

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147007.4 - phase: 0
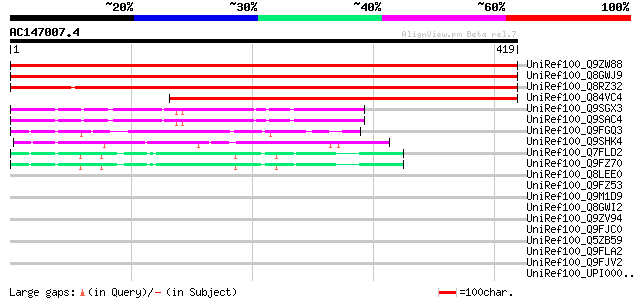
(419 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ZW88 F5A8.10 protein [Arabidopsis thaliana] 540 e-152
UniRef100_Q8GWJ9 Hypothetical protein At1g67190/F5A8_10 [Arabido... 536 e-151
UniRef100_Q8RZ32 P0489B03.11 protein [Oryza sativa] 497 e-139
UniRef100_Q84VC4 Hypothetical protein [Oryza sativa] 333 4e-90
UniRef100_Q9SGX3 F20B24.20 [Arabidopsis thaliana] 69 2e-10
UniRef100_Q9SAC4 T16B5.8 [Arabidopsis thaliana] 69 2e-10
UniRef100_Q9FGQ3 Arabidopsis thaliana genomic DNA, chromosome 5,... 69 2e-10
UniRef100_Q9SHK4 F12K11.6 [Arabidopsis thaliana] 54 1e-05
UniRef100_Q7FLD2 Hypothetical protein At1g13570/F13B4_26 [Arabid... 51 7e-05
UniRef100_Q9FZ70 F13B4.6 protein [Arabidopsis thaliana] 50 9e-05
UniRef100_Q8LEE0 Hypothetical protein [Arabidopsis thaliana] 47 0.001
UniRef100_Q9FZ53 F6I1.5 protein [Arabidopsis thaliana] 46 0.002
UniRef100_Q9M1D9 Hypothetical protein T2O9.20 [Arabidopsis thali... 45 0.003
UniRef100_Q8GWI2 Hypothetical protein At3g60040/T2O9_20 [Arabido... 45 0.003
UniRef100_Q9ZV94 F9K20.20 protein [Arabidopsis thaliana] 45 0.004
UniRef100_Q9FJC0 Heat shock transcription factor HSF30-like prot... 44 0.007
UniRef100_Q5ZB59 Ribosomal RNA apurinic site specific lyase-like... 44 0.011
UniRef100_Q9FLA2 Similarity to heat shock transcription factor [... 43 0.015
UniRef100_Q9FJV2 Emb|CAB89229.1 [Arabidopsis thaliana] 43 0.015
UniRef100_UPI00002B2087 UPI00002B2087 UniRef100 entry 43 0.019
>UniRef100_Q9ZW88 F5A8.10 protein [Arabidopsis thaliana]
Length = 419
Score = 540 bits (1390), Expect = e-152
Identities = 263/419 (62%), Positives = 327/419 (77%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
M+ LPVEV+G ILS LG ARDV+IAS TC+KWREA + HLQTLSFN++DWP YR++ ++
Sbjct: 1 MDYLPVEVIGNILSRLGGARDVVIASATCRKWREACRKHLQTLSFNSADWPFYRDLTTNR 60
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFNII 120
LE+LIT T+FQT GLQ L+I MDD ++FS VIAWLMYTRD+LR L YNVRT+PN NI+
Sbjct: 61 LEILITQTIFQTMGLQGLSIMMDDANKFSAATVIAWLMYTRDTLRRLSYNVRTTPNVNIL 120
Query: 121 EKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSACPKLETLS 180
E C R+ LE L LA N I+GVEP + +FPCLKSLSLS+VSISALDL+LLLSACP +E+L
Sbjct: 121 EICGRQKLEALVLAHNSITGVEPSFQRFPCLKSLSLSYVSISALDLNLLLSACPMIESLE 180
Query: 181 IVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADMLENLHLKDCSFDAFELINK 240
+V EIAMSD++ +IELSS +LK + + S DK IL AD +E LH+KDC + FELI
Sbjct: 181 LVSLEIAMSDAQVTIELSSPTLKSVYFDGISLDKFILEADSIEFLHMKDCVLELFELIGN 240
Query: 241 GTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLKKLRLWSVVF 300
GTLK KLDDVSVIHLDI + +E+LE+VDV +F +W FY MIS++ KLKKLRLW VVF
Sbjct: 241 GTLKHFKLDDVSVIHLDIMETSESLEVVDVNHFTMVWPKFYQMISRSQKLKKLRLWDVVF 300
Query: 301 DDEDEVVDIETISVCFPRLTHLSLSYDLKDGVLHYGLQGLSFLMNVVVLELGWTTISDLF 360
DD+DE++D+E+I+ F LTHLSLSYDLKDG HY LQG + L NV VLELGWT I+D+F
Sbjct: 301 DDDDEIIDVESIAAGFSHLTHLSLSYDLKDGAAHYSLQGTTQLENVTVLELGWTVINDVF 360
Query: 361 SVWVAGLLEGCPNLKKMVIYGYVAEIKTHEECQTFTKFTEFMIQLGRKYSHIKFEFEYE 419
S+WV LL CPNLKK++IYG V+E KT +CQ FT ++QL RKY H++ +FEYE
Sbjct: 361 SIWVEELLRRCPNLKKLIIYGVVSETKTQGDCQILATFTWSIVQLMRKYIHVEVQFEYE 419
>UniRef100_Q8GWJ9 Hypothetical protein At1g67190/F5A8_10 [Arabidopsis thaliana]
