

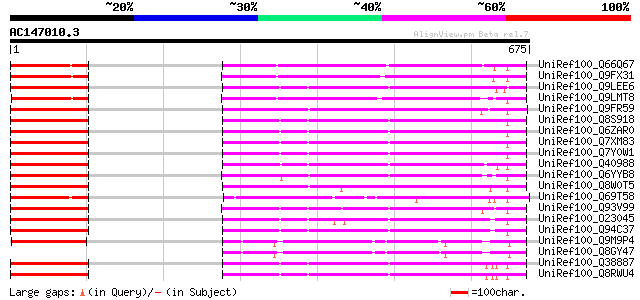
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.3 - phase: 0 /pseudo
(675 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q66Q67 Homeodomain protein HOX3 [Gossypium hirsutum] 307 8e-82
UniRef100_Q9FX31 Homeobox protein, putative [Arabidopsis thaliana] 292 3e-77
UniRef100_Q9LEE6 OCL5 protein [Zea mays] 284 5e-75
UniRef100_Q9LMT8 F2H15.14 protein [Arabidopsis thaliana] 280 8e-74
UniRef100_Q9FR59 Homeobox 1 [Picea abies] 278 4e-73
UniRef100_Q8S918 Roc1 [Oryza sativa] 277 6e-73
UniRef100_Q6ZAR0 Roc1 [Oryza sativa] 277 6e-73
UniRef100_Q7XM83 OSJNBb0060E08.16 protein [Oryza sativa] 275 2e-72
UniRef100_Q7Y0W1 GL2-type homeodomain protein [Oryza sativa] 274 7e-72
UniRef100_Q40988 Homeobox protein [Phalaenopsis sp. SM9108] 273 9e-72
UniRef100_Q6YYB8 Putative OCL5 protein [Oryza sativa] 268 3e-70
UniRef100_Q8W0T5 OCL5 protein [Sorghum bicolor] 266 1e-69
UniRef100_Q69T58 Putative homeobox [Oryza sativa] 264 7e-69
UniRef100_Q93V99 Protodermal factor2 [Arabidopsis thaliana] 264 7e-69
UniRef100_O23045 YUP8H12.16 protein [Arabidopsis thaliana] 263 1e-68
UniRef100_Q94C37 At1g05230/YUP8H12_16 [Arabidopsis thaliana] 263 1e-68
UniRef100_Q9M9P4 T17B22.5 protein [Arabidopsis thaliana] 263 2e-68
UniRef100_Q8GY47 Hypothetical protein At3g03260/T17B22_5 [Arabid... 262 2e-68
UniRef100_Q38887 Meristem L1 layer homeobox protein [Arabidopsis... 258 3e-67
UniRef100_Q8RWU4 Putative L1-specific homeobox gene ATML1/ovule-... 256 2e-66
>UniRef100_Q66Q67 Homeodomain protein HOX3 [Gossypium hirsutum]
Length = 713
Score = 307 bits (786), Expect = 8e-82
Identities = 174/405 (42%), Positives = 243/405 (59%), Gaps = 16/405 (3%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
LFPTIV+ A+TI+V G G+ +L LM EE+ +LSPLV REF +RYC++++ G+W
Sbjct: 310 LFPTIVSIAKTIEVISPGMLGTHRCSLQLMYEELQVLSPLVPTREFYTLRYCQQIEQGLW 369
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
I +VS+D P A R + PSGC+I++MP+G VTW+E VE+EDK H +YRDL
Sbjct: 370 AIVNVSYDL--PQFASQCRSHRLPSGCLIQDMPNGYSKVTWLERVEIEDKTPIHRLYRDL 427
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
V + +GAE W+ LQRMCE V + + GVI + EGR S++KLA RMV F
Sbjct: 428 VHSGSAFGAERWLTTLQRMCEWFACLRVSSTSTRDLGGVIPSPEGRRSMMKLAQRMVNNF 487
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
C + L+ + GVRV++ ++D QPNG V++AATT WLP+ Q VF
Sbjct: 488 CTSVGTSNSHRSTTLSGSNEVGVRVTVHKSSDP--GQPNGIVLSAATTFWLPVSPQNVFN 545
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIKSFIPTQRQMVILQESFTSSV 577
F KD R QW+ LS GN + E+AHI+NG + GNCISV+++F + M+ILQES S
Sbjct: 546 FFKDERTRPQWDVLSNGNAVQEVAHIANGSHPGNCISVLRAFNTSHNNMLILQESCIDSS 605
Query: 578 GSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPFGASKS-------I 630
GS V+Y P+D ++VA+ GED +P+LP G + +L GAS S
Sbjct: 606 GSLVVYCPVDLPAINVAMSGEDPSYIPLLPSGFTITP--DGHLEQGDGASTSSSTGHGRS 663
Query: 631 EDGSLITLAAQTYAI---GQVLNVDSLNDINSQLASTILNVKDAL 672
GSLIT+A Q LN+DS+ +N+ +A+T+ +K AL
Sbjct: 664 SGGSLITVAFQILVSSLPSAKLNLDSVTIVNNLIANTVQQIKAAL 708
Score = 115 bits (288), Expect = 4e-24
Identities = 58/101 (57%), Positives = 70/101 (68%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPDE QR QL+ ++GL P+QIK WFQN+R +K QHER N LR ENDKIR EN+
Sbjct: 47 KECPHPDEKQRLQLSRELGLAPRQIKFWFQNRRTQMKAQHERADNSALRAENDKIRCENI 106
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
I+E LK IC CGG P +D Q+M+ ENAQLK+E
Sbjct: 107 AIREALKNVICPSCGGPP-ANEDSYFDDQKMRMENAQLKEE 146
>UniRef100_Q9FX31 Homeobox protein, putative [Arabidopsis thaliana]
Length = 722
Score = 292 bits (747), Expect = 3e-77
Identities = 158/405 (39%), Positives = 242/405 (59%), Gaps = 15/405 (3%)
Query: 276 TCLFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPG 335
T LFP+I+ ++T+ V G G+ +GAL L+ EEM +LSPLV REF +RYC++ + G
Sbjct: 316 TELFPSIIAASKTLAVISSGMGGTHEGALHLLYEEMEVLSPLVATREFCELRYCQQTEQG 375
Query: 336 VWVITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYR 395
W++ +VS+D P S+ ++ PSGC+I++MP+G VTWVEH+E E+K H +YR
Sbjct: 376 SWIVVNVSYDL--PQFVSHSQSYRFPSGCLIQDMPNGYSKVTWVEHIETEEKELVHELYR 433
Query: 396 DLVGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVK 455
+++ +GA+ W+ LQRMCER +V A + GVI + EG+ S+++LA RM+
Sbjct: 434 EIIHRGIAFGADRWVTTLQRMCERFASLSVPASSSRDLGGVILSPEGKRSMMRLAQRMIS 493
Query: 456 MFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKV 515
+C ++ ++ + G+RV T + +PNGTV+ AATT WLP Q V
Sbjct: 494 NYCLSVSRSNNTRSTVVSELNEVGIRV-----TAHKSPEPNGTVLCAATTFWLPNSPQNV 548
Query: 516 FEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIKSFIPT-QRQMVILQESFT 574
F FLKD R QW+ LS GN + E+AHISNG + GNCISV++ T M+ILQES T
Sbjct: 549 FNFLKDERTRPQWDVLSNGNAVQEVAHISNGSHPGNCISVLRGSNATHSNNMLILQESST 608
Query: 575 SSVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPFGAS----KSI 630
S G++V+Y+P+D +++A+ GED +P+L G + + + GAS ++
Sbjct: 609 DSSGAFVVYSPVDLAALNIAMSGEDPSYIPLLSSGFTISPDGNGSNSEQGGASTSSGRAS 668
Query: 631 EDGSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
GSLIT+ Q LN++S+ +N+ + +T+ +K AL
Sbjct: 669 ASGSLITVGFQIMVSNLPTAKLNMESVETVNNLIGTTVHQIKTAL 713
Score = 110 bits (274), Expect = 2e-22
Identities = 55/101 (54%), Positives = 70/101 (68%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPDE QR QL+ ++GL P+QIK WFQN+R LK QHER N L+ ENDKIR EN+
