

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147002.5 + phase: 0
(143 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
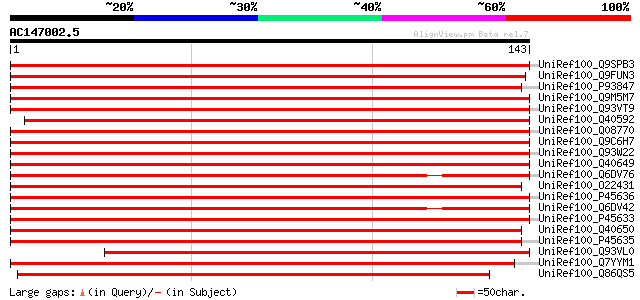
UniRef100_Q9SPB3 60S ribosomal protein L10 [Vitis riparia] 258 1e-68
UniRef100_Q9FUN3 QM-like protein [Elaeis guineensis var. tenera] 257 3e-68
UniRef100_P93847 60S ribosomal protein L10 [Solanum melongena] 254 2e-67
UniRef100_Q9M5M7 60S ribosomal protein L10 [Euphorbia esula] 253 6e-67
UniRef100_Q93VT9 At1g14320/F14L17_28 [Arabidopsis thaliana] 251 2e-66
UniRef100_Q40592 60S ribosomal protein L10 [Nicotiana tabacum] 250 4e-66
UniRef100_Q08770 60S ribosomal protein L10 [Arabidopsis thaliana] 250 4e-66
UniRef100_Q9C6H7 60S ribosomal protein L10, putative [Arabidopsi... 243 6e-64
UniRef100_Q93W22 At1g66580/T12I7_3 [Arabidopsis thaliana] 243 6e-64
UniRef100_Q40649 60S ribosomal protein L10-3 [Oryza sativa] 242 1e-63
UniRef100_Q6DV76 QM family protein [Caragana jubata] 241 2e-63
UniRef100_O22431 60S ribosomal protein L10 [Pinus taeda] 241 3e-63
UniRef100_P45636 60S ribosomal protein L10-2 [Oryza sativa] 240 4e-63
UniRef100_Q6DV42 QM-like protein [Camellia sinensis] 240 5e-63
UniRef100_P45633 60S ribosomal protein L10 [Zea mays] 236 8e-62
UniRef100_Q40650 Wilms' tumor-related protein QM [Oryza sativa] 234 2e-61
UniRef100_P45635 60S ribosomal protein L10-1 [Oryza sativa] 231 3e-60
UniRef100_Q93VL0 At1g14320/F14L17_28 [Arabidopsis thaliana] 206 1e-52
UniRef100_Q7YYM1 Ribsomal protein L10, probable [Cryptosporidium... 202 1e-51
UniRef100_Q86QS5 Ribosomal protein L10 [Branchiostoma belcheri t... 191 2e-48
>UniRef100_Q9SPB3 60S ribosomal protein L10 [Vitis riparia]
Length = 220
Score = 258 bits (660), Expect = 1e-68
Identities = 122/143 (85%), Positives = 132/143 (91%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+K+AGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP GTCARVSIGQVL
Sbjct: 76 MTKYAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGTCARVSIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK GN HAQEALRRAKFKFP RQKIIVSRKWGFTK + +Y+K KS+NRI+PDGV
Sbjct: 136 LSVRCKDGNSHHAQEALRRAKFKFPARQKIIVSRKWGFTKFNRTDYIKWKSQNRILPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK+LGCHGPLANRQPG+AF+ A
Sbjct: 196 NAKLLGCHGPLANRQPGKAFINA 218
>UniRef100_Q9FUN3 QM-like protein [Elaeis guineensis var. tenera]
Length = 224
Score = 257 bits (657), Expect = 3e-68
Identities = 122/142 (85%), Positives = 131/142 (91%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+K+AGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP GTCARV IGQVL
Sbjct: 76 MAKYAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGTCARVMIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK GN +HAQEALRRAKFKFPGRQKII+SRKWGFTK + +YL+ KSENRIVPDGV
Sbjct: 136 LSVRCKDGNSNHAQEALRRAKFKFPGRQKIIISRKWGFTKFNRTDYLRWKSENRIVPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFLP 142
NAK+LGCHG L+ RQPGRAFLP
Sbjct: 196 NAKLLGCHGRLSARQPGRAFLP 217
>UniRef100_P93847 60S ribosomal protein L10 [Solanum melongena]
Length = 219
Score = 254 bits (650), Expect = 2e-67
Identities = 124/141 (87%), Positives = 129/141 (90%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+K AGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP G CARV+IGQVL
Sbjct: 76 MTKSAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGVCARVAIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK GN +HAQEALRRAKFKFPGRQKIIVSRKWGFTK S +YLK KSENRIVPDGV
Sbjct: 136 LSVRCKDGNSNHAQEALRRAKFKFPGRQKIIVSRKWGFTKFSRTDYLKYKSENRIVPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFL 141
NAK+LG HGPLA RQPGRAFL
Sbjct: 196 NAKLLGNHGPLAARQPGRAFL 216
>UniRef100_Q9M5M7 60S ribosomal protein L10 [Euphorbia esula]
Length = 220
Score = 253 bits (646), Expect = 6e-67
