124
++R+IDTPGL P+ DQ N KIL SVK++ K+ PPDIVLY+DRLD SRD +D+PLL+T
Sbjct: 529 QIRIIDTPGLRPNVMDQGSNRKILASVKKYTKRCPPDIVLYVDRLDSLSRDLNDLPLLKT 588
Query: 125 ITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDMRLM 184
IT + G IWFNAIV LTHAASAPP+G NG P +Y++ + QRSH++QQ+IRQAAGDMRLM
Sbjct: 589 ITSVLGSSIWFNAIVALTHAASAPPEGLNGAPMTYEVLMAQRSHIIQQSIRQAAGDMRLM 648
Query: 185 NPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPREKPY 244
NPV+LVENH +CR N GQ+VLPNGQ W+ Q+LLL ++SKIL+EAN+LLKLQD K +
Sbjct: 649 NPVALVENHPSCRRNREGQKVLPNGQSWRHQMLLLCYSSKILSEANSLLKLQDPNPGKLF 708
Query: 245 TARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSD---SGDETDPDDLPP 301
R R+PPLPFLLSSLLQSR KL DQ +E + DLD+ SD DE + D LPP
Sbjct: 709 GFRFRSPPLPFLLSSLLQSRAHPKLSPDQGGNEGDSDIDLDDYSDIEQDEDEEEYDQLPP 768
Query: 302 FKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSDY 361
FKPLTK+Q+ L++ QK AY DE +YR KL KKQ K E ++ K MK+ ++ D+ +
Sbjct: 769 FKPLTKSQLARLTKEQKNAYFDEYDYRVKLLQKKQWKDEIRRLKEMKKRGKT--DMDAYG 826
Query: 362 VENVEEESG---GAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWD 418
N+ E+ +V VP+PDM LP SFD D PT+RYR L+ ++ L RPVL+ HGWD
Sbjct: 827 YANIAGENDLDPPPENVSVPLPDMVLPPSFDCDNPTYRYRFLEPTSTVLARPVLDAHGWD 886
Query: 419 HDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQ 478
HD GY+G++VE L +K P + + QVTKDKK+ ++ ++ + S K GE ++ GFD+Q
Sbjct: 887 HDCGYDGVSVEETLALLNKFPANMAVQVTKDKKEFSIHLDSSISAKLGEDASSLAGFDIQ 946
Query: 479 TVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGG 538
TVG+ LAY LR ETKF N +NK T G S T LGD ++ G+KVED+L KR LV + G
Sbjct: 947 TVGRQLAYILRGETKFKNIKKNKTTGGFSVTFLGDIVATGLKVEDQLSLGKRLALVASTG 1006
Query: 539 AMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTN 598
AM + D AYG +LEA+L+DK+YP+G+SLSTLGLS+M W DLA+G NLQSQ IGR +
Sbjct: 1007 AMRAQGDTAYGANLEARLKDKDYPIGQSLSTLGLSLMKWRRDLALGANLQSQFSIGRGSK 1066
Query: 599 LVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKV 637
+V R LNN+ +GQI++R ++SEQ+QIAL+GLIP+ +
Sbjct: 1067 MVVRLGLNNKLSGQITVRTSTSEQVQIALLGLIPVAASI 1105
>UniRef100_Q9LKR1 Chloroplast protein import component Toc159 [Pisum sativum]
Length = 1469
Score = 660 bits (1704), Expect = 0.0
Identities = 337/659 (51%), Positives = 465/659 (70%), Gaps = 18/659 (2%)
Query: 2 AEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMV 61
A +LE+ G++ FS I+VLGK+GVGKS+TINSIF E K + A+ T V ++VGMV
Sbjct: 814 ASRLEAEGRDDFAFSLNILVLGKTGVGKSATINSIFGETKTSFSAYGPATTSVTEIVGMV 873
Query: 62 QGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPL 121
G+++RV DTPGL S +Q +N K+L +VK+ KK+PPDIVLY+DRLD+Q+RD +D+P+
Sbjct: 874 DGVEIRVFDTPGLKSSAFEQSYNRKVLSTVKKLTKKSPPDIVLYVDRLDLQTRDMNDLPM 933
Query: 122 LRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDM 181
LR++T GP IW N IV LTHAASAPPDGP+G+P SYD+FV QRSH+VQQAI QA GD+
Sbjct: 934 LRSVTSALGPTIWRNVIVTLTHAASAPPDGPSGSPLSYDVFVAQRSHIVQQAIGQAVGDL 993
Query: 182 RLMNP-----VSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQ 236
RLMNP VSLVENH +CR N GQ+VLPNGQ WKP LLLL ++ KIL+EA + K Q
Sbjct: 994 RLMNPNLMNPVSLVENHPSCRKNRDGQKVLPNGQSWKPLLLLLCYSMKILSEATNISKTQ 1053
Query: 237 DNP-REKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETD 295
+ + + R+RAPPLP+LLS LLQSR KLP+ D + ++ + SDS E
Sbjct: 1054 EAADNRRLFGFRSRAPPLPYLLSWLLQSRAHPKLPDQAGIDNGDSDIEMADLSDSDGEEG 1113
Query: 296 PDD---LPPFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAE 352
D+ LPPFKPL K+QI L+ Q+KAYL+E +YR KL KKQ + E K+ M++M +
Sbjct: 1114 EDEYDQLPPFKPLKKSQIAKLNGEQRKAYLEEYDYRVKLLQKKQWREELKR---MRDMKK 1170
Query: 353 SVKDLPSDYVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVL 412
K+ +DY+E +EE+G A+VPVP+PDM LP SFDSD P +RYR L+ ++Q L RPVL
Sbjct: 1171 RGKNGENDYMEE-DEENGSPAAVPVPLPDMVLPQSFDSDNPAYRYRFLEPNSQLLTRPVL 1229