Sbjct: 52 KECPHPDEKQRNQLSRELGLAPRQIKFWFQNRRTQLKAQHERADNSALKAENDKIRCENI 111
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
I+E LK IC +CGG P +D Q+++ ENA L++E
Sbjct: 112 AIREALKHAICPNCGGPPV-SEDPYFDEQKLRIENAHLREE 151
>UniRef100_Q9LEE6 OCL5 protein [Zea mays]
Length = 795
Score = 284 bits (727), Expect = 5e-75
Identities = 165/404 (40%), Positives = 241/404 (58%), Gaps = 15/404 (3%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F +IV++A T +V G GS +GAL +M+ E + SPLV RE +RYCK G W
Sbjct: 390 VFSSIVSRASTHEVLSTGVAGSYNGALQVMSMEFQVPSPLVPTRESYFVRYCKNNPDGTW 449
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ DVS DS RP+ P+ + + PSGC+I+EMP+G VTWVEHVEV+D+ H +YR L
Sbjct: 450 AVVDVSLDSLRPS--PVIKCRRRPSGCLIQEMPNGYSKVTWVEHVEVDDR-SVHNLYRPL 506
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
V +GA+ W+ L R CER + IP + +GVI ++EGR S++KLA+RMV F
Sbjct: 507 VNSGLAFGAKRWVGTLDRQCERLASAMASNIPNGD-LGVITSIEGRKSMLKLAERMVASF 565
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
C +T + L+ VRV R + DD +P G V+ AAT+ WLP+P ++VF+
Sbjct: 566 CGGVTASAAHQWTTLSGSGAEDVRVMTRKSVDDPG-RPPGIVLNAATSFWLPVPPKRVFD 624
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESFTS 575
FL+D T RS+W+ LS G + E+AHI+NG HGNC+S+++ S Q M+ILQES T
Sbjct: 625 FLRDETSRSEWDILSNGGAVQEMAHIANGRDHGNCVSLLRVNSANSNQSNMLILQESCTD 684
Query: 576 SVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPFGASKSIE---D 632
+ GSYV+YAP+D M+V L G D + +LP G + A + ++
Sbjct: 685 ASGSYVVYAPVDVVAMNVVLNGGDPDYVALLPSGFAILPDGPPPAGAAPSHGEGLDAGGG 744
Query: 633 GSLITLAAQ----TYAIGQVLNVDSLNDINSQLASTILNVKDAL 672
GSL+T+A Q + G+ L++ S+ +NS +A T+ +K A+
Sbjct: 745 GSLLTVAFQILVDSVPTGK-LSLGSVATVNSLIACTVERIKAAV 787
Score = 102 bits (254), Expect = 4e-20
Identities = 49/101 (48%), Positives = 66/101 (64%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ ++GLEP Q+K WFQNKR +K+QHER N LR ENDK+R EN+
Sbjct: 124 KECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKNQHERHENAQLRAENDKLRAENM 183
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ KE L C CGG P + + ++ ENA+L+ E
Sbjct: 184 RYKEALGTASCPSCGG-PAALGEMSFDEHHLRLENARLRDE 223
>UniRef100_Q9LMT8 F2H15.14 protein [Arabidopsis thaliana]
Length = 687
Score = 280 bits (717), Expect = 8e-74
Identities = 163/404 (40%), Positives = 237/404 (58%), Gaps = 25/404 (6%)
Query: 276 TCLFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPG 335
T LFP+IV ++T+ V G RG+ AL LM EE+ +LSPLV REF ++RYC++++ G
Sbjct: 296 TELFPSIVASSKTLAVISSGLRGNHGDALHLMIEELQVLSPLVTTREFCVLRYCQQIEHG 355
Query: 336 VWVITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYR 395
W I +VS++ P SR ++ PSGC+I++M +G VTWVEH E E++ H +++
Sbjct: 356 TWAIVNVSYEF--PQFISQSRSYRFPSGCLIQDMSNGYSKVTWVEHGEFEEQEPIHEMFK 413
Query: 396 DLVGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVK 455
D+V + +GAE WI LQRMCER A + GVI + EG+ S+++LA RMV
Sbjct: 414 DIVHKGLAFGAERWIATLQRMCERFTNLLEPATSSLDLGGVIPSPEGKRSIMRLAHRMVS 473
Query: 456 MFCECLTMPGQVELNHLT-LDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQK 514
FC + ++ LD G IR T+ +PNG V+ AAT+ WLP+ Q
Sbjct: 474 NFCLSVGTSNNTRSTVVSGLDEFG-----IRVTSHKSRHEPNGMVLCAATSFWLPISPQN 528
Query: 515 VFEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIKSF--IPTQRQMVILQES 572
VF FLKD R QW+ LS GN + E+AHI+NG GNCISV++ F +Q M+ILQES
Sbjct: 529 VFNFLKDERTRPQWDVLSNGNSVQEVAHITNGSNPGNCISVLRGFNASSSQNNMLILQES 588
Query: 573 -FTSSVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPFGASKSIE 631
SS + VIY P+D +++A+ G+D+ +PILP G + +P G+SK
Sbjct: 589 CIDSSSAALVIYTPVDLPALNIAMSGQDTSYIPILPSGFAI---------SPDGSSKG-- 637
Query: 632 DGSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
GSLIT+ Q G LN++S+ +N+ + +T+ +K L
Sbjct: 638 GGSLITVGFQIMVSGLQPAKLNMESMETVNNLINTTVHQIKTTL 681
Score = 105 bits (261), Expect = 6e-21
Identities = 53/100 (53%), Positives = 67/100 (67%), Gaps = 1/100 (1%)
Query: 3 ECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENLK 62
EC HPDE QR QL+ ++GL P+QIK WFQN+R K QHER N L+ ENDKIR EN+
Sbjct: 42 ECQHPDEKQRNQLSRELGLAPRQIKFWFQNRRTQKKAQHERADNCALKEENDKIRCENIA 101
Query: 63 IKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
I+E +K IC CG SP +D Q+++ ENAQL+ E
Sbjct: 102 IREAIKHAICPSCGDSPV-NEDSYFDEQKLRIENAQLRDE 140
>UniRef100_Q9FR59 Homeobox 1 [Picea abies]
Length = 763
Score = 278 bits (711), Expect = 4e-73
Identities = 161/404 (39%), Positives = 238/404 (58%), Gaps = 13/404 (3%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+FP IV++ T+ VF G G+ +GAL +M+ E + SPLV RE +RYCK+ +W
Sbjct: 361 MFPGIVSRPFTVDVFSTGVAGNYNGALQVMHAEFQVPSPLVPTREIYFVRYCKQHSDSIW 420
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ DVS DS R N++ + R + PSGC+I+EMP+ VTWVEHVE +D+ H++YR L
Sbjct: 421 AVVDVSLDSLRGNSSSVIRCRRRPSGCLIQEMPNSYSKVTWVEHVEADDRA-VHHIYRQL 479
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
V +GA+ WI LQR CER IP + +GVI + EGR S++KLA+RMV F
Sbjct: 480 VNSGMAFGAKRWIATLQRQCERLASVLASNIPARD-LGVIPSPEGRKSILKLAERMVTSF 538
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
C ++ L+ VRV R + DD +P G +++AAT+LWLP+P +KVF+
Sbjct: 539 CAGVSASTAHTWTTLSGSGAEDVRVMTRKSI-DDPGRPPGIILSAATSLWLPVPPKKVFD 597
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESFTS 575