Identities = 121/143 (84%), Positives = 132/143 (91%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+KFAGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP G CARV+IGQVL
Sbjct: 76 MTKFAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGVCARVAIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N +AQEALRRAKFKFPGRQKIIVSRKWGFTK++ A+Y +LKSENRI+PDGV
Sbjct: 136 LSVRCKDNNSHNAQEALRRAKFKFPGRQKIIVSRKWGFTKINRADYPRLKSENRILPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK+LGCHG LANR+PGRAF+ A
Sbjct: 196 NAKLLGCHGRLANRKPGRAFIDA 218
>UniRef100_Q93VT9 At1g14320/F14L17_28 [Arabidopsis thaliana]
Length = 220
Score = 251 bits (642), Expect = 2e-66
Identities = 120/143 (83%), Positives = 128/143 (88%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M K AGKD FHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGAFGK LGTCARV+IGQVL
Sbjct: 76 MVKSAGKDAFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKALGTCARVAIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK +G HAQEALRRAKFKFPGRQKIIVSRKWGFTK + A++ KL+ E R+VPDGV
Sbjct: 136 LSVRCKDAHGHHAQEALRRAKFKFPGRQKIIVSRKWGFTKFNRADFTKLRQEKRVVPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK L CHGPLANRQPG AFLPA
Sbjct: 196 NAKFLSCHGPLANRQPGSAFLPA 218
>UniRef100_Q40592 60S ribosomal protein L10 [Nicotiana tabacum]
Length = 150
Score = 250 bits (639), Expect = 4e-66
Identities = 122/139 (87%), Positives = 126/139 (89%)
Query: 5 AGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVLLSVR 64
AGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP G CARV+IGQVLLSVR
Sbjct: 9 AGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGVCARVAIGQVLLSVR 68
Query: 65 CKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGVNAKV 124
CK GN +HAQEALRRAKFKFP RQKIIVSRKWGFTK S +YLK KSENRIVPDGVNAK+
Sbjct: 69 CKDGNANHAQEALRRAKFKFPRRQKIIVSRKWGFTKFSRTDYLKYKSENRIVPDGVNAKL 128
Query: 125 LGCHGPLANRQPGRAFLPA 143
LGCHG LA RQPGRAFL A
Sbjct: 129 LGCHGRLAARQPGRAFLEA 147
>UniRef100_Q08770 60S ribosomal protein L10 [Arabidopsis thaliana]
Length = 221
Score = 250 bits (639), Expect = 4e-66
Identities = 121/143 (84%), Positives = 127/143 (88%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M K AGKD FHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGAFGK LGTCARV+IGQVL
Sbjct: 76 MVKSAGKDAFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKALGTCARVAIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK +G HAQEALRRAKFKFPGRQKIIVSRKWGFTK + A+Y KL+ E RIVPDGV
Sbjct: 136 LSVRCKDAHGHHAQEALRRAKFKFPGRQKIIVSRKWGFTKFNRADYTKLRQEKRIVPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK L CHGPLANRQPG AFL A
Sbjct: 196 NAKFLSCHGPLANRQPGSAFLSA 218
>UniRef100_Q9C6H7 60S ribosomal protein L10, putative [Arabidopsis thaliana]
Length = 184
Score = 243 bits (620), Expect = 6e-64
Identities = 119/143 (83%), Positives = 126/143 (87%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M K AGKD FHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGAFGK LGTCARV+IGQVL
Sbjct: 39 MVKSAGKDAFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKALGTCARVAIGQVL 98
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK +G HAQEALRRAKFKFPGRQKIIVSRKWGFTK + AEY KL++ RIVPDGV
Sbjct: 99 LSVRCKDNHGVHAQEALRRAKFKFPGRQKIIVSRKWGFTKFNRAEYTKLRAMKRIVPDGV 158
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK L HGPLANRQPG AF+ A
Sbjct: 159 NAKFLSNHGPLANRQPGSAFISA 181
>UniRef100_Q93W22 At1g66580/T12I7_3 [Arabidopsis thaliana]
Length = 221
Score = 243 bits (620), Expect = 6e-64
Identities = 119/143 (83%), Positives = 126/143 (87%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M K AGKD FHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGAFGK LGTCARV+IGQVL