Query: 413 ETHGWDHDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATS 472
+TH WDHD GY+G+N+E + +K P + + QVTKDK+D ++ ++ + + K+GE +T
Sbjct: 1230 DTHSWDHDCGYDGVNIENSMAIINKFPAAVTVQVTKDKQDFSIHLDSSVAAKHGENGSTM 1289
Query: 473 LGFDMQTVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFK 532
GFD+Q +GK LAY +R ETKF NF RNK AG+S T LG+ +S GVK+ED++ KR
Sbjct: 1290 AGFDIQNIGKQLAYIVRGETKFKNFKRNKTAAGVSVTFLGENVSTGVKLEDQIALGKRLV 1349
Query: 533 LVIAGGAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIP 592
LV + G + ++D AYG ++E +LR+ ++P+G+ S+L LS++ W GDLA+G N QSQI
Sbjct: 1350 LVGSTGTVRSQNDSAYGANVEVRLREADFPVGQDQSSLSLSLVQWRGDLALGANFQSQIS 1409
Query: 593 IGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVI-----GYSQNCSL 646
+GR + RA LNN+ +GQI++R +SS+QLQIALI ++P+ K + G ++N S+
Sbjct: 1410 LGRSYKMAVRAGLNNKLSGQINVRTSSSDQLQIALIAILPVAKAIYKNFWPGVTENYSI 1468
>UniRef100_Q7DLK2 Chloroplast outer envelope protein 86 [Pisum sativum]
Length = 879
Score = 660 bits (1704), Expect = 0.0
Identities = 337/659 (51%), Positives = 465/659 (70%), Gaps = 18/659 (2%)
Query: 2 AEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMV 61
A +LE+ G++ FS I+VLGK+GVGKS+TINSIF E K + A+ T V ++VGMV
Sbjct: 224 ASRLEAEGRDDFAFSLNILVLGKTGVGKSATINSIFGETKTSFSAYGPATTSVTEIVGMV 283
Query: 62 QGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPL 121
G+++RV DTPGL S +Q +N K+L +VK+ KK+PPDIVLY+DRLD+Q+RD +D+P+
Sbjct: 284 DGVEIRVFDTPGLKSSAFEQSYNRKVLSTVKKLTKKSPPDIVLYVDRLDLQTRDMNDLPM 343
Query: 122 LRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDM 181
LR++T GP IW N IV LTHAASAPPDGP+G+P SYD+FV QRSH+VQQAI QA GD+
Sbjct: 344 LRSVTSALGPTIWRNVIVTLTHAASAPPDGPSGSPLSYDVFVAQRSHIVQQAIGQAVGDL 403
Query: 182 RLMNP-----VSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQ 236
RLMNP VSLVENH +CR N GQ+VLPNGQ WKP LLLL ++ KIL+EA + K Q
Sbjct: 404 RLMNPNLMNPVSLVENHPSCRKNRDGQKVLPNGQSWKPLLLLLCYSMKILSEATNISKTQ 463
Query: 237 DNP-REKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETD 295
+ + + R+RAPPLP+LLS LLQSR KLP+ D + ++ + SDS E
Sbjct: 464 EAADNRRLFGFRSRAPPLPYLLSWLLQSRAHPKLPDQAGIDNGDSDIEMADLSDSDGEEG 523
Query: 296 PDD---LPPFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAE 352
D+ LPPFKPL K+QI L+ Q+KAYL+E +YR KL KKQ + E K+ M++M +
Sbjct: 524 EDEYDQLPPFKPLKKSQIAKLNGEQRKAYLEEYDYRVKLLQKKQWREELKR---MRDMKK 580
Query: 353 SVKDLPSDYVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVL 412
K+ +DY+E +EE+G A+VPVP+PDM LP SFDSD P +RYR L+ ++Q L RPVL
Sbjct: 581 RGKNGENDYMEE-DEENGSPAAVPVPLPDMVLPQSFDSDNPAYRYRFLEPNSQLLTRPVL 639
Query: 413 ETHGWDHDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATS 472
+TH WDHD GY+G+N+E + +K P + + QVTKDK+D ++ ++ + + K+GE +T
Sbjct: 640 DTHSWDHDCGYDGVNIENSMAIINKFPAAVTVQVTKDKQDFSIHLDSSVAAKHGENGSTM 699
Query: 473 LGFDMQTVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFK 532
GFD+Q +GK LAY +R ETKF NF RNK AG+S T LG+ +S GVK+ED++ KR
Sbjct: 700 AGFDIQNIGKQLAYIVRGETKFKNFKRNKTAAGVSVTFLGENVSTGVKLEDQIALGKRLV 759
Query: 533 LVIAGGAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIP 592
LV + G + ++D AYG ++E +LR+ ++P+G+ S+L LS++ W GDLA+G N QSQI
Sbjct: 760 LVGSTGTVRSQNDSAYGANVEVRLREADFPVGQDQSSLSLSLVQWRGDLALGANFQSQIS 819
Query: 593 IGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVI-----GYSQNCSL 646
+GR + RA LNN+ +GQI++R +SS+QLQIALI ++P+ K + G ++N S+
Sbjct: 820 LGRSYKMAVRAGLNNKLSGQINVRTSSSDQLQIALIAILPVAKAIYKNFWPGVTENYSI 878
>UniRef100_Q41010 GTP-binding protein [Pisum sativum]
Length = 879
Score = 654 bits (1687), Expect = 0.0
Identities = 335/659 (50%), Positives = 462/659 (69%), Gaps = 18/659 (2%)
Query: 2 AEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMV 61