FL+D R++W+ LS G + E+ HI+NG GNC+S+++ + Q M+ILQES T
Sbjct: 598 FLRDENSRNEWDILSNGGLVQEVDHIANGQDPGNCVSLLRVNTVNSNQSNMLILQESCTD 657
Query: 576 SVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIV---CSKNQANLNAPFGASKSIED 632
+ GS+VIYAP+D M+V L G D + +LP G + K A N+ G +
Sbjct: 658 ASGSFVIYAPVDIVAMNVVLSGGDPDYVALLPSGFAILPDSPKCMAVTNS--GINDLGTG 715
Query: 633 GSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDALM 673
GSL+T+A Q L++ S+ +NS ++ T+ +K A+M
Sbjct: 716 GSLLTVAFQILVDSVPTAKLSLGSVATVNSLISCTVDRIKAAVM 759
Score = 103 bits (257), Expect = 2e-20
Identities = 48/101 (47%), Positives = 70/101 (68%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ ++GLEP Q+K WFQNKR +K QHER N LR EN+K+R+EN+
Sbjct: 104 KECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKTQHERHENTQLRSENEKLRSENM 163
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ +E L C +CGG P + + Q+++ ENA+L++E
Sbjct: 164 RYREALNNASCPNCGG-PAALGEMSFDEQQLRMENARLREE 203
>UniRef100_Q8S918 Roc1 [Oryza sativa]
Length = 784
Score = 277 bits (709), Expect = 6e-73
Identities = 164/402 (40%), Positives = 235/402 (57%), Gaps = 12/402 (2%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F +IV++A T +V G G+ +GAL +M+ E + SPLV RE +RYCK G W
Sbjct: 381 VFSSIVSRASTHEVLSTGVAGNYNGALQVMSMEFQVPSPLVPTRESYFVRYCKNNSDGTW 440
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ DVS DS RP+ P+ + + PSGC+I+EMP+G VTWVEHVEV+D H +Y+ L
Sbjct: 441 AVVDVSLDSLRPS--PVQKCRRRPSGCLIQEMPNGYSKVTWVEHVEVDDS-SVHNIYKPL 497
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
V +GA+ W+ L R CER + IP + +GVI ++EGR S++KLA+RMV F
Sbjct: 498 VNSGLAFGAKRWVGTLDRQCERLASAMASNIPNGD-LGVITSVEGRKSMLKLAERMVASF 556
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
C +T + L+ VRV R + DD +P G V+ AAT+ WLP+P VF+
Sbjct: 557 CGGVTASVAHQWTTLSGSGAEDVRVMTRKSVDDPG-RPPGIVLNAATSFWLPVPPAAVFD 615
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESFTS 575
FL+D T RS+W+ LS G + E+AHI+NG HGN +S+++ S Q M+ILQES T
Sbjct: 616 FLRDETSRSEWDILSNGGAVQEMAHIANGRDHGNSVSLLRVNSANSNQSNMLILQESCTD 675
Query: 576 SVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKN-QANLNAPFGASKS-IEDG 633
+ GSYV+YAP+D M+V L G D + +LP G + N A G + S G
Sbjct: 676 ASGSYVVYAPVDIVAMNVVLNGGDPDYVALLPSGFAILPDGPSGNAQAAVGENGSGSGGG 735
Query: 634 SLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
SL+T+A Q L++ S+ +NS +A T+ +K A+
Sbjct: 736 SLLTVAFQILVDSVPTAKLSLGSVATVNSLIACTVERIKAAV 777
Score = 103 bits (258), Expect = 1e-20
Identities = 49/101 (48%), Positives = 68/101 (66%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ ++GLEP Q+K WFQNKR +K+QHER N LR ENDK+R EN+
Sbjct: 126 KECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKNQHERHENAQLRAENDKLRAENM 185
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ KE L + C +CGG P + + ++ ENA+L+ E
Sbjct: 186 RYKEALSSASCPNCGG-PAALGEMSFDEHHLRVENARLRDE 225
>UniRef100_Q6ZAR0 Roc1 [Oryza sativa]
Length = 784
Score = 277 bits (709), Expect = 6e-73
Identities = 164/402 (40%), Positives = 235/402 (57%), Gaps = 12/402 (2%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F +IV++A T +V G G+ +GAL +M+ E + SPLV RE +RYCK G W
Sbjct: 381 VFSSIVSRASTHEVLSTGVAGNYNGALQVMSMEFQVPSPLVPTRESYFVRYCKNNSDGTW 440
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ DVS DS RP+ P+ + + PSGC+I+EMP+G VTWVEHVEV+D H +Y+ L
Sbjct: 441 AVVDVSLDSLRPS--PVQKCRRRPSGCLIQEMPNGYSKVTWVEHVEVDDS-SVHNIYKPL 497
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
V +GA+ W+ L R CER + IP + +GVI ++EGR S++KLA+RMV F
Sbjct: 498 VNSGLAFGAKRWVGTLDRQCERLASAMASNIPNGD-LGVITSVEGRKSMLKLAERMVASF 556
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
C +T + L+ VRV R + DD +P G V+ AAT+ WLP+P VF+
Sbjct: 557 CGGVTASVAHQWTTLSGSGAEDVRVMTRKSVDDPG-RPPGIVLNAATSFWLPVPPAAVFD 615
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESFTS 575
FL+D T RS+W+ LS G + E+AHI+NG HGN +S+++ S Q M+ILQES T
Sbjct: 616 FLRDETSRSEWDILSNGGAVQEMAHIANGRDHGNSVSLLRVNSANSNQSNMLILQESCTD 675
Query: 576 SVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKN-QANLNAPFGASKS-IEDG 633
+ GSYV+YAP+D M+V L G D + +LP G + N A G + S G
Sbjct: 676 ASGSYVVYAPVDIVAMNVVLNGGDPDYVALLPSGFAILPDGPSGNAQAAVGENGSGSGGG 735
Query: 634 SLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
SL+T+A Q L++ S+ +NS +A T+ +K A+
Sbjct: 736 SLLTVAFQILVDSVPTAKLSLGSVATVNSLIACTVERIKAAV 777
Score = 103 bits (258), Expect = 1e-20
Identities = 49/101 (48%), Positives = 68/101 (66%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ ++GLEP Q+K WFQNKR +K+QHER N LR ENDK+R EN+
Sbjct: 126 KECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKNQHERHENAQLRAENDKLRAENM 185
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ KE L + C +CGG P + + ++ ENA+L+ E
Sbjct: 186 RYKEALSSASCPNCGG-PAALGEMSFDEHHLRVENARLRDE 225
>UniRef100_Q7XM83 OSJNBb0060E08.16 protein [Oryza sativa]
Length = 781
Score = 275 bits (704), Expect = 2e-72
Identities = 162/401 (40%), Positives = 234/401 (57%), Gaps = 11/401 (2%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F IV++A T++V G G+ +GAL +M+ E + SPLV RE +RYCK+ G W
Sbjct: 379 VFSNIVSRAITLEVLSTGVAGNYNGALQVMSVEFQVPSPLVPTRESYFVRYCKQNADGTW 438
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ DVS DS RP+ P+ + + PSGC+I+EMP+G VTWVEHVEV+D+ H +Y+ L