Length = 419
Score = 536 bits (1381), Expect = e-151
Identities = 262/419 (62%), Positives = 326/419 (77%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
M+ LPVEV+G ILS LG ARDV+IAS TC+KWREA + HLQTLSFN++DWP YR++ ++
Sbjct: 1 MDYLPVEVIGNILSRLGGARDVVIASATCRKWREACRKHLQTLSFNSADWPFYRDLTTNR 60
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFNII 120
LE+LIT T+FQT GLQ L+I MDD ++FS VIAWLM TRD+LR L YNVRT+PN NI+
Sbjct: 61 LEILITQTIFQTMGLQGLSIMMDDANKFSAATVIAWLMCTRDTLRRLSYNVRTTPNVNIL 120
Query: 121 EKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSACPKLETLS 180
E C R+ LE L LA N I+GVEP + +FPCLKSLSLS+VSISALDL+LLLSACP +E+L
Sbjct: 121 EICGRQKLEALVLAHNSITGVEPSFQRFPCLKSLSLSYVSISALDLNLLLSACPMIESLE 180
Query: 181 IVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADMLENLHLKDCSFDAFELINK 240
+V EIAMSD++ +IELSS +LK + + S DK IL AD +E LH+KDC + FELI
Sbjct: 181 LVSLEIAMSDAQVTIELSSPTLKSVYFDGISLDKFILEADSIEFLHMKDCVLELFELIGN 240
Query: 241 GTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLKKLRLWSVVF 300
GTLK KLDDVSVIHLDI + +E+LE+VDV +F +W FY MIS++ KLKKLRLW VVF
Sbjct: 241 GTLKHFKLDDVSVIHLDIMETSESLEVVDVNHFTMVWPKFYQMISRSQKLKKLRLWDVVF 300
Query: 301 DDEDEVVDIETISVCFPRLTHLSLSYDLKDGVLHYGLQGLSFLMNVVVLELGWTTISDLF 360
DD+DE++D+E+I+ F LTHLSLSYDLKDG HY LQG + L NV VLELGWT I+D+F
Sbjct: 301 DDDDEIIDVESIAAGFSHLTHLSLSYDLKDGAAHYSLQGTTQLENVTVLELGWTVINDVF 360
Query: 361 SVWVAGLLEGCPNLKKMVIYGYVAEIKTHEECQTFTKFTEFMIQLGRKYSHIKFEFEYE 419
S+WV LL CPNLKK++IYG V+E KT +CQ FT ++QL RKY H++ +FEYE
Sbjct: 361 SIWVEELLRRCPNLKKLIIYGVVSETKTQGDCQILATFTWSIVQLMRKYIHVEVQFEYE 419
>UniRef100_Q8RZ32 P0489B03.11 protein [Oryza sativa]
Length = 417
Score = 497 bits (1280), Expect = e-139
Identities = 249/419 (59%), Positives = 318/419 (75%), Gaps = 2/419 (0%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
ME LPVEV+G IL+HL +ARDV++AS C+KWR A + HL +LSFN+ D+P R++ +
Sbjct: 1 MEHLPVEVIGNILAHLSAARDVMVASGVCRKWRTACRKHLHSLSFNSDDFP--RDMTTRQ 58
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFNII 120
LE++IT T+FQT GLQCL+I +D HEFS PVIAWLMYTR++LR L YNVRT+PN NI+
Sbjct: 59 LEIVITQTIFQTMGLQCLSIHIDRTHEFSAAPVIAWLMYTRETLRSLSYNVRTNPNVNIL 118
Query: 121 EKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSACPKLETLS 180
EKC R+ LEVL L N I+GVEP Y +F CLKSLSL VSISALDLSLL++ACPK+E+L+
Sbjct: 119 EKCGRQKLEVLDLDHNTITGVEPSYQRFTCLKSLSLRHVSISALDLSLLVAACPKIESLA 178
Query: 181 IVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADMLENLHLKDCSFDAFELINK 240
+ E+ SD ++++EL+S +LK F +S DK+IL D LE L+L + D FELI K
Sbjct: 179 LDFLEVVTSDPQSTMELTSHTLKSLFAKSVGVDKIILDTDNLEVLNLNALNLDLFELIGK 238
Query: 241 GTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLKKLRLWSVVF 300
GTLK LK+DDVSV H+DIG++T++LE+VDV NF + Y+MIS+AS L+ LR W VVF
Sbjct: 239 GTLKHLKIDDVSVTHMDIGESTDHLEVVDVSNFTIVRPKLYSMISRASNLRMLRFWGVVF 298
Query: 301 DDEDEVVDIETISVCFPRLTHLSLSYDLKDGVLHYGLQGLSFLMNVVVLELGWTTISDLF 360
DDEDE+VD ETI+V FP L HLSLSY+L+DG+LHY LQG S L NV VLELGWT IS+ F
Sbjct: 299 DDEDEIVDSETIAVSFPLLRHLSLSYELRDGLLHYSLQGSSPLDNVSVLELGWTVISEHF 358
Query: 361 SVWVAGLLEGCPNLKKMVIYGYVAEIKTHEECQTFTKFTEFMIQLGRKYSHIKFEFEYE 419
WV G++E CPNLKK+VI+G ++E KT EE Q FT F++ L RKY H+ +FEYE
Sbjct: 359 GPWVFGMIERCPNLKKLVIHGVLSEAKTREERQMLASFTSFIVCLMRKYVHVDVQFEYE 417
>UniRef100_Q84VC4 Hypothetical protein [Oryza sativa]