A +LE+ G++ FS I+VLGK+GVGKS+TINSIF E K + A+ T V ++VGMV
Sbjct: 224 ASRLEAEGRDDFAFSLNILVLGKTGVGKSATINSIFGETKTSFSAYGPATTSVTEIVGMV 283
Query: 62 QGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPL 121
G+++RV DTPGL S +Q +N K+L +VK+ KK+PPDIVLY+DRLD+Q+RD +D+P+
Sbjct: 284 DGVEIRVFDTPGLKSSAFEQSYNRKVLSTVKKLTKKSPPDIVLYVDRLDLQTRDMNDLPM 343
Query: 122 LRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDM 181
LR++T GP IW N IV LTHAASAPPD G+P SYD+FV QRSH+VQQAI QA GD+
Sbjct: 344 LRSVTSALGPTIWRNVIVTLTHAASAPPDEQQGSPLSYDVFVAQRSHIVQQAIGQAVGDL 403
Query: 182 RLMNP-----VSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQ 236
RLMNP VSLVENH +CR N GQ+VLPNGQ WKP LLLL ++ KIL+EA + K Q
Sbjct: 404 RLMNPNLMNPVSLVENHPSCRKNRDGQKVLPNGQSWKPLLLLLCYSMKILSEATNISKTQ 463
Query: 237 DNP-REKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETD 295
+ + + R+RAPPLP+LLS LLQSR KLP+ D + ++ + SDS E
Sbjct: 464 EAADNRRLFGFRSRAPPLPYLLSWLLQSRAHPKLPDQAGIDNGDSDIEMADLSDSDGEEG 523
Query: 296 PDD---LPPFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAE 352
D+ LPPFKPL K+QI L+ Q+KAYL+E +YR KL KKQ + E K+ M++M +
Sbjct: 524 EDEYDQLPPFKPLKKSQIAKLNGEQRKAYLEEYDYRVKLLQKKQWREELKR---MRDMKK 580
Query: 353 SVKDLPSDYVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVL 412
K+ +DY+E +EE+G A+VPVP+PDM LP SFDSD P +RYR L+ ++Q L RPVL
Sbjct: 581 RGKNGENDYMEE-DEENGSPAAVPVPLPDMVLPQSFDSDNPAYRYRFLEPNSQLLTRPVL 639
Query: 413 ETHGWDHDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATS 472
+TH WDHD GY+G+N+E + +K P + + QVTKDK+D ++ ++ + + K+GE +T
Sbjct: 640 DTHSWDHDCGYDGVNIENSMAIINKFPAAVTVQVTKDKQDFSIHLDSSVAAKHGENGSTM 699
Query: 473 LGFDMQTVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFK 532
GFD+Q +GK LAY +R ETKF NF RNK AG+S T LG+ +S GVK+ED++ KR
Sbjct: 700 AGFDIQNIGKQLAYIVRGETKFKNFKRNKTAAGVSVTFLGENVSTGVKLEDQIALGKRLV 759
Query: 533 LVIAGGAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIP 592
LV + G + ++D AYG ++E +LR+ ++P+G+ S+L LS++ W GDLA+G N QSQI
Sbjct: 760 LVGSTGTVRSQNDSAYGANVEVRLREADFPVGQDQSSLSLSLVQWRGDLALGANFQSQIS 819
Query: 593 IGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVI-----GYSQNCSL 646
+GR + RA LNN+ +GQI++R +SS+QLQIALI ++P+ K + G ++N S+
Sbjct: 820 LGRSYKMAVRAGLNNKLSGQINVRTSSSDQLQIALIAILPVAKAIYKNFWPGVTENYSI 878
>UniRef100_O22774 Putative chloroplast outer envelope 86-like protein [Arabidopsis
thaliana]
Length = 865
Score = 646 bits (1667), Expect = 0.0
Identities = 332/647 (51%), Positives = 445/647 (68%), Gaps = 11/647 (1%)
Query: 2 AEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMV 61
A + E+ G E L FS I+VLGK+GVGKS+TINSI + DAF + T V+++ G V
Sbjct: 203 AVESEAEGNEELIFSLNILVLGKAGVGKSATINSILGNQIASIDAFGLSTTSVREISGTV 262
Query: 62 QGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPL 121
G+K+ IDTPGL + DQ N K+L SVK+ +KK PPDIVLY+DRLD Q+RD +++PL
Sbjct: 263 NGVKITFIDTPGLKSAAMDQSTNAKMLSSVKKVMKKCPPDIVLYVDRLDTQTRDLNNLPL 322
Query: 122 LRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDM 181
LRTIT G IW NAIV LTHAASAPPDGP+GTP SYD+FV Q SH+VQQ+I QA GD+
Sbjct: 323 LRTITASLGTSIWKNAIVTLTHAASAPPDGPSGTPLSYDVFVAQCSHIVQQSIGQAVGDL 382
Query: 182 RLMNP-----VSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQ 236
RLMNP VSLVENH CR N G +VLPNGQ W+ QLLLL ++ K+L+E N+LL+ Q
Sbjct: 383 RLMNPSLMNPVSLVENHPLCRKNREGVKVLPNGQTWRSQLLLLCYSLKVLSETNSLLRPQ 442
Query: 237 DN-PREKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDS----G 291
+ K + R R+PPLP+LLS LLQSR KLP DQ D + ++D+ SDS G
Sbjct: 443 EPLDHRKVFGFRVRSPPLPYLLSWLLQSRAHPKLPGDQGGDSVDSDIEIDDVSDSEQEDG 502
Query: 292 DETDPDDLPPFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMA 351
++ + D LPPFKPL K Q+ LS Q+KAY +E +YR KL KKQ + E K+ K MK+