Sbjct: 439 AVVDVSLDSLRPS--PVLKCRRRPSGCLIQEMPNGYSKVTWVEHVEVDDR-SVHNIYKLL 495
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
V +GA W+ L R CER IP + IGVI + EGR S++KLA+RMV F
Sbjct: 496 VNSGLAFGARRWVGTLDRQCERLASVMASNIPTSD-IGVITSSEGRKSMLKLAERMVVSF 554
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
C +T + L+ VRV R + DD +P G V+ AAT+ WLP+P ++VF+
Sbjct: 555 CGGVTASVAHQWTTLSGSGAEDVRVMTRKSVDDPG-RPPGIVLNAATSFWLPVPPKRVFD 613
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESFTS 575
FL+D + RS+W+ LS G + E+AHI+NG GNC+S+++ S Q M+ILQES T
Sbjct: 614 FLRDESSRSEWDILSNGGIVQEMAHIANGRDQGNCVSLLRVNSSNSNQSNMLILQESCTD 673
Query: 576 SVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPFGASKSI-EDGS 634
+ GSYVIYAP+D M+V L G D + +LP G + A+ + GS
Sbjct: 674 ASGSYVIYAPVDVVAMNVVLNGGDPDYVALLPSGFAILPDGPAHDGGDGDGGVGVGSGGS 733
Query: 635 LITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
L+T+A Q L++ S+ +NS +A T+ +K A+
Sbjct: 734 LLTVAFQILVDSVPTAKLSLGSVATVNSLIACTVERIKAAV 774
Score = 101 bits (252), Expect = 6e-20
Identities = 47/101 (46%), Positives = 69/101 (67%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ ++GLEP Q+K WFQNKR +K+QHER N LR +N+K+R EN+
Sbjct: 121 KECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKNQHERHENSQLRSDNEKLRAENM 180
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ KE L + C +CGG P + + ++ ENA+L++E
Sbjct: 181 RYKEALSSASCPNCGG-PAALGEMSFDEHHLRIENARLREE 220
>UniRef100_Q7Y0W1 GL2-type homeodomain protein [Oryza sativa]
Length = 783
Score = 274 bits (700), Expect = 7e-72
Identities = 161/401 (40%), Positives = 233/401 (57%), Gaps = 11/401 (2%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F IV++A T++V G G+ +GAL +M+ E + SPLV RE +RYCK+ G W
Sbjct: 381 VFSNIVSRAITLEVLSTGVAGNYNGALQVMSVEFQVPSPLVPTRESYFVRYCKQNADGTW 440
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ DVS DS RP+ P+ + + PSGC+I+EMP+G VTWVEHVEV+D+ H +Y+ L
Sbjct: 441 AVVDVSLDSLRPS--PVLKCRRRPSGCLIQEMPNGYSKVTWVEHVEVDDR-SVHNIYKLL 497
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
V +GA W+ L R CER IP + IGVI + EGR S++KLA+RMV F
Sbjct: 498 VNSGLAFGARRWVGTLDRQCERLASVMASNIPTSD-IGVITSSEGRKSMLKLAERMVVSF 556
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
C +T + L+ VRV R + DD +P G V+ A T+ WLP+P+++VF
Sbjct: 557 CGGVTASVAHQWTTLSGSGAEDVRVMTRKSVDDPG-RPPGIVLNAVTSFWLPVPSKRVFH 615
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESFTS 575
FL+D + RS+W+ LS G + E+AHI+NG GNC+S+++ S Q M+ILQES T
Sbjct: 616 FLRDESSRSEWDILSNGGIVQEMAHIANGRDQGNCVSLLRVNSSNSNQSNMLILQESCTD 675
Query: 576 SVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPFGASKSI-EDGS 634
+ GSYVIYAP+D M+V L G D + +LP G + A+ + GS
Sbjct: 676 ASGSYVIYAPVDVVAMNVVLNGGDPDYVALLPSGFAILPDGPAHDGGDGDGGVGVGSGGS 735
Query: 635 LITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
L+T+A Q L++ S+ +NS +A T+ +K A+
Sbjct: 736 LLTVAFQILVDSVPTAKLSLGSVATVNSLIACTVERIKAAV 776
Score = 101 bits (252), Expect = 6e-20
Identities = 47/101 (46%), Positives = 69/101 (67%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ ++GLEP Q+K WFQNKR +K+QHER N LR +N+K+R EN+
Sbjct: 123 KECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKNQHERHENSQLRSDNEKLRAENM 182
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ KE L + C +CGG P + + ++ ENA+L++E
Sbjct: 183 RYKEALSSASCPNCGG-PAALGEMSFDEHHLRIENARLREE 222
>UniRef100_Q40988 Homeobox protein [Phalaenopsis sp. SM9108]
Length = 768
Score = 273 bits (699), Expect = 9e-72
Identities = 162/405 (40%), Positives = 232/405 (57%), Gaps = 17/405 (4%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F IV++ T++V G G+ +GAL +M E + SPLV RE +RYCK+ G W
Sbjct: 365 MFSGIVSRGMTLEVLSTGVAGNYNGALQVMTAEFQVPSPLVPTRESYFVRYCKQHPDGTW 424
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ DVS DS RP++ + R + PSGC+I+EMP+G V WVEH EV+D+ H +Y+ L
Sbjct: 425 AVVDVSLDSLRPSSL-MMRCRRRPSGCLIQEMPNGYSKVIWVEHFEVDDR-SVHSIYKPL 482
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
V +GA+ W+ L R CER +IP E IGVI T EGR S++KLA+RMV F
Sbjct: 483 VNSGIAFGAKRWVSTLDRQCERLASVMASSIPSGE-IGVITTSEGRKSMLKLAERMVLSF 541
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
C ++ + L+ VRV R + DD +P G V+ AAT+ WLP+ ++VF+
Sbjct: 542 CGGVSASTTHQWTTLSGSGAEDVRVMTRKSVDDPG-RPPGIVLNAATSFWLPVSPKRVFD 600
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESFTS 575
FL+D + RS+W+ LS G + E+AHI+NG HGNC+S+++ S Q M+ILQES T
Sbjct: 601 FLRDESSRSEWDILSNGGVVQEMAHIANGRDHGNCVSLLRVNSTNSNQSNMLILQESCTD 660
Query: 576 SVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPFGASKSIED--- 632
GSYVIYAP+D M+V L G D + +LP G + N G I +
Sbjct: 661 PTGSYVIYAPVDVVAMNVVLNGGDPDYVALLPSGFAILPDGS---NGVHGGGSGIGEVGS 717
Query: 633 --GSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
GSL+T+A Q L++ S+ +NS +A T+ +K A+
Sbjct: 718 GGGSLLTVAFQILVDSIPTAKLSLGSVATVNSLIACTVERIKAAV 762
Score = 104 bits (260), Expect = 7e-21
Identities = 50/101 (49%), Positives = 68/101 (66%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR L+ ++GLEP Q+K WFQNKR +K QH+R+ N LR ENDK+RNENL
Sbjct: 109 KECPHPDDKQRKALSKELGLEPLQVKFWFQNKRTQMKTQHDRQENSQLRAENDKLRNENL 168
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ KE L C +CGG P + + ++ ENA+L++E