Length = 288
Score = 333 bits (855), Expect = 4e-90
Identities = 168/287 (58%), Positives = 214/287 (74%)
Query: 133 LASNPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSACPKLETLSIVCPEIAMSDSE 192
L N I+GVEP Y +F CLKSLSL VSISALDLSLL++ACPK+E+L++ E+ SD +
Sbjct: 2 LDHNTITGVEPSYQRFTCLKSLSLRHVSISALDLSLLVAACPKIESLALDFLEVVTSDPQ 61
Query: 193 ASIELSSSSLKDFFVESYSFDKLILVADMLENLHLKDCSFDAFELINKGTLKVLKLDDVS 252
+++EL+S +LK F +S DK+IL D LE L+L + D FELI KGTLK LK+DDVS
Sbjct: 62 STMELTSHTLKSLFAKSVGVDKIILDTDNLEVLNLNALNLDLFELIGKGTLKHLKIDDVS 121
Query: 253 VIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLKKLRLWSVVFDDEDEVVDIETI 312
V H+DIG++T++LE+VDV NF + Y+MIS+AS L+ LR W VVFDDEDE+VD ETI
Sbjct: 122 VTHMDIGESTDHLEVVDVSNFTIVRPKLYSMISRASNLRMLRFWGVVFDDEDEIVDSETI 181
Query: 313 SVCFPRLTHLSLSYDLKDGVLHYGLQGLSFLMNVVVLELGWTTISDLFSVWVAGLLEGCP 372
+V FP L HLSLSY+L+DG+LHY LQG S L NV VLELGWT IS+ F WV G++E CP
Sbjct: 182 AVSFPLLRHLSLSYELRDGLLHYSLQGSSPLDNVSVLELGWTVISEHFGPWVFGMIERCP 241
Query: 373 NLKKMVIYGYVAEIKTHEECQTFTKFTEFMIQLGRKYSHIKFEFEYE 419
NLKK+VI+G ++E KT EE Q FT F++ L RKY H+ +FEYE
Sbjct: 242 NLKKLVIHGVLSEAKTREERQMLASFTSFIVCLMRKYVHVDVQFEYE 288
>UniRef100_Q9SGX3 F20B24.20 [Arabidopsis thaliana]
Length = 457
Score = 69.3 bits (168), Expect = 2e-10
Identities = 72/302 (23%), Positives = 142/302 (46%), Gaps = 22/302 (7%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
M+ LP ++ ILS+L SARDV + K+W+E+ + ++++ F+ + + E S
Sbjct: 7 MDSLPDAILQYILSYLTSARDVAACNCVSKRWKESTDS-VKSVVFHRNSFESIMETDDS- 64
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFNII 120
+ ++ + ++ L+ L ++ F+ + + +W+M+ SLR L + + ++
Sbjct: 65 -DSIVRKMISSSRRLEELVVY----SPFTSSGLASWMMHVSSSLRLLELRMDNLASEEVV 119
Query: 121 EKCSRKTLEVLTLASN----PISGV----EPKYHKFPCLKSLSLSFVSISALDLSLLLSA 172
+ K L+ + +A N + GV PK+ FP L+SL + + LS L A
Sbjct: 120 VEGPLK-LDCIGVAKNLEILKLWGVLMMSPPKWDMFPNLRSLEIVGAKMDDSSLSHALRA 178
Query: 173 CPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADMLENLHLKDCSF 232
CP L L ++ E S S L L DF+ + + L+L + L +L ++ CS+
Sbjct: 179 CPNLSNLLLLACEGVKSISIDLPYLEHCKL-DFYGQGNTL--LVLTSQRLVSLDVQGCSW 235
Query: 233 DAFELINKGTLKVLKLDDVS-VIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLK 291
+ LK L + V+ +++ +N +LE + + + W ++ +A +K
Sbjct: 236 --IRVPETKFLKNLSISSVTGRVYMVDFNNLSSLEALSIRGVQWCWDAICMILQQARDVK 293
Query: 292 KL 293
L
Sbjct: 294 HL 295
>UniRef100_Q9SAC4 T16B5.8 [Arabidopsis thaliana]
Length = 418
Score = 69.3 bits (168), Expect = 2e-10
Identities = 72/302 (23%), Positives = 142/302 (46%), Gaps = 22/302 (7%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
M+ LP ++ ILS+L SARDV + K+W+E+ + ++++ F+ + + E S
Sbjct: 1 MDSLPDAILQYILSYLTSARDVAACNCVSKRWKESTDS-VKSVVFHRNSFESIMETDDS- 58
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFNII 120
+ ++ + ++ L+ L ++ F+ + + +W+M+ SLR L + + ++
Sbjct: 59 -DSIVRKMISSSRRLEELVVY----SPFTSSGLASWMMHVSSSLRLLELRMDNLASEEVV 113
Query: 121 EKCSRKTLEVLTLASN----PISGV----EPKYHKFPCLKSLSLSFVSISALDLSLLLSA 172
+ K L+ + +A N + GV PK+ FP L+SL + + LS L A
Sbjct: 114 VEGPLK-LDCIGVAKNLEILKLWGVLMMSPPKWDMFPNLRSLEIVGAKMDDSSLSHALRA 172
Query: 173 CPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADMLENLHLKDCSF 232
CP L L ++ E S S L L DF+ + + L+L + L +L ++ CS+
Sbjct: 173 CPNLSNLLLLACEGVKSISIDLPYLEHCKL-DFYGQGNTL--LVLTSQRLVSLDVQGCSW 229