Sbjct: 503 EDDEYDQLPPFKPLRKTQLAKLSNEQRKAYFEEYDYRVKLLQKKQWREELKRMKEMKKNG 562
Query: 352 ESVKDLPSDYV-ENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRP 410
+ + + Y E + E+G A+VPVP+PDM LP SFDSD +RYR+L+ ++Q L RP
Sbjct: 563 KKLGESEFGYPGEEDDPENGAPAAVPVPLPDMVLPPSFDSDNSAYRYRYLEPTSQLLTRP 622
Query: 411 VLETHGWDHDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKA 470
VL+THGWDHD GY+G+N E L + P + + QVTKDKK+ N+ ++ + S K+GE +
Sbjct: 623 VLDTHGWDHDCGYDGVNAEHSLALASRFPATATVQVTKDKKEFNIHLDSSVSAKHGENGS 682
Query: 471 TSLGFDMQTVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKR 530
T GFD+Q VGK LAY +R ETKF N +NK T G S T LG+ ++ GVK+ED++ KR
Sbjct: 683 TMAGFDIQNVGKQLAYVVRGETKFKNLRKNKTTVGGSVTFLGENIATGVKLEDQIALGKR 742
Query: 531 FKLVIAGGAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQ 590
LV + G M + D AYG +LE +LR+ ++P+G+ S+ GLS++ W GDLA+G NLQSQ
Sbjct: 743 LVLVGSTGTMRSQGDSAYGANLEVRLREADFPIGQDQSSFGLSLVKWRGDLALGANLQSQ 802
Query: 591 IPIGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKV 637
+ +GR + + RA LNN+ +GQI++R +SS+QLQIAL ++P+ +
Sbjct: 803 VSVGRNSKIALRAGLNNKMSGQITVRTSSSDQLQIALTAILPIAMSI 849
>UniRef100_O81283 T14P8.24 protein [Arabidopsis thaliana]
Length = 1503
Score = 646 bits (1667), Expect = 0.0
Identities = 332/647 (51%), Positives = 445/647 (68%), Gaps = 11/647 (1%)
Query: 2 AEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMV 61
A + E+ G E L FS I+VLGK+GVGKS+TINSI + DAF + T V+++ G V
Sbjct: 841 AVESEAEGNEELIFSLNILVLGKAGVGKSATINSILGNQIASIDAFGLSTTSVREISGTV 900
Query: 62 QGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPL 121
G+K+ IDTPGL + DQ N K+L SVK+ +KK PPDIVLY+DRLD Q+RD +++PL
Sbjct: 901 NGVKITFIDTPGLKSAAMDQSTNAKMLSSVKKVMKKCPPDIVLYVDRLDTQTRDLNNLPL 960
Query: 122 LRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDM 181
LRTIT G IW NAIV LTHAASAPPDGP+GTP SYD+FV Q SH+VQQ+I QA GD+
Sbjct: 961 LRTITASLGTSIWKNAIVTLTHAASAPPDGPSGTPLSYDVFVAQCSHIVQQSIGQAVGDL 1020
Query: 182 RLMNP-----VSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQ 236
RLMNP VSLVENH CR N G +VLPNGQ W+ QLLLL ++ K+L+E N+LL+ Q
Sbjct: 1021 RLMNPSLMNPVSLVENHPLCRKNREGVKVLPNGQTWRSQLLLLCYSLKVLSETNSLLRPQ 1080
Query: 237 DN-PREKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDS----G 291
+ K + R R+PPLP+LLS LLQSR KLP DQ D + ++D+ SDS G
Sbjct: 1081 EPLDHRKVFGFRVRSPPLPYLLSWLLQSRAHPKLPGDQGGDSVDSDIEIDDVSDSEQEDG 1140
Query: 292 DETDPDDLPPFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMA 351
++ + D LPPFKPL K Q+ LS Q+KAY +E +YR KL KKQ + E K+ K MK+
Sbjct: 1141 EDDEYDQLPPFKPLRKTQLAKLSNEQRKAYFEEYDYRVKLLQKKQWREELKRMKEMKKNG 1200
Query: 352 ESVKDLPSDYV-ENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRP 410
+ + + Y E + E+G A+VPVP+PDM LP SFDSD +RYR+L+ ++Q L RP
Sbjct: 1201 KKLGESEFGYPGEEDDPENGAPAAVPVPLPDMVLPPSFDSDNSAYRYRYLEPTSQLLTRP 1260
Query: 411 VLETHGWDHDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKA 470
VL+THGWDHD GY+G+N E L + P + + QVTKDKK+ N+ ++ + S K+GE +
Sbjct: 1261 VLDTHGWDHDCGYDGVNAEHSLALASRFPATATVQVTKDKKEFNIHLDSSVSAKHGENGS 1320
Query: 471 TSLGFDMQTVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKR 530
T GFD+Q VGK LAY +R ETKF N +NK T G S T LG+ ++ GVK+ED++ KR
Sbjct: 1321 TMAGFDIQNVGKQLAYVVRGETKFKNLRKNKTTVGGSVTFLGENIATGVKLEDQIALGKR 1380
Query: 531 FKLVIAGGAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQ 590
LV + G M + D AYG +LE +LR+ ++P+G+ S+ GLS++ W GDLA+G NLQSQ
Sbjct: 1381 LVLVGSTGTMRSQGDSAYGANLEVRLREADFPIGQDQSSFGLSLVKWRGDLALGANLQSQ 1440
Query: 591 IPIGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKV 637
+ +GR + + RA LNN+ +GQI++R +SS+QLQIAL ++P+ +
Sbjct: 1441 VSVGRNSKIALRAGLNNKMSGQITVRTSSSDQLQIALTAILPIAMSI 1487
>UniRef100_Q75LI9 Putative GTP-binding protein, having alternative splicing products