Sbjct: 169 RYKEALSNASCPNCGG-PATLGEMSFDEHHLRIENARLREE 208
>UniRef100_Q6YYB8 Putative OCL5 protein [Oryza sativa]
Length = 828
Score = 268 bits (686), Expect = 3e-70
Identities = 158/408 (38%), Positives = 230/408 (55%), Gaps = 24/408 (5%)
Query: 276 TCLFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPG 335
T LF +IV++A T++V G G+ +GAL LM+ E + SPLV RE +RYCK+ G
Sbjct: 422 TALFSSIVSRAATLEVLSTGVAGNHNGALQLMSAEFQMPSPLVPTRETQFLRYCKQHPDG 481
Query: 336 VWVITDVSFDSSRPNTA-----PLSRGWKH-PSGCIIREMPHGGCLVTWVEHVEVEDKIH 389
W + DVS D R +RG + PSGC+I+EMP+G VTWVEHVE +D++
Sbjct: 482 TWAVVDVSLDGLRAGAGGGCQPAAARGHRRRPSGCLIQEMPNGYSKVTWVEHVEADDQM- 540
Query: 390 THYVYRDLVGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKL 449
H +Y+ +V +GA W+ L+R CER + + GVI T EGR S++KL
Sbjct: 541 VHNLYKPVVNSGMAFGARRWVATLERQCERLASAMASNVASSGDAGVITTSEGRRSMLKL 600
Query: 450 ADRMVKMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLP 509
A+RMV FC +T + L+ VRV R + DD +P G ++ AAT+ WLP
Sbjct: 601 AERMVASFCGGVTASTTHQWTTLSGSGAEDVRVMTRKSVDDPG-RPPGIILNAATSFWLP 659
Query: 510 LPAQKVFEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMV 567
+P +VF+FL+D + RS+W+ LS G + E+AHI+NG HGN +S+++ + Q M+
Sbjct: 660 VPPSRVFDFLRDDSTRSEWDILSNGGVVQEMAHIANGRDHGNAVSLLRVNNANSNQSNML 719
Query: 568 ILQESFTSSVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPFGAS 627
ILQE T + GSYVIYAP+D M+V L G D + +LP G + + P G
Sbjct: 720 ILQECCTDATGSYVIYAPVDVVAMNVVLNGGDPDYVALLPSGFAILP------DGPDGGG 773
Query: 628 KSIEDGSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
GSL+T+A Q L++ S+ +NS +A T+ +K A+
Sbjct: 774 -----GSLLTVAFQILVDSVPTAKLSLGSVATVNSLIACTVERIKAAI 816
Score = 99.8 bits (247), Expect = 2e-19
Identities = 48/101 (47%), Positives = 66/101 (64%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ ++GLEP Q+K WFQNKR +K QHER N LR EN+K+R EN+
Sbjct: 187 KECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKTQHERHENNALRAENEKLRAENM 246
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ KE L C +CGG P + + ++ ENA+L+ E
Sbjct: 247 RYKEALANASCPNCGG-PAAIGEMSFDEHHLRLENARLRDE 286
>UniRef100_Q8W0T5 OCL5 protein [Sorghum bicolor]
Length = 803
Score = 266 bits (680), Expect = 1e-69
Identities = 157/416 (37%), Positives = 229/416 (54%), Gaps = 23/416 (5%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
LF +IV++A T++V G G+ +GAL LM E + SPLV RE +RYCK+ G W
Sbjct: 377 LFSSIVSRAATLEVLSTGVAGNYNGALQLMTAEFQMPSPLVPTRESQFLRYCKQHTDGSW 436
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ DVS + R + +RG + PSGC+I+EMP+G VTWVEHVE +D + H +YR L
Sbjct: 437 AVVDVSVEGLRASGQAGARGRRRPSGCLIQEMPNGYSRVTWVEHVEADDMM-VHDLYRPL 495
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPV-----EETIGVIQTLEGRNSVIKLADR 452
V +GA W L+R CER + +P + +GV+ ++EGR S+++LA+R
Sbjct: 496 VCSGLAFGARRWAAALERQCERLASAMASGVPAGPSSGGDAVGVVTSVEGRRSMLRLAER 555
Query: 453 MVKMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPA 512
MV FC +T + L+ VRV R + DD +P G ++ AAT+ WLP+P
Sbjct: 556 MVTSFCGGVTASTTHQWTKLSGSGAEDVRVMTRKSV-DDPGRPPGIILNAATSFWLPVPP 614
Query: 513 QKVFEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQ 570
+VF FL+D RS+W+ LS G + E+AHI+NG HGN +S+++ + Q M+ILQ
Sbjct: 615 ARVFGFLRDDATRSEWDILSNGGDVQEMAHIANGRDHGNAVSLLRVNNANSNQSNMLILQ 674
Query: 571 ESFTSSVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAP------- 623
E T + GSYVIYAP+D M+V L G D + +LP G + AP
Sbjct: 675 ECCTDATGSYVIYAPVDVVAMNVVLNGGDPDYVALLPSGFAILPDGSGAGGAPPGFAVLP 734
Query: 624 ----FGASKSIEDGSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
G GSL+T+A Q L++ S+ +NS +A T+ +K A+
Sbjct: 735 DGPGAGGGGGGGGGSLLTVAFQILVDSVPTAKLSLGSVATVNSLIACTVERIKAAV 790
Score = 89.4 bits (220), Expect = 3e-16
Identities = 45/102 (44%), Positives = 64/102 (62%), Gaps = 1/102 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KE HPD+ QR +L+ ++GLEP Q+K WFQNKR +K Q ER N LR EN+K+R EN
Sbjct: 119 KEYPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKTQQERHENMQLRAENEKLRAENA 178
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQ-EMKQENAQLKQE 102
+ K+ L C +CGG + +F + ++ ENA+L+ E
Sbjct: 179 RYKDALANASCPNCGGPATAVIGEMSFDEHHLRIENARLRDE 220
>UniRef100_Q69T58 Putative homeobox [Oryza sativa]
Length = 734
Score = 264 bits (674), Expect = 7e-69
Identities = 165/441 (37%), Positives = 239/441 (53%), Gaps = 52/441 (11%)
Query: 279 FPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVWV 338
FP+IV+KA TI V + G R +L+LM EE+HI++P V RE N +RYC++++ G+W
Sbjct: 296 FPSIVSKAHTIDVL-VNGMGGRSESLILMYEELHIMTPAVPTREVNFVRYCRQIEQGLWA 354
Query: 339 ITDVSFDSSRPNT--APLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRD 396
I DVS D R AP R + PSGC+I +M +G VTWVEH+EVE+K + +YRD
Sbjct: 355 IADVSVDLQRDAHFGAPPPRSRRLPSGCLIADMANGYSKVTWVEHMEVEEKSPINVLYRD 414
Query: 397 LVGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKM 456
LV +GA W+ LQR CER +++ A+ V I + T EG+ S++KL+ RMV
Sbjct: 415 LVLSGAAFGAHRWLAALQRACERY--ASLVALGVPHHIAGV-TPEGKRSMMKLSQRMVNS 471
Query: 457 FCECLTMPGQVELNHLTLDSIGGVRVSIRATT--DDDASQPNGTVVTAATTLWLPLPAQK 514
FC L G +++ T S G VS+R T D QPNG V++AAT++WLP+P
Sbjct: 472 FCSSL---GASQMHQWTTLS-GSNEVSVRVTMHRSTDPGQPNGVVLSAATSIWLPVPCDH 527
Query: 515 VFEFLKDPTKRSQ------------------------WNGLSCGNPMHEIAHISNGPYHG 550