Query: 233 DAFELINKGTLKVLKLDDVS-VIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLK 291
+ LK L + V+ +++ +N +LE + + + W ++ +A +K
Sbjct: 230 --IRVPETKFLKNLSISSVTGRVYMVDFNNLSSLEALSIRGVQWCWDAICMILQQARDVK 287
Query: 292 KL 293
L
Sbjct: 288 HL 289
>UniRef100_Q9FGQ3 Arabidopsis thaliana genomic DNA, chromosome 5, TAC clone:K6O8
[Arabidopsis thaliana]
Length = 444
Score = 69.3 bits (168), Expect = 2e-10
Identities = 78/306 (25%), Positives = 135/306 (43%), Gaps = 51/306 (16%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICS-- 58
+ LP ++ +ILSHL S +D + S+ +WR WQ + L F++ + + E S
Sbjct: 20 LSQLPDHLICVILSHL-STKDAVRTSILSTRWRNLWQL-VPVLDFDSRELRSFSEFVSFA 77
Query: 59 ---------SHLEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRY 109
S+++ L C ++ G LT ++D + TR ++ +
Sbjct: 78 GSFFYLHKDSYIQKLRVC-IYDLAGNYYLTSWID--------------LVTRHRIQHIDI 122
Query: 110 NVRTSPNFNIIEKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSI-SALDLSL 168
+V T F +I ++ L + ++ V ++ PCLK L L FV+ + L
Sbjct: 123 SVFTCSGFGVIPLSLYTCDTLVHLKLSRVTMVNVEFVSLPCLKILDLDFVNFTNETTLDK 182
Query: 169 LLSACPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSFDK---LILVADMLENL 225
++S P LE L+IV + D+ I++ S +LK + FD+ L++ +L+ L
Sbjct: 183 IISCSPVLEELTIV---KSSEDNVKIIQVRSQTLKRVEIHR-RFDRHNGLVIDTPLLQFL 238
Query: 226 HLKDCSFDAFELINKG-TLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMI 284
+K S + E IN G T KV D+ V LD D + M ++F+ I
Sbjct: 239 SIKAHSIKSIEFINLGFTTKV----DIDVNLLDPNDLSNR----------SMTRDFFTTI 284
Query: 285 SKASKL 290
S+ L
Sbjct: 285 SRVRSL 290
>UniRef100_Q9SHK4 F12K11.6 [Arabidopsis thaliana]
Length = 2025
Score = 53.5 bits (127), Expect = 1e-05
Identities = 87/344 (25%), Positives = 143/344 (41%), Gaps = 42/344 (12%)
Query: 4 LPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSHLEM 63
LP E++G ILS L + + + S+ KKWR ++ + TL F+ S + + S
Sbjct: 431 LPDEILGKILSLLAT-KQAVSTSVLSKKWRTLFKL-VDTLEFDDSVSGMGEQEASYVFPE 488
Query: 64 LITCTLFQTKGLQC----------LTIFMDDEHEFS-VTPVIAWLMYTRDSLRELRYNVR 112
+ +T LQC + DDE + V I+ ++ S LR N R
Sbjct: 489 SFKDLVDRTVALQCDYPIRKLSLKCHVGRDDEQRKACVGRWISNVVGRGVSEVVLRINDR 548
Query: 113 TSPNFNIIEKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSL---SLSFVSISALDLSLL 169
+F + + KTL LTL + G P Y P LK L S+ F L ++L
Sbjct: 549 -GLHFLSPQLLTCKTLVKLTLGTRLFLGKLPSYVSLPSLKFLFIHSVFFDDFGELS-NVL 606
Query: 170 LSACPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVE-SYSFDKLILV-ADMLENLHL 227
L+ CP +E L + + +SS +LK V Y F+ +I LE L
Sbjct: 607 LAGCPVVEALYL-----NQNGESMPYTISSPTLKRLSVHYEYHFESVISFDLPNLEYLDY 661
Query: 228 KDCSFDAFELINKGTLKVLKLDDVSVIHLDIGDNTE------NLEIVDV----------- 270
D + + +N +L L+ H++ D T+ N+EI+ +
Sbjct: 662 SDYALYGYPQVNLESLVEAYLNLDKAEHVESPDVTKLIMGIRNVEILSLSPDSVGVIYSC 721
Query: 271 CNFIFMWQNFYNMISKASKLKKLRLWSVVFDDEDEVVDIETISV 314
C + + F N++S + KK R W ++ D + +ET+ +
Sbjct: 722 CKYGLLLPVFNNLVSLSFGTKKTRAWKLLADILKQSPKLETLII 765
>UniRef100_Q7FLD2 Hypothetical protein At1g13570/F13B4_26 [Arabidopsis thaliana]
Length = 416
Score = 50.8 bits (120), Expect = 7e-05
Identities = 84/343 (24%), Positives = 138/343 (39%), Gaps = 48/343 (13%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREIC--S 58
+ DLP ++ IL+ L S RD + S+ KWR W T L L F+ + C