[Oryza sativa]
Length = 1206
Score = 545 bits (1405), Expect = e-153
Identities = 288/629 (45%), Positives = 409/629 (64%), Gaps = 11/629 (1%)
Query: 2 AEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMV 61
A QLE+ G E L+FSC ++VLGK+GVGKS+TINSIF E K T AF T V+++ G+V
Sbjct: 570 ALQLEAEGTEDLEFSCNVLVLGKTGVGKSATINSIFGEDKSKTSAFLPATTAVKEISGVV 629
Query: 62 QGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPL 121
G+K RV+DTPGL + D+ N K+L+SVK++IK+ PPD+VLY+DR+D Q +D +++ L
Sbjct: 630 GGVKFRVVDTPGLGTTHMDEKSNRKVLNSVKKYIKRCPPDVVLYVDRIDTQRQDANNLSL 689
Query: 122 LRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDM 181
L+ IT + G IW I+ LTH+A+APP+GP+G P +Y+MFVTQR+H +QQ+IRQA D
Sbjct: 690 LQCITSVLGSSIWSKTIITLTHSAAAPPEGPSGIPLNYEMFVTQRTHAIQQSIRQATNDP 749
Query: 182 RLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPRE 241
R N +LVENH CR NT G++VLPNG +W+ LLLL ++ K + E N+L +P
Sbjct: 750 RFENTSALVENHHLCRRNTEGEKVLPNGLIWRRLLLLLCYSVKTV-ETNSLSARVASPAN 808
Query: 242 KPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLPP 301
++ R R PPLP LSSLLQSR + DQ D + D DE + +E D D LPP
Sbjct: 809 L-FSLRFRMPPLPHFLSSLLQSREHPRCAADQ----DVGDIDPDELINEDEEDDYDQLPP 863
Query: 302 FKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSDY 361
FKPL+K+Q+ LS+ Q+K Y DE +YR KL KKQLK + ++ K MK + L +
Sbjct: 864 FKPLSKSQVAKLSKEQQKLYFDEYDYRTKLLEKKQLKEQLRRLKEMKIEGNNHDVLGDN- 922
Query: 362 VENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWDHDV 421
+N ++E SV MPD +LP+SFDSD P +RYR LD +LVR + GWDHD
Sbjct: 923 -DNPDDEYETERSV---MPDWALPSSFDSDDPAYRYRCLDPKPNFLVRAITNPDGWDHDC 978
Query: 422 GYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQTVG 481
G++G++++ + P S QV KDK+++ + + + S K+ E ++ GFD+QT+
Sbjct: 979 GFDGVSLQYSLDAANAFPASLWVQVNKDKRESTIHLVSSISAKHRENVSSLAGFDIQTIM 1038
Query: 482 KDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGAMT 541
LAYTLR E+KF N +N T GLS T LGD + G K EDKL R L+ GA++
Sbjct: 1039 DQLAYTLRGESKFKNSKKNTTTGGLSMTFLGDTMVTGAKFEDKLSVGDRLTLLANTGAVS 1098
Query: 542 GRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLVA 601
R D AYG ++EA LR+K+Y +G+ L+ LG S++ WH + ++ L SQ +GR +N+
Sbjct: 1099 IRGDTAYGVNMEATLREKDYLMGQDLAILGASLVRWHKEWSMAAKLDSQFSMGRASNVAV 1158
Query: 602 RANLNNRGAGQISIRLNSSEQLQIALIGL 630
+L N+ G++SI+ N+SEQL+IAL+G+
Sbjct: 1159 HVDLTNKLTGRVSIKANTSEQLKIALLGV 1187
>UniRef100_Q6S5G3 Chloroplast import receptor Toc90 [Arabidopsis thaliana]
Length = 793
Score = 486 bits (1252), Expect = e-136
Identities = 274/645 (42%), Positives = 390/645 (59%), Gaps = 23/645 (3%)
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
+A + ES+G LDFS I+VLGK+GVGKS+TINSIF + K TDAF GT ++++V+G
Sbjct: 151 LAREQESSGIPELDFSLRILVLGKTGVGKSATINSIFGQPKSETDAFRPGTDRIEEVMGT 210
Query: 61 VQGIKVRVIDTPGLLP-SWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDM 119
V G+KV IDTPG P S S N KIL S+KR++KK PPD+VLYLDRLDM +SD
Sbjct: 211 VSGVKVTFIDTPGFHPLSSSSTRKNRKILLSIKRYVKKRPPDVVLYLDRLDMIDMRYSDF 270
Query: 120 PLLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAG 179
LL+ IT+IFG IW N I+V+TH+A A +G NG +Y+ +V QR VVQ I QA
Sbjct: 271 SLLQLITEIFGAAIWLNTILVMTHSA-ATTEGRNGQSVNYESYVGQRMDVVQHYIHQAVS 329
Query: 180 DMRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDN- 238
D +L NPV LVENH +C+ N AG+ VLPNG VWKPQ + L +K+L + +LL+ +D+
Sbjct: 330 DTKLENPVLLVENHPSCKKNLAGEYVLPNGVVWKPQFMFLCVCTKVLGDVQSLLRFRDSI 389
Query: 239 PREKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDD 298
+P + R + LP LLS L+ R E + + LN DL+E E + D
Sbjct: 390 GLGQPSSTRTAS--LPHLLSVFLRRRLSSGADETEKEIDKLLNLDLEE------EDEYDQ 441
Query: 299 LPPFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLP 358
LP + L K++ LS++QKK YLDE++YRE L++KKQLK E ++R+ K + E