VF F++D RSQ W+ LS GN + E++ I NG G
Sbjct: 528 VFAFVRDENTRSQVSHPLSPPLISLTHSLCPPLLLLQWDVLSHGNQVQEVSRIPNGSNPG 587
Query: 551 NCISVIKSFIPTQRQMVILQESFTSSVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGL 610
NCIS+++ +Q M+ILQES T + GS V+Y+PID +V + GED +P+LP G
Sbjct: 588 NCISLLRGLNASQNSMLILQESCTDASGSLVVYSPIDIPAANVVMSGEDPSSIPLLPSGF 647
Query: 611 IVCSKNQANLNA--------PFGASK-----SIEDGSLITLAAQTYAI---GQVLNVDSL 654
+ + A P A++ GS++T+A Q LN +S+
Sbjct: 648 TILPDGRPGSAAGASTSSAGPLAAARGGGGGGAGGGSVVTVAFQILVSSLPSSKLNAESV 707
Query: 655 NDINSQLASTILNVKDALMSS 675
+N + +T+ +K AL S
Sbjct: 708 ATVNGLITTTVEQIKAALNCS 728
Score = 111 bits (278), Expect = 6e-23
Identities = 56/101 (55%), Positives = 71/101 (69%), Gaps = 2/101 (1%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPDE QR QL+ ++GLEP+QIK WFQN+R +K QHER N LR ENDKIR EN+
Sbjct: 35 KECPHPDENQRAQLSRELGLEPRQIKFWFQNRRTQMKAQHERADNCFLRAENDKIRCENI 94
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
I+E LK IC CGG P+ + Q+++ ENA+LK+E
Sbjct: 95 AIREALKNVICPTCGGP--PVGEDYFDEQKLRMENARLKEE 133
>UniRef100_Q93V99 Protodermal factor2 [Arabidopsis thaliana]
Length = 743
Score = 264 bits (674), Expect = 7e-69
Identities = 158/410 (38%), Positives = 236/410 (57%), Gaps = 19/410 (4%)
Query: 276 TCLFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPG 335
+C+F IV++A T++V G G+ +GAL +M E + SPLV RE +RYCK+ G
Sbjct: 332 SCVFSGIVSRALTLEVLSTGVAGNYNGALQVMTAEFQVPSPLVPTRENYFVRYCKQHSDG 391
Query: 336 VWVITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYR 395
W + DVS DS RP+T P+ R + PSGC+I+E+P+G VTW+EH+EV+D+ H +Y+
Sbjct: 392 SWAVVDVSLDSLRPST-PILRTRRRPSGCLIQELPNGYSKVTWIEHMEVDDR-SVHNMYK 449
Query: 396 DLVGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVK 455
LV +GA+ W+ L+R CER S IP + + VI + EGR S++KLA+RMV
Sbjct: 450 PLVQSGLAFGAKRWVATLERQCERLASSMASNIPGD--LSVITSPEGRKSMLKLAERMVM 507
Query: 456 MFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKV 515
FC + ++ VRV R + DD +P G V++AAT+ W+P+ ++V
Sbjct: 508 SFCSGVGASTAHAWTTMSTTGSDDVRVMTRKSMDDPG-RPPGIVLSAATSFWIPVAPKRV 566
Query: 516 FEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESF 573
F+FL+D R +W+ LS G + E+AHI+NG GNC+S+++ S +Q M+ILQES
Sbjct: 567 FDFLRDENSRKEWDILSNGGMVQEMAHIANGHEPGNCVSLLRVNSGNSSQSNMLILQESC 626
Query: 574 TSSVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVC--------SKNQANLNAPFG 625
T + GSYVIYAP+D M+V L G D + +LP G + NQ
Sbjct: 627 TDASGSYVIYAPVDIVAMNVVLSGGDPDYVALLPSGFAILPDGSVGGGDGNQHQEMVSTT 686
Query: 626 ASKSIEDGSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
+S S GSL+T+A Q L++ S+ +NS + T+ +K A+
Sbjct: 687 SSGSC-GGSLLTVAFQILVDSVPTAKLSLGSVATVNSLIKCTVERIKAAV 735
Score = 92.4 bits (228), Expect = 4e-17
Identities = 46/101 (45%), Positives = 64/101 (62%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ + LEP Q+K WFQNKR +K Q ER N L+ +NDK+R EN
Sbjct: 82 KECPHPDDKQRKELSRDLNLEPLQVKFWFQNKRTQMKAQSERHENQILKSDNDKLRAENN 141
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ KE L C +CGG P + + Q ++ ENA+L++E
Sbjct: 142 RYKEALSNATCPNCGG-PAAIGEMSFDEQHLRIENARLREE 181
>UniRef100_O23045 YUP8H12.16 protein [Arabidopsis thaliana]
Length = 749
Score = 263 bits (672), Expect = 1e-68
Identities = 153/411 (37%), Positives = 236/411 (57%), Gaps = 25/411 (6%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F +V++A T+ V G G+ +GAL +M+ E + SPLV RE RYCK+ G W
Sbjct: 343 IFAGMVSRAMTLAVLSTGVAGNYNGALQVMSAEFQVPSPLVPTRETYFARYCKQQGDGSW 402
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ D+S DS +PN P +R + SGC+I+E+P+G VTWVEHVEV+D+ H +Y+ +
Sbjct: 403 AVVDISLDSLQPN--PPARCRRRASGCLIQELPNGYSKVTWVEHVEVDDR-GVHNLYKHM 459
Query: 398 VGEYNLYGAESWIKELQRMCERS---LGSNVEAIPVEET----------IGVIQTLEGRN 444
V + +GA+ W+ L R CER + +N+ + V E+ VI EGR
Sbjct: 460 VSTGHAFGAKRWVAILDRQCERLASVMATNISSGEVGESESESQFYINEYAVITNQEGRR 519
Query: 445 SVIKLADRMVKMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAAT 504
S++KLA+RMV FC ++ L+ VRV R + DD +P G V++AAT
Sbjct: 520 SMLKLAERMVISFCAGVSASTAHTWTTLSGTGAEDVRVMTRKSVDDPG-RPPGIVLSAAT 578
Query: 505 TLWLPLPAQKVFEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIKSFIPTQR 564
+ W+P+P ++VF+FL+D R++W+ LS G + E+AHI+NG GNC+S+++S +Q
Sbjct: 579 SFWIPVPPKRVFDFLRDENSRNEWDILSNGGVVQEMAHIANGRDTGNCVSLLRSANSSQS 638
Query: 565 QMVILQESFTSSVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPF 624
M+ILQES T S+VIYAP+D M++ L G D + +LP G + AN AP
Sbjct: 639 NMLILQESCTDPTASFVIYAPVDIVAMNIVLNGGDPDYVALLPSGFAILPDGNANSGAPG 698
Query: 625 GASKSIEDGSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
G + GSL+T+A Q L++ S+ +N+ +A T+ +K ++
Sbjct: 699 G-----DGGSLLTVAFQILVDSVPTAKLSLGSVATVNNLIACTVERIKASM 744
Score = 100 bits (248), Expect = 2e-19
Identities = 47/101 (46%), Positives = 68/101 (66%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR QL+ ++ LEP Q+K WFQNKR +K+ HER N LR EN+K+RN+NL
Sbjct: 100 KECPHPDDKQRKQLSRELNLEPLQVKFWFQNKRTQMKNHHERHENSHLRAENEKLRNDNL 159
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ +E L C +CGG P + + +++ ENA+L++E
Sbjct: 160 RYREALANASCPNCGG-PTAIGEMSFDEHQLRLENARLREE 199
>UniRef100_Q94C37 At1g05230/YUP8H12_16 [Arabidopsis thaliana]
Length = 721
Score = 263 bits (672), Expect = 1e-68