Sbjct: 8 ISDLPQSIIENILTRL-SIRDAIRTSVLSSKWRYKWST-LTDLVFDEKCVSPSNDRCVVE 65
Query: 59 SHLEMLITCTLFQTKG----LQCLTIFMDDEHEFSVTPVIAWLMY-TRDSLRELRYNVRT 113
++L IT L +G Q T F + WL++ +R+ ++EL +
Sbjct: 66 TNLVRFITGVLLLHQGPIHKFQLSTSFKQCRPDID-----QWLLFLSRNGIKELVLKLGE 120
Query: 114 SPNFNIIEKCSRKTLEVLTLASNPISGVEPKYHK-FPCLKSLSLSFVSISALDLSLLLSA 172
F + C L++ L P+Y K F LKSL+L + ++ + L+S
Sbjct: 121 G-EFRV-PACLFNCLKLTCLELCHCEFDPPQYFKGFSYLKSLNLHQIPVAPEVIESLISG 178
Query: 173 CPKLETLSIVCPE---IAMSDSEASIELSSSSLKDFFVESYSFDKLILVA-------DML 222
CP LE LS+ + +++S KD F+E + KL+ ++ D+
Sbjct: 179 CPLLEFLSLSYFDSLVLSISAPNLMYLYLDGEFKDIFLE--NTPKLVAISVSMYMHEDVT 236
Query: 223 ENLHLKDCSFDAFELINKGTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYN 282
+ D + F L L+ L +L IGD+ L + +
Sbjct: 237 DFEQSSDYNLVKF-LGGVPLLEKLVGYIYFTKYLSIGDDPGRLPLTYI------------ 283
Query: 283 MISKASKLKKLRLWSVVFDDEDEVVDIETISVCFPRLTHLSLS 325
LK + L+ V F+D DEV+ + + P L L +S
Sbjct: 284 ------HLKTIELYQVCFEDADEVLVLLRLVTHSPNLKELKVS 320
>UniRef100_Q9FZ70 F13B4.6 protein [Arabidopsis thaliana]
Length = 416
Score = 50.4 bits (119), Expect = 9e-05
Identities = 84/343 (24%), Positives = 138/343 (39%), Gaps = 48/343 (13%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREIC--S 58
+ DLP ++ IL+ L S RD + S+ KWR W T L L F+ + C
Sbjct: 8 ISDLPQSIIENILTRL-SIRDAIRTSVLSSKWRYKWST-LTDLVFDEKCVSPSNDRCVVE 65
Query: 59 SHLEMLITCTLFQTKG----LQCLTIFMDDEHEFSVTPVIAWLMY-TRDSLRELRYNVRT 113
++L IT L +G Q T F + WL++ +R+ ++EL +
Sbjct: 66 TNLVRFITGVLLLHQGPIHKFQLSTSFKQCRPDID-----QWLLFLSRNGIKELVLKLGE 120
Query: 114 SPNFNIIEKCSRKTLEVLTLASNPISGVEPKYHK-FPCLKSLSLSFVSISALDLSLLLSA 172
F + C L++ L P+Y K F LKSL+L + ++ + L+S
Sbjct: 121 G-EFRV-PACLFNCLKLTCLELCHCEFDPPQYFKGFSYLKSLNLHQILVAPEVIESLISG 178
Query: 173 CPKLETLSIVCPE---IAMSDSEASIELSSSSLKDFFVESYSFDKLILVA-------DML 222
CP LE LS+ + +++S KD F+E + KL+ ++ D+
Sbjct: 179 CPLLEFLSLSYFDSLVLSISAPNLMYLYLDGEFKDIFLE--NTPKLVAISVSMYMHEDVT 236
Query: 223 ENLHLKDCSFDAFELINKGTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYN 282
+ D + F L L+ L +L IGD+ L + +
Sbjct: 237 DFEQSSDYNLVKF-LGGVPLLEKLVGYIYFTKYLSIGDDPGRLPLTYI------------ 283
Query: 283 MISKASKLKKLRLWSVVFDDEDEVVDIETISVCFPRLTHLSLS 325
LK + L+ V F+D DEV+ + + P L L +S
Sbjct: 284 ------HLKTIELYQVCFEDADEVLVLLRLVTHSPNLKELKVS 320
>UniRef100_Q8LEE0 Hypothetical protein [Arabidopsis thaliana]
Length = 459
Score = 47.0 bits (110), Expect = 0.001
Identities = 45/136 (33%), Positives = 68/136 (49%), Gaps = 14/136 (10%)
Query: 126 KTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISA-LDLSLLLSACPKLETLSIVCP 184
KTL L L + ++ +P PCLK + L V S + L L+S CP LE L++V
Sbjct: 141 KTLVTLKLRDSALN--DPGLVSMPCLKFMILREVRWSGTMSLEKLVSGCPVLEELTLVRT 198
Query: 185 EIAMSDSE--ASIELSSSSLKDFFV--------ESYSFDKLILV-ADMLENLHLKDCSFD 233
I D + A + S SLK F+V S DK++ + A LEN+ LK+ FD
Sbjct: 199 LICDIDDDELAVTRVRSRSLKTFYVPLAYGVGCRSRVPDKVLEIDAPGLENMTLKEDHFD 258
Query: 234 AFELINKGTLKVLKLD 249
+ N +L +++L+
Sbjct: 259 GIVVKNLTSLFMIELN 274
>UniRef100_Q9FZ53 F6I1.5 protein [Arabidopsis thaliana]
Length = 434
Score = 45.8 bits (107), Expect = 0.002
Identities = 83/378 (21%), Positives = 150/378 (38%), Gaps = 45/378 (11%)
Query: 21 DVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSHLEMLITCTLFQTKGL----- 75
+V+ SL C++WR WQ+ L L + Y + M + + G