Sbjct: 442 LPTIRILGKSRFEKLSKSQKKEYLDELDYRETLYLKKQLKEECRRRRDEKLVEE------ 495
Query: 359 SDYVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWD 418
+ +E+ E+ A VP+PDM+ P SFDSD P HRYR + + +QWLVRPV + GWD
Sbjct: 496 -ENLEDTEQRDQAA----VPLPDMAGPDSFDSDFPAHRYRCVSAGDQWLVRPVYDPQGWD 550
Query: 419 HDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKAT-SLGFDM 477
DVG++G+N+E + + S +GQV++DK+ +Q E ++ + T S+ D+
Sbjct: 551 RDVGFDGINIETAAKINRNLFASATGQVSRDKQRFTIQSETNAAYTRNFREQTFSVAVDL 610
Query: 478 QTVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAG 537
Q+ G+DL Y+ + TK F N G+ T G G K+ED L+ KR KL
Sbjct: 611 QSSGEDLVYSFQGGTKLQTFKHNTTDVGVGLTSFGGKYYVGGKLEDTLLVGKRVKLTANA 670
Query: 538 GAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYT 597
G M G A GGS EA +R ++YP+ L ++ + + +L + LQ+Q R T
Sbjct: 671 GQMRGSGQTANGGSFEACIRGRDYPVRNEQIGLTMTALSFKRELVLNYGLQTQFRPARGT 730
Query: 598 NLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVIGYSQ 642
N+ N+NNR G+I+++LNSSE +IALI + + K ++ S+
Sbjct: 731 NIDVNINMNNRKMGKINVKLNSSEHWEIALISALTMFKALVRRSK 775
>UniRef100_Q945M4 AT4g02510/T10P11_19 [Arabidopsis thaliana]
Length = 481
Score = 446 bits (1147), Expect = e-123
Identities = 225/461 (48%), Positives = 312/461 (66%), Gaps = 6/461 (1%)
Query: 183 LMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDN-PRE 241
LMNPVSLVENH CR N G +VLPNGQ W+ QLLLL ++ K+L+E N+LL+ Q+
Sbjct: 5 LMNPVSLVENHPLCRKNREGVKVLPNGQTWRSQLLLLCYSLKVLSETNSLLRPQEPLDHR 64
Query: 242 KPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDS----GDETDPD 297
K + R R+PPLP+LLS LLQSR KLP DQ D + ++D+ SDS G++ + D
Sbjct: 65 KVFGFRVRSPPLPYLLSWLLQSRAHPKLPGDQGGDSVDSDIEIDDVSDSEQEDGEDDEYD 124
Query: 298 DLPPFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDL 357
LPPFKPL K Q+ LS Q+KAY +E +YR KL KKQ + E K+ K MK+ + + +
Sbjct: 125 QLPPFKPLRKTQLAKLSNEQRKAYFEEYDYRVKLLQKKQWREELKRMKEMKKNGKKLGES 184
Query: 358 PSDYV-ENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHG 416
Y E + E+G A+VPVP+PDM LP SFDSD +RYR+L+ ++Q L RPVL+THG
Sbjct: 185 EFGYPGEEDDPENGAPAAVPVPLPDMVLPPSFDSDNSAYRYRYLEPTSQLLTRPVLDTHG 244
Query: 417 WDHDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFD 476
WDHD GY+G+N E L + P + + QVTKDKK+ N+ ++ + S K+GE +T GFD
Sbjct: 245 WDHDCGYDGVNAEHSLALASRFPATATVQVTKDKKEFNIHLDSSVSAKHGENGSTMAGFD 304
Query: 477 MQTVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIA 536
+Q VGK LAY +R ETKF N +NK T G S T LG+ ++ GVK+ED++ KR LV +
Sbjct: 305 IQNVGKQLAYVVRGETKFKNLRKNKTTVGGSVTFLGENIATGVKLEDQIALGKRLVLVGS 364
Query: 537 GGAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRY 596
G M + D AYG +LE +LR+ ++P+G+ S+ GLS++ W GDLA+G NLQSQ+ +GR
Sbjct: 365 TGTMRSQGDSAYGANLEVRLREADFPIGQDQSSFGLSLVKWRGDLALGANLQSQVSVGRN 424
Query: 597 TNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKV 637
+ + RA LNN+ +GQI++R +SS+QLQIAL ++P+ +
Sbjct: 425 SKIALRAGLNNKMSGQITVRTSSSDQLQIALTAILPIAMSI 465
>UniRef100_Q5ZBJ3 Putative OEP86=outer envelope protein [Oryza sativa]
Length = 801
Score = 368 bits (944), Expect = e-100
Identities = 227/627 (36%), Positives = 336/627 (53%), Gaps = 61/627 (9%)
Query: 10 QEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVRVI 69
Q+ L FSC I+VLGK GVGKS+ INSI E K +AF T V+ V +V GIKV +I
Sbjct: 218 QKDLSFSCNILVLGKIGVGKSTVINSIMGEEKNKINAFDGATTNVRLVSSVVDGIKVNII 277
Query: 70 DTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIF 129
DTPGL + DQ N+KIL +V + KK PPDI+LY+DRLD S F D+PLL+TIT I
Sbjct: 278 DTPGLRTNVMDQGWNKKILSTVNSYTKKCPPDIILYVDRLDSWSNHFDDIPLLKTITTIL 337
Query: 130 GPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDMRLMNPVSL 189
G IW N +V THA S PPD NG P +Y+ F+ QRSH+VQQ+I+QA GDM L+N S