Identities = 153/400 (38%), Positives = 232/400 (57%), Gaps = 15/400 (3%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F +V++A T+ V G G+ +GAL +M+ E + SPLV RE RYCK+ G W
Sbjct: 327 IFAGMVSRAMTLAVLSTGVAGNYNGALQVMSAEFQVPSPLVPTRETYFARYCKQQGDGSW 386
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ D+S DS +PN P +R + SGC+I+E+P+G VTWVEHVEV+D+ H +Y+ +
Sbjct: 387 AVVDISLDSLQPN--PPARCRRRASGCLIQELPNGYSKVTWVEHVEVDDR-GVHNLYKHM 443
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
V + +GA+ W+ L R CER I E +GVI EGR S++KLA+RMV F
Sbjct: 444 VSTGHAFGAKRWVAILDRQCERLASVMATNISSGE-VGVITNQEGRRSMLKLAERMVISF 502
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
C ++ L+ VRV R + DD +P G V++AAT+ W+P+P ++VF+
Sbjct: 503 CAGVSASTAHTWTTLSGTGAEDVRVMTRKSVDDPG-RPPGIVLSAATSFWIPVPPKRVFD 561
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESFTS 575
FL+D R++W+ LS G + E+AHI+NG GNC+S+++ S +Q M+ILQES T
Sbjct: 562 FLRDENSRNEWDILSNGGVVQEMAHIANGRDTGNCVSLLRVNSANSSQSNMLILQESCTD 621
Query: 576 SVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPFGASKSIEDGSL 635
S+VIYAP+D M++ L G D + +LP G + AN AP G + GSL
Sbjct: 622 PTASFVIYAPVDIVAMNIVLNGGDPDYVALLPSGFAILPDGNANSGAPGG-----DGGSL 676
Query: 636 ITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
+T+A Q L++ S+ +N+ +A T+ +K ++
Sbjct: 677 LTVAFQILVDSVPTAKLSLGSVATVNNLIACTVERIKASM 716
Score = 100 bits (248), Expect = 2e-19
Identities = 47/101 (46%), Positives = 68/101 (66%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR QL+ ++ LEP Q+K WFQNKR +K+ HER N LR EN+K+RN+NL
Sbjct: 84 KECPHPDDKQRKQLSRELNLEPLQVKFWFQNKRTQMKNHHERHENSHLRAENEKLRNDNL 143
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ +E L C +CGG P + + +++ ENA+L++E
Sbjct: 144 RYREALANASCPNCGG-PTAIGEMSFDEHQLRLENARLREE 183
>UniRef100_Q9M9P4 T17B22.5 protein [Arabidopsis thaliana]
Length = 695
Score = 263 bits (671), Expect = 2e-68
Identities = 163/422 (38%), Positives = 245/422 (57%), Gaps = 51/422 (12%)
Query: 278 LFPTIVTKAETIKVFEIGS--RGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPG 335
LFPTIV KA TI V G RG+ + L +M E++HILSPLV REF ++R C++++ G
Sbjct: 293 LFPTIVNKANTIHVLGSGLPIRGNCN-VLQVMWEQLHILSPLVPAREFMVVRCCQEIEKG 351
Query: 336 VWVITDVS----FDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTH 391
+W+I DVS FD + +K PSGC+I+ +P V W+EHVEV+ K+ TH
Sbjct: 352 IWIIADVSHRANFDFGN------AACYKRPSGCLIQALPDAHSKVMWIEHVEVDHKLDTH 405
Query: 392 YVYRDLVGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLAD 451
+YRDL+ + YGA+ WI L+RMCER S+++ +P + VI T E R SV+KL +
Sbjct: 406 KIYRDLLSGGSGYGAKRWIVTLERMCERMALSSIQTLPPSDRSEVITTGEARRSVMKLGE 465
Query: 452 RMVKMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLP 511
RMVK F E LTM G+++ S GVRVSIR +A QP G VV+A+++L +PL
Sbjct: 466 RMVKNFNEMLTMSGKIDFPQ---QSKNGVRVSIRMNI--EAGQPPGIVVSASSSLAIPLT 520
Query: 512 AQKVFEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIKSFIPTQRQ------ 565
+VF FL++ R QW+ LS G ++EIA I G NC+++++ PT +
Sbjct: 521 PLQVFAFLQNLDTRQQWDILSYGTVVNEIARIVTGSSETNCVTILR---PTHEENNDKMV 577
Query: 566 --------MVILQESFTSSVGSYVIYAPIDRKTMDVALRGE-DSKELPILPYGLIVCSKN 616
M++LQ+ + ++G ++YAP+D TM A+ GE D +PILP G ++ S
Sbjct: 578 VQDSCKDDMLMLQDCYMDALGGMIVYAPMDMATMHFAVSGEVDPSHIPILPSGFVISSD- 636
Query: 617 QANLNAPFGASKSIED-GSLITLAAQTYAIGQV-----LNVDSLNDINSQLASTILNVKD 670
G ++ED G+L+T+A Q G+ +N S++ +++ ++STI +K
Sbjct: 637 --------GRRSTVEDGGTLLTVAFQILVSGKANRSREVNEKSVDTVSALISSTIQRIKG 688
Query: 671 AL 672
L
Sbjct: 689 LL 690
Score = 96.7 bits (239), Expect = 2e-18
Identities = 46/98 (46%), Positives = 66/98 (66%)
Query: 3 ECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENLK 62
EC HPDE QR QL ++ LEP QIK WFQNKR K Q +R TN LR EN+ ++++N
Sbjct: 42 ECPHPDERQRNQLCRELKLEPDQIKFWFQNKRTQSKTQEDRSTNVLLRGENETLQSDNEA 101
Query: 63 IKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLK 100
+ + LK+ +C CGG PF ++ + +Q+++ ENA+LK
Sbjct: 102 MLDALKSVLCPACGGPPFGREERGHNLQKLRFENARLK 139
>UniRef100_Q8GY47 Hypothetical protein At3g03260/T17B22_5 [Arabidopsis thaliana]
Length = 517
Score = 262 bits (670), Expect = 2e-68
Identities = 163/422 (38%), Positives = 245/422 (57%), Gaps = 49/422 (11%)
Query: 278 LFPTIVTKAETIKVFEIGS--RGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPG 335
LFPTIV KA TI V G RG+ + L +M E++HILSPLV REF ++R C++++ G
Sbjct: 113 LFPTIVNKANTIHVLGSGLPIRGNCN-VLQVMWEQLHILSPLVPAREFMVVRCCQEIEKG 171
Query: 336 VWVITDVS----FDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTH 391
+W+I DVS FD + +K PSGC+I+ +P V W+EHVEV+ K+ TH
Sbjct: 172 IWIIADVSHRANFDFGN------AACYKRPSGCLIQALPDAHSKVMWIEHVEVDHKLDTH 225
Query: 392 YVYRDLVGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLAD 451
+YRDL+ + YGA+ WI L+RMCER S+++ +P + VI T E R SV+KL +
Sbjct: 226 KIYRDLLSGGSGYGAKRWIVTLERMCERMALSSIQTLPPSDRSEVITTGEARRSVMKLGE 285
Query: 452 RMVKMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLP 511
RMVK F E LTM G+++ S GVRVSIR +A QP G VV+A+++L +PL
Sbjct: 286 RMVKNFNEMLTMSGKIDFPQ---QSKNGVRVSIRMNI--EAGQPPGIVVSASSSLAIPLT 340
Query: 512 AQKVFEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIKSFIPTQRQ------ 565
+VF FL++ R QW+ LS G ++EIA I G NC+++++ PT +
Sbjct: 341 PLQVFAFLQNLDTRQQWDILSYGTVVNEIARIVTGSSETNCVTILRVH-PTHEENNDKMV 399