Sbjct: 32 EVVKTSLICRRWRYVWQS-LPGLDLVINGSKNYDKFDFLERFMFLQRVKLRYVGYGHNCR 90
Query: 76 QCLTIFMDD--EHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFNIIEKCSRKTLEVLTL 133
++ M++ +H+ V + Y D + E+ + TS C R L
Sbjct: 91 NMTSMMMNNVIKHKIQHLDVGSNRRYVYDRV-EIPPTIYTS--------CERLVFLKLHR 141
Query: 134 ASNPISGVEPKYHKFPCLKSLSLSFVS-ISALDLSLLLSACPKLETLSIVCPEIAMSDSE 192
A+ P S P PCLK + L ++ + +LD+ L+S CP LETL++ D
Sbjct: 142 ANLPKS---PDSVSLPCLKIMDLQKINFVDSLDMEKLVSVCPALETLTM--------DKM 190
Query: 193 ASIELSSSSLKDFFVES----YSFDKLILVADMLENLHLKDCSFDAFELINKGTLKVLKL 248
++SS SL F + + Y ++++ L+ L L + + ++ +L L
Sbjct: 191 YGAKVSSQSLLSFCLTNNETGYLKTQVVMQTPKLKYLKLNRQFIQRIVINDLSSIVMLNL 250
Query: 249 DDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLKKLRLWSVVFDDEDEVVD 308
DDV+ + + + V F Y SK+ L K SV+ + V
Sbjct: 251 DDVAYFGETLLSILKLISCVRDLTISFDILQDYRHFSKSKSLPKFHKLSVLSVKDMAVGS 310
Query: 309 IETISVCFPRLTHLSLSYDLKDGVLHYGLQGLSF-------LMNVVVLELGWTTISDLFS 361
E++ + +L L G Y G++F L ++ +E+ ++D+
Sbjct: 311 WESLLIFLESCQNLK---SLVMGFRDYN-WGINFSDVPQCVLSSLEFVEVKAREVADMKK 366
Query: 362 VWVAGLLEGCPNLKKMVI 379
+W + +E LKK +
Sbjct: 367 LW-SYFMENSTVLKKFTL 383
>UniRef100_Q9M1D9 Hypothetical protein T2O9.20 [Arabidopsis thaliana]
Length = 838
Score = 45.4 bits (106), Expect = 0.003
Identities = 88/353 (24%), Positives = 144/353 (39%), Gaps = 49/353 (13%)
Query: 4 LPVEVVGIILSHLGSARDVLIASLTCKKWREAWQT--HLQTL-SFNTSDWPLYREICSSH 60
LP EV+G ILS L + + SL KKWR ++ HL+ SF+ H
Sbjct: 237 LPDEVLGKILS-LIPTKQAVSTSLLAKKWRTIFRLVDHLELDDSFSLQAVKDQTPGLRKH 295
Query: 61 LEMLIT----CTLFQTKGLQC--------LTIFMDDEHEFSVTPVIAWLM-YTRDSLREL 107
+ + T + +T LQC L + E V W+ + EL
Sbjct: 296 VRFVFTEDFKIFVDRTLALQCDYPIKNFSLKCHVSKYDERQKACVGRWISNVVGRGVFEL 355
Query: 108 RYNVRTSPNFNIIEK--CSRKTLEVLTLASNPISGVEPKYHKFPCLKSL---SLSFVSIS 162
++ P + + + KTL LTL + G P Y P LKSL ++ F I
Sbjct: 356 DLRMK-DPGIHFLPPHLVASKTLVKLTLGTQLCLGQLPSYVSLPSLKSLFIDTIVFYDIE 414
Query: 163 ALDLSLLLSACPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADM- 221
L +LL+ CP LE LS+ + + +SS +LK V+ + D + + M
Sbjct: 415 DL-CCVLLAGCPVLEELSVHHHDFIATPH----TISSPTLKRLSVDYHCPDDVDSASHMS 469
Query: 222 -----LENLHLKDCSFDAFELINKGTLKVLKLD---DVSVIHLDIGD------NTENLEI 267
L L C+ + IN +L KLD + V+ +D+ D N ++L +
Sbjct: 470 FDLPKLVYLEYSHCALGEYWQINLESLVEAKLDLGLERKVLRMDVTDLIIGIRNVQSLHL 529
Query: 268 -VDVCNFIFMWQN-----FYNMISKASKLKKLRLWSVVFDDEDEVVDIETISV 314
D + I+ + F N+++ + K R W ++ + + +ET+ V
Sbjct: 530 SPDSVHVIYSYCRHGLPVFKNLVNLSFGSKNKRGWRLLANLLKQSTKLETLIV 582
>UniRef100_Q8GWI2 Hypothetical protein At3g60040/T2O9_20 [Arabidopsis thaliana]
Length = 449
Score = 45.4 bits (106), Expect = 0.003
Identities = 88/353 (24%), Positives = 144/353 (39%), Gaps = 49/353 (13%)
Query: 4 LPVEVVGIILSHLGSARDVLIASLTCKKWREAWQT--HLQTL-SFNTSDWPLYREICSSH 60
LP EV+G ILS L + + SL KKWR ++ HL+ SF+ H
Sbjct: 18 LPDEVLGKILS-LIPTKQAVSTSLLAKKWRTIFRLVDHLELDDSFSLQAVKDQTPGLRKH 76
Query: 61 LEMLIT----CTLFQTKGLQC--------LTIFMDDEHEFSVTPVIAWLM-YTRDSLREL 107
+ + T + +T LQC L + E V W+ + EL
Sbjct: 77 VRFVFTEDFKIFVDRTLALQCDYPIKNFSLKCHVSKYDERQKACVGRWISNVVGRGVFEL 136
Query: 108 RYNVRTSPNFNIIEK--CSRKTLEVLTLASNPISGVEPKYHKFPCLKSL---SLSFVSIS 162
++ P + + + KTL LTL + G P Y P LKSL ++ F I
Sbjct: 137 DLRMK-DPGIHFLPPHLVASKTLVKLTLGTQLCLGQLPSYVSLPSLKSLFIDTIVFYDIE 195