Sbjct: 338 GTSIWVNTVVTFTHANSIPPDNSNGDPMTYETFIAQRSHIVQQSIQQATGDMCLINAFSF 397
Query: 190 VENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPREKPYTARAR 249
VEN+ C+ N G++VLP Q W+ LL+L +++K P+ +P
Sbjct: 398 VENYLYCKRNCQGKKVLPTIQNWRKYLLILCYSTK--------------PKYQPKA---- 439
Query: 250 APPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLPPFKPLTKAQ 309
S L ED + D ++ D DE + LP PL KAQ
Sbjct: 440 -------------SIHHKGLKEDSSIEVDDYSEVCD------DEYEYGQLPTLWPLMKAQ 480
Query: 310 IRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSDYVENVEEES 369
L + + K DE Y KL Q + +P D N +++
Sbjct: 481 FDELMKDKNK---DECAYHVKLIQGMQFN------------GVTQGSMPCDNDLNPLQKN 525
Query: 370 GGAASVPVPMPDMSLPASFD-SDTPTHRYRHLDSSNQWLVRPVLETHGWDHDVGYEGLNV 428
+ P+ +M + SFD D PTH+Y L+ ++ + VL H WDH+ ++G ++
Sbjct: 526 RMS-----PILNMVIEPSFDFDDPPTHQYNLLEPTSIITRKHVLGAHTWDHEYNFDGASL 580
Query: 429 ERLFVL--KDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQTVGKDLAY 486
E+ VL K + + +KD K + + + K+ + + LG+++Q K LAY
Sbjct: 581 EKTLVLHKPTKCFEATLVEFSKDMKKSRIHFNSSFRSKHVDDASHCLGYNIQNAWKKLAY 640
Query: 487 TLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGAMTGRDDV 546
+ ET ++K GLS LGD + GVK+ED + + L+++ G M + +
Sbjct: 641 CIWGETT-TKDTKHKTVGGLSVMFLGDTMLTGVKIEDYISVGESLALLVSIGTMQAKGNT 699
Query: 547 AYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLVARANLN 606
AYG ++E++L+ K YP+ R + GLS++ H +A+G NLQSQ + R++ + LN
Sbjct: 700 AYGVNMESRLKIKYYPINRLMLFFGLSLIKLHSAIALGINLQSQYLLRRHSKMALHIGLN 759
Query: 607 NRGAGQISIRLNSSEQLQIALIGLIPL 633
GQI++++++S+ +QIAL+GL+PL
Sbjct: 760 TLRNGQINLKMSTSKMVQIALLGLVPL 786
>UniRef100_Q6RJP1 Chloroplast Toc34-1 [Physcomitrella patens]
Length = 350
Score = 145 bits (365), Expect = 5e-33
Identities = 111/321 (34%), Positives = 152/321 (46%), Gaps = 47/321 (14%)
Query: 16 SCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVRVIDTPGLL 75
S T++V+GK GVGKSST+NSI E AF T + G + +IDTPGL+
Sbjct: 37 SLTVLVVGKGGVGKSSTVNSIVGERVTIVSAFQSETLRPLQCARSRAGFTLNIIDTPGLV 96
Query: 76 PSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIFGPPIWF 135
N++ L +KRF+ D+VLY+DRLD D D ++R + FGP W
Sbjct: 97 EGGCI---NDQALDIIKRFLLSKTIDVVLYVDRLDGYRVDNLDRQVIRGLARSFGPNFWR 153
Query: 136 NAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAG---DMRLMNPVSLVEN 192
A++VLTHA +P DG N Y FV +RS +Q AIRQ AG D + + P +LVEN
Sbjct: 154 LAVIVLTHAQFSPSDGVN-----YTEFVEKRSAALQAAIRQEAGLKKDEKEV-PYALVEN 207
Query: 193 HSACRTNTAGQRVLPNGQVWKPQLL--LLSFASKILAEANALLKLQDNPREKPY------ 244
C TN G+++LPNG VW P L+ ++ A+ KL D P +
Sbjct: 208 SGRCNTNDGGEKILPNGTVWLPALVERIVEVATGTTDSVFVDKKLIDGPNANEWGKWWIP 267
Query: 245 ---TARARAPPLPFL------LSSLLQSRPQLKLPEDQFSDEDSLNDD--LDE------- 286
A+ +P L+ RPQ ++ ++F SL DD +DE
Sbjct: 268 VLAVAQYFLVVMPIRKAIEKDLAEEKNQRPQWEIRAEEFRRNTSLYDDDQVDEQAISQAL 327
Query: 287 ---------PSDSGDETDPDD 298
PS DE D DD
Sbjct: 328 EQDDGLSGQPSSFVDEDDDDD 348
>UniRef100_Q6RJN9 Chloroplast Toc34-3 [Physcomitrella patens]
Length = 350
Score = 144 bits (364), Expect = 6e-33
Identities = 86/204 (42%), Positives = 113/204 (55%), Gaps = 10/204 (4%)
Query: 16 SCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVRVIDTPGLL 75
S T++V+GK GVGKSST+NSI E AF T + G + VIDTPGL+
Sbjct: 37 SLTVLVVGKGGVGKSSTVNSIIGERVTVVSAFQSETLRPLQCARTRAGFTLNVIDTPGLI 96
Query: 76 PSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIFGPPIWF 135
N++ L +KRF+ D+VLY+DRLD D D ++R + FGP W
Sbjct: 97 EGGCI---NDQALDIIKRFLLNKTIDVVLYVDRLDGYRVDNLDKQVIRALARSFGPNFWR 153
Query: 136 NAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDMRLMNPVS--LVENH 193
AI+ LTHA +PPDG + Y FV RS ++ AIRQ AG + +S LVEN
Sbjct: 154 IAIIALTHAQLSPPDGVD-----YTEFVNNRSAALRAAIRQEAGFKKSEGEISYMLVENS 208
Query: 194 SACRTNTAGQRVLPNGQVWKPQLL 217