Query: 566 --------MVILQESFTSSVGSYVIYAPIDRKTMDVALRGE-DSKELPILPYGLIVCSKN 616
M++LQ+ + ++G ++YAP+D TM A+ GE D +PILP G ++ S
Sbjct: 400 VQDSCKDDMLMLQDCYMDALGGMIVYAPMDMATMHFAVSGEVDPSHIPILPSGFVISSD- 458
Query: 617 QANLNAPFGASKSIED-GSLITLAAQTYAIGQV-----LNVDSLNDINSQLASTILNVKD 670
G ++ED G+L+T+A Q G+ +N S++ +++ ++STI +K
Sbjct: 459 --------GRRSTVEDGGTLLTVAFQILVSGKANRSREVNEKSVDTVSALISSTIQRIKG 510
Query: 671 AL 672
L
Sbjct: 511 LL 512
>UniRef100_Q38887 Meristem L1 layer homeobox protein [Arabidopsis thaliana]
Length = 718
Score = 258 bits (660), Expect = 3e-67
Identities = 161/420 (38%), Positives = 237/420 (56%), Gaps = 30/420 (7%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F IV++A T++V G RG+ +GAL +M E + SPLV RE +RYCK+ G+W
Sbjct: 299 VFCGIVSRALTLEVLSTGVRGNYNGALQVMTAEFQVPSPLVPTRENYFVRYCKQHSDGIW 358
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ DVS DS RP+ P++R + PSGC+I+E+ +G VTWVEH+EV+D+ H +Y+ L
Sbjct: 359 AVVDVSLDSLRPS--PITRSRRRPSGCLIQELQNGYSKVTWVEHIEVDDR-SVHNMYKPL 415
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
V +GA+ W+ L R CER S IP + + VI + EGR S++KLA+RMV F
Sbjct: 416 VNTGLAFGAKRWVATLDRQCERLASSMASNIPACD-LSVITSPEGRKSMLKLAERMVMSF 474
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
C + L+ VRV R + DD +P G V++AAT+ W+P+ ++VF+
Sbjct: 475 CTGVGASTADAWTTLSTTGSDDVRVMTRKSMDDPG-RPPGIVLSAATSFWIPVAPKRVFD 533
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESFTS 575
FL+D RS+W+ LS G + E+AHI+NG GN +S+++ S Q M+ILQES T
Sbjct: 534 FLRDENSRSEWDILSNGGLVQEMAHIANGRDPGNSVSLLRVNSGNSGQSNMLILQESCTD 593
Query: 576 SVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQA-----NLNAPFGA---- 626
+ GSYVIYAP+D M+V L G D + +LP G + A + NA GA
Sbjct: 594 ASGSYVIYAPVDIIAMNVVLSGGDPDYVALLPSGFAILPDGSARGGGGSANASAGAGVEG 653
Query: 627 ---SKSIE--------DGSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
++E GSL+T+A Q L++ S+ +NS + T+ +K AL
Sbjct: 654 GGEGNNLEVVTTTGSCGGSLLTVAFQILVDSVPTAKLSLGSVATVNSLIKCTVERIKAAL 713
Score = 96.7 bits (239), Expect = 2e-18
Identities = 47/101 (46%), Positives = 66/101 (64%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ ++ LEP Q+K WFQNKR +K QHER N L+ ENDK+R EN
Sbjct: 38 KECPHPDDKQRKELSRELSLEPLQVKFWFQNKRTQMKAQHERHENQILKSENDKLRAENN 97
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ K+ L C +CGG P + + Q ++ ENA+L++E
Sbjct: 98 RYKDALSNATCPNCGG-PAAIGEMSFDEQHLRIENARLREE 137
>UniRef100_Q8RWU4 Putative L1-specific homeobox gene ATML1/ovule-specific homeobox
protein A20 [Arabidopsis thaliana]
Length = 762
Score = 256 bits (654), Expect = 2e-66
Identities = 160/420 (38%), Positives = 236/420 (56%), Gaps = 30/420 (7%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F IV++A T++V G G+ +GAL +M E + SPLV RE +RYCK+ G+W
Sbjct: 343 VFCGIVSRALTLEVLSTGVAGNYNGALQVMTAEFQVPSPLVPTRENYFVRYCKQHSDGIW 402
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
+ DVS DS RP+ P++R + PSGC+I+E+ +G VTWVEH+EV+D+ H +Y+ L
Sbjct: 403 AVVDVSLDSLRPS--PITRSRRRPSGCLIQELQNGYSKVTWVEHIEVDDR-SVHNMYKPL 459
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
V +GA+ W+ L R CER S IP + + VI + EGR S++KLA+RMV F
Sbjct: 460 VNTGLAFGAKRWVATLDRQCERLASSMASNIPACD-LSVITSPEGRKSMLKLAERMVMSF 518
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
C + L+ VRV R + DD +P G V++AAT+ W+P+ ++VF+
Sbjct: 519 CTGVGASTAHAWTTLSTTGSDDVRVMTRKSMDDPG-RPPGIVLSAATSFWIPVAPKRVFD 577
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESFTS 575
FL+D RS+W+ LS G + E+AHI+NG GN +S+++ S Q M+ILQES T
Sbjct: 578 FLRDENSRSEWDILSNGGLVQEMAHIANGRDPGNSVSLLRVNSGNSGQSNMLILQESCTD 637
Query: 576 SVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQA-----NLNAPFGA---- 626
+ GSYVIYAP+D M+V L G D + +LP G + A + NA GA
Sbjct: 638 ASGSYVIYAPVDIIAMNVVLSGGDPDYVALLPSGFAILPDGSARGGGGSANASAGAGVEG 697
Query: 627 ---SKSIE--------DGSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
++E GSL+T+A Q L++ S+ +NS + T+ +K AL
Sbjct: 698 GGEGNNLEVVTTTGSCGGSLLTVAFQILVDSVPTAKLSLGSVATVNSLIKCTVERIKAAL 757
Score = 96.7 bits (239), Expect = 2e-18
Identities = 47/101 (46%), Positives = 66/101 (64%), Gaps = 1/101 (0%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ ++ LEP Q+K WFQNKR +K QHER N L+ ENDK+R EN
Sbjct: 82 KECPHPDDKQRKELSRELSLEPLQVKFWFQNKRTQMKAQHERHENQILKSENDKLRAENN 141
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ K+ L C +CGG P + + Q ++ ENA+L++E
Sbjct: 142 RYKDALSNATCPNCGG-PAAIGEMSFDEQHLRIENARLREE 181
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.330 0.142 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,092,427,625
Number of Sequences: 2790947
Number of extensions: 44644402
Number of successful extensions: 138420
Number of sequences better than 10.0: 1154
Number of HSP's better than 10.0 without gapping: 967
Number of HSP's successfully gapped in prelim test: 187
Number of HSP's that attempted gapping in prelim test: 136954
Number of HSP's gapped (non-prelim): 1310
length of query: 675
length of database: 848,049,833
effective HSP length: 134
effective length of query: 541
effective length of database: 474,062,935
effective search space: 256468047835
effective search space used: 256468047835
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 78 (34.7 bits)
Medicago: description of AC147010.3