Query: 163 ALDLSLLLSACPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADM- 221
L +LL+ CP LE LS+ + + +SS +LK V+ + D + + M
Sbjct: 196 DL-CCVLLAGCPVLEELSVHHHDFIATPH----TISSPTLKRLSVDYHCPDDVDSASHMS 250
Query: 222 -----LENLHLKDCSFDAFELINKGTLKVLKLD---DVSVIHLDIGD------NTENLEI 267
L L C+ + IN +L KLD + V+ +D+ D N ++L +
Sbjct: 251 FDLPKLVYLEYSHCALGEYWQINLESLVEAKLDLGLERKVLRMDVTDLIIGIRNVQSLHL 310
Query: 268 -VDVCNFIFMWQN-----FYNMISKASKLKKLRLWSVVFDDEDEVVDIETISV 314
D + I+ + F N+++ + K R W ++ + + +ET+ V
Sbjct: 311 SPDSVHVIYSYCRHGLPVFKNLVNLSFGSKNKRGWRLLANLLKQSTKLETLIV 363
>UniRef100_Q9ZV94 F9K20.20 protein [Arabidopsis thaliana]
Length = 452
Score = 45.1 bits (105), Expect = 0.004
Identities = 66/260 (25%), Positives = 116/260 (44%), Gaps = 21/260 (8%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
+ +LP ++ I+ ++ + +DV+ +S+ K+WR W+ ++ L+ + + Y S
Sbjct: 18 LSNLPESLICQIMLNIPT-KDVVKSSVLSKRWRNLWR-YVPGLNVEYNQFLDYNAFVSFV 75
Query: 61 LEMLITCTLFQTKGLQCLTIFMD-DEHEFSVTPVIAWLMYTRD-SLRELRYNVRTSPNFN 118
L L + Q + D DE E +++ V W+ D L+ L T N
Sbjct: 76 DRFL---ALDRESCFQSFRLRYDCDEEERTISNVKRWINIVVDQKLKVLDVLDYTWGNEE 132
Query: 119 IIEKCSRKTLEVLTLASNPISGV---EPKYHKFPCLKSLSLSFVSIS-ALDLSLLLSACP 174
+ S T E +L S + V PK P +K + L V S AL L ++S+C
Sbjct: 133 VQIPPSVYTCE--SLVSLKLCNVILPNPKVISLPLVKVIELDIVKFSNALVLEKIISSCS 190
Query: 175 KLETLSIVCPEIAMSDSEASIELSSSSLKDF-----FVESYSFDKLILVADMLENLHLKD 229
LE+L I + D + +SS SL F + + ++ + A LE L++ D
Sbjct: 191 ALESLIISRSSV---DDINVLRVSSRSLLSFKHIGNCSDGWDELEVAIDAPKLEYLNISD 247
Query: 230 CSFDAFELINKGTLKVLKLD 249
S F++ N G+L K++
Sbjct: 248 HSTAKFKMKNSGSLVEAKIN 267
>UniRef100_Q9FJC0 Heat shock transcription factor HSF30-like protein [Arabidopsis
thaliana]
Length = 454
Score = 44.3 bits (103), Expect = 0.007
Identities = 57/242 (23%), Positives = 111/242 (45%), Gaps = 19/242 (7%)
Query: 12 ILSHLGSARDVLIASLTCKKWREAWQT--HLQTLSFNTSDWPLYREICSSHLEMLITCTL 69
IL++L + +DV+ S+ +WR W L+ S + SD+ + C + + +
Sbjct: 35 ILNYLPT-KDVVKTSVLSTRWRSLWLLVPSLELDSRDFSDFNTFVSFCDRYFDSNRVLCI 93
Query: 70 FQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRY-NVRTSPNFNIIEKCSRKTL 128
+ K LTI ++E F + +W+ + R++++ +V+ P F+ I K
Sbjct: 94 NKLK----LTISENEEDGFYLK---SWI--DAAAKRKIQHLDVQFLPQFHKIHFNLYKCE 144
Query: 129 EVLTLASNPISGVEPKYHKFPCLKSLSL-SFVSISALDLSLLLSACPKLETLSIVCPEIA 187
+++L +S + + FPC+K++ L V + L+S CP LE L++ I
Sbjct: 145 ALVSLRLFEVSLDKGRIFSFPCMKTMHLEDNVYPNEATFKELISCCPVLEDLTV----II 200
Query: 188 MSDSEASIELSSSSLK-DFFVESYSFDKLILVADMLENLHLKDCSFDAFELINKGTLKVL 246
++ S SLK F+ + +++ A +L +L + D +F + N G+ L
Sbjct: 201 YGMDRKVFQVHSQSLKSSSFLHEVALSGVVIDAPLLCSLRINDNVSKSFIVNNLGSNDKL 260
Query: 247 KL 248
L
Sbjct: 261 DL 262
>UniRef100_Q5ZB59 Ribosomal RNA apurinic site specific lyase-like [Oryza sativa]
Length = 543
Score = 43.5 bits (101), Expect = 0.011
Identities = 33/83 (39%), Positives = 46/83 (54%), Gaps = 5/83 (6%)
Query: 146 HKFPCLKSLSLSFVSISALDLSLLLSACPKLETLSIVCPEIAMSDSEASIELSSSSLK-D 204
H FP L+ L L F+SI A DL LL P LETL++V + S S A I + S +L+
Sbjct: 249 HVFPHLRELGLCFISIDAQDLDGLLQCSPVLETLALV----SNSYSPAHIRVRSRTLRCV 304
Query: 205 FFVESYSFDKLILVADMLENLHL 227
F S + + ++VA L+ L L
Sbjct: 305 LFWMSLAQEIALVVAPRLDRLIL 327
>UniRef100_Q9FLA2 Similarity to heat shock transcription factor [Arabidopsis
thaliana]