C TN+ G++VLPNG VW P L+
Sbjct: 209 GRCNTNSEGEKVLPNGSVWLPALV 232
>UniRef100_Q9M465 Toc34-2 protein [Zea mays]
Length = 326
Score = 136 bits (342), Expect = 2e-30
Identities = 81/207 (39%), Positives = 115/207 (55%), Gaps = 17/207 (8%)
Query: 18 TIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVRVIDTPGLLPS 77
TI+V+GK GVGKSST+NSI E + AF + G + +IDTPGL+
Sbjct: 39 TILVMGKGGVGKSSTVNSIVGERVASVSAFQSEGLRPMMCSRTRAGFTLNIIDTPGLIEG 98
Query: 78 WSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIFGPPIWFNA 137
NE+ + +KRF+ D++LY+DRLD D D ++R IT+ FG IW +
Sbjct: 99 GYI---NEQAVDIIKRFLLGKTIDVLLYVDRLDAYRMDTLDEQVIRAITNSFGKDIWRRS 155
Query: 138 IVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAA-------GDMRLMNPVSLV 190
+VVLTHA +PPDG Y+ F T+RS + + I A GD L P++LV
Sbjct: 156 LVVLTHAQLSPPDG-----IDYNDFFTRRSEALLRYIHSGAGINKREYGDFPL--PIALV 208
Query: 191 ENHSACRTNTAGQRVLPNGQVWKPQLL 217
EN C+TN G+++LP+G +W P L+
Sbjct: 209 ENSGRCKTNEHGEKILPDGTLWVPNLM 235
>UniRef100_Q41009 Translocase of chloroplast 34 [Pisum sativum]
Length = 310
Score = 135 bits (339), Expect = 5e-30
Identities = 97/275 (35%), Positives = 137/275 (49%), Gaps = 25/275 (9%)
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
+ E L + QE ++ S TI+V+GK GVGKSST+NSI E + F + V
Sbjct: 25 LLELLGNLKQEDVN-SLTILVMGKGGVGKSSTVNSIIGERVVSISPFQSEGPRPVMVSRS 83
Query: 61 VQGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMP 120
G + +IDTPGL+ N+ L+ +K F+ D++LY+DRLD D D
Sbjct: 84 RAGFTLNIIDTPGLIEGGYI---NDMALNIIKSFLLDKTIDVLLYVDRLDAYRVDNLDKL 140
Query: 121 LLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAG- 179
+ + ITD FG IW AIV LTHA +PPDG YD F ++RS + Q +R A
Sbjct: 141 VAKAITDSFGKGIWNKAIVALTHAQFSPPDG-----LPYDEFFSKRSEALLQVVRSGASL 195
Query: 180 --DMRLMN-PVSLVENHSACRTNTAGQRVLPNGQVWKPQL------LLLSFASKILAEAN 230
D + + PV L+EN C N + ++VLPNG W P L + L+ + I + N
Sbjct: 196 KKDAQASDIPVVLIENSGRCNKNDSDEKVLPNGIAWIPHLVQTITEVALNKSESIFVDKN 255
Query: 231 ALLKLQDNPREKPYTARARAPPLPFLLSSLLQSRP 265
+ N R K + PL F L L ++P
Sbjct: 256 LIDGPNPNQRGKLWI------PLIFALQYLFLAKP 284
>UniRef100_Q9SBX0 Toc34-1 protein [Zea mays]
Length = 326
Score = 133 bits (335), Expect = 1e-29
Identities = 79/205 (38%), Positives = 113/205 (54%), Gaps = 13/205 (6%)
Query: 18 TIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVRVIDTPGLLPS 77
TI+V+GK GVGKSST+NSI E AF + G + +IDTPGL+
Sbjct: 39 TILVMGKGGVGKSSTVNSIVGERIATVSAFQSEGLRPMMWSRTRAGFTLNIIDTPGLIEG 98
Query: 78 WSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIFGPPIWFNA 137
NE+ + +KRF+ D++LY+DRLD D D ++R IT+ FG IW +
Sbjct: 99 GYI---NEQAVDIIKRFLLGKTIDVLLYVDRLDAYRMDTLDGQVIRAITNSFGKDIWRRS 155
Query: 138 IVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDMR-----LMNPVSLVEN 192
+VVLTHA +PPDG Y+ F T+RS + + I AG + P++LVEN
Sbjct: 156 LVVLTHAQLSPPDG-----IEYNDFFTRRSEALLRYIHSGAGIKKREYGDFPLPIALVEN 210
Query: 193 HSACRTNTAGQRVLPNGQVWKPQLL 217
C+TN G+++LP+G W P+L+
Sbjct: 211 SGRCKTNEHGEKILPDGTPWVPKLM 235
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,119,671,765
Number of Sequences: 2790947
Number of extensions: 49321584
Number of successful extensions: 194387
Number of sequences better than 10.0: 651
Number of HSP's better than 10.0 without gapping: 106
Number of HSP's successfully gapped in prelim test: 548
Number of HSP's that attempted gapping in prelim test: 193420
Number of HSP's gapped (non-prelim): 1024
length of query: 663
length of database: 848,049,833
effective HSP length: 134
effective length of query: 529
effective length of database: 474,062,935
effective search space: 250779292615
effective search space used: 250779292615
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC147010.6