Length = 438
Score = 43.1 bits (100), Expect = 0.015
Identities = 62/277 (22%), Positives = 110/277 (39%), Gaps = 27/277 (9%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
+ +LP ++ IL +L + + S+ +WR+ W + L N D+P + S
Sbjct: 6 ISELPDGLLNHILMYL-HIEESIRTSVLSSRWRKLW-LKVPGLDVNVHDFPADGNLFESL 63
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRD-SLRELRYNVRTSPNFNI 119
++ + + LQ + + + + + W+ D ++ L P + I
Sbjct: 64 MDKFLEVNRGR---LQNFKLNYESNLYYLMDRFVPWIATVVDRGIQHLDVTATDCPPWTI 120
Query: 120 ----IEKCSRKTLEVLTLASNPISGVEPKYH-KFPCLKSLSLSFVSISALDLSLLLSACP 174
C KTL L L + + PK+ PCLK + L + S L L+S CP
Sbjct: 121 DFMPANICKSKTLVSLKLVNVGLD--TPKFVVSLPCLKIMHLEDIFYSPLIAENLISGCP 178
Query: 175 KLETLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSFD--------KLILVADMLENLH 226
LE L+IV D + + S +LK+F +FD + + A L+ +
Sbjct: 179 VLEDLTIVRNH---EDFLNFLRVMSQTLKNF---RLTFDWGMGSNDFSVEIDAPGLKYMS 232
Query: 227 LKDCSFDAFELINKGTLKVLKLDDVSVIHLDIGDNTE 263
+D D + N +L + LD + +G E
Sbjct: 233 FRDSQSDRIVVKNLSSLVKIDLDTEFNLKFGLGSPLE 269
>UniRef100_Q9FJV2 Emb|CAB89229.1 [Arabidopsis thaliana]
Length = 477
Score = 43.1 bits (100), Expect = 0.015
Identities = 50/184 (27%), Positives = 83/184 (44%), Gaps = 9/184 (4%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
+ DLP E++ ILS L + L L ++W+ W+ + ++F+ D P S
Sbjct: 7 LSDLPDELLLKILSALPMFKVTLATRLISRRWKGPWKL-VPDVTFDDDDIPFK----SFE 61
Query: 61 LEMLITCTLFQTKGLQCL-TIFMDDEHEFSVTPVIAWL-MYTRDSLRELRYNVRTSPNFN 118
M F + Q L + + ++S + + W+ + S+RELR ++
Sbjct: 62 TFMSFVYGSFLSNDAQILDRLHLKLNQKYSASDINFWVQVAVNRSVRELRIDLFGKTLEL 121
Query: 119 IIEKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALD-LSLLLSACPKLE 177
CS TL+ LTL I V P + + P LK+L L V S+ ++ +L CP LE
Sbjct: 122 PCCLCSCITLKELTLHDLCIK-VVPAWFRLPSLKTLHLLSVKFSSDGFVASILRICPVLE 180
Query: 178 TLSI 181
L +
Sbjct: 181 RLVV 184
>UniRef100_UPI00002B2087 UPI00002B2087 UniRef100 entry
Length = 263
Score = 42.7 bits (99), Expect = 0.019
Identities = 49/180 (27%), Positives = 82/180 (45%), Gaps = 16/180 (8%)
Query: 203 KDFFVESYSFDKLILVADMLENLHLKDCSFDAFELINKGT-LKVLKLDDVSVIHLDIGDN 261
K+F + KL LV + + L L F E + + T LK L L++ + +
Sbjct: 7 KEFLRDHCKEHKLYLVPWLNDTLFLHFKGFSTIENLEEYTGLKCLWLENNRIKCI----- 61
Query: 262 TENLEIVDVCNFIFMWQNFYNMISKASKLKKLRLWSVVFDDEDEVVDIETISVCFPRLTH 321
ENL+ +F+ QN + + LK L + +V + + IE IS C P LT
Sbjct: 62 -ENLDSQTKLRCLFLQQNLIYKLENMASLKSLHILNV---SNNYIRTIEHIS-CLPDLTT 116
Query: 322 LSLSYDLKDGVLHYGLQGLSFLMNVVVLELGWTTISDLFSVWVAGLLEGCPNLKKMVIYG 381
L ++++ V +Q LS + + VL+L + + D V +L+ PNLK + + G
Sbjct: 117 LQIAHNKLKTV--DDIQHLSQCLAISVLDLSYNLLCD---PGVLTVLQAMPNLKVLNLIG 171
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 677,764,378
Number of Sequences: 2790947
Number of extensions: 27168373
Number of successful extensions: 64189
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 110
Number of HSP's that attempted gapping in prelim test: 64110
Number of HSP's gapped (non-prelim): 171
length of query: 419
length of database: 848,049,833
effective HSP length: 130
effective length of query: 289
effective length of database: 485,226,723
effective search space: 140230522947
effective search space used: 140230522947
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 76 (33.9 bits)
Medicago: description of AC147007.4