Sbjct: 76 MVKSAGKDAFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKALGTCARVAIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK +G HAQEALRRAKFKFPGRQKIIVSRKWGFTK + AEY KL++ RIVPDGV
Sbjct: 136 LSVRCKDNHGVHAQEALRRAKFKFPGRQKIIVSRKWGFTKFNRAEYTKLRAMKRIVPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK L HGPLANRQPG AF+ A
Sbjct: 196 NAKFLSNHGPLANRQPGSAFISA 218
>UniRef100_Q40649 60S ribosomal protein L10-3 [Oryza sativa]
Length = 219
Score = 242 bits (617), Expect = 1e-63
Identities = 118/143 (82%), Positives = 125/143 (86%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+K AGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP GTCARV IGQVL
Sbjct: 76 MTKSAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGTCARVDIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N HA EALRRAKFKFPGRQKII SRKWGFTK S EY++LKSE RI+PDGV
Sbjct: 136 LSVRCKPNNAVHASEALRRAKFKFPGRQKIIESRKWGFTKFSRDEYVRLKSEGRIMPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK+LGCHG L+ R PG+AFL A
Sbjct: 196 NAKLLGCHGRLSARAPGKAFLSA 218
>UniRef100_Q6DV76 QM family protein [Caragana jubata]
Length = 216
Score = 241 bits (615), Expect = 2e-63
Identities = 118/143 (82%), Positives = 127/143 (88%), Gaps = 4/143 (2%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
++KFAGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP GTCARV+IGQVL
Sbjct: 76 LTKFAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGTCARVAIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK GN HAQEALRRAKFKFPGRQKIIVSRKWGFTK + +YL+ KSENRI V
Sbjct: 136 LSVRCKDGNSHHAQEALRRAKFKFPGRQKIIVSRKWGFTKYNRTDYLRWKSENRI----V 191
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
+ + LGCHGPLANRQPGRAF+ A
Sbjct: 192 SMQPLGCHGPLANRQPGRAFINA 214
>UniRef100_O22431 60S ribosomal protein L10 [Pinus taeda]
Length = 228
Score = 241 bits (614), Expect = 3e-63
Identities = 115/141 (81%), Positives = 128/141 (90%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M KFAGKD FHLRVRVHPFHVLR NKMLSCAGADRLQTGMRGAFGKP GTCARV+IGQVL
Sbjct: 76 MVKFAGKDGFHLRVRVHPFHVLRSNKMLSCAGADRLQTGMRGAFGKPQGTCARVAIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVR + + +HAQEALRRAKFKFPGR+KIIV+RKWGFTK + A+YLK K+ENRIVPDGV
Sbjct: 136 LSVRSRDNHSNHAQEALRRAKFKFPGREKIIVNRKWGFTKYTRADYLKWKTENRIVPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFL 141
N K+LGC GPL+NR+PG+AFL
Sbjct: 196 NPKLLGCRGPLSNRKPGQAFL 216
>UniRef100_P45636 60S ribosomal protein L10-2 [Oryza sativa]
Length = 218
Score = 240 bits (613), Expect = 4e-63
Identities = 117/143 (81%), Positives = 124/143 (85%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+K AGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP GTCARV IGQVL
Sbjct: 75 MTKSAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGTCARVDIGQVL 134
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N HA EALRRAKFKFPGRQKII SRKWGFTK S EY++LKSE RI+PDGV
Sbjct: 135 LSVRCKPNNAVHASEALRRAKFKFPGRQKIIESRKWGFTKFSRDEYVRLKSEGRIMPDGV 194
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK+LGCHG L+ R PG+AF A
Sbjct: 195 NAKLLGCHGRLSARAPGKAFFSA 217
>UniRef100_Q6DV42 QM-like protein [Camellia sinensis]
Length = 216
Score = 240 bits (612), Expect = 5e-63
Identities = 118/143 (82%), Positives = 126/143 (87%), Gaps = 4/143 (2%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+KFAGKD FHLRVRVHPFHVLRIN MLSCAGADRLQTGMRGAFGKP GTCARV+IGQVL
Sbjct: 76 MTKFAGKDAFHLRVRVHPFHVLRINTMLSCAGADRLQTGMRGAFGKPQGTCARVAIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK GN HAQEALRRAKFKFPGRQKIIVSRKWGFTK + +YL+ KSENRI V
Sbjct: 136 LSVRCKDGNSHHAQEALRRAKFKFPGRQKIIVSRKWGFTKYNRTDYLRWKSENRI----V 191
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
+ + LGCHGPLANRQPGRAF+ A
Sbjct: 192 SMQPLGCHGPLANRQPGRAFINA 214
>UniRef100_P45633 60S ribosomal protein L10 [Zea mays]
Length = 220
Score = 236 bits (602), Expect = 8e-62
Identities = 117/143 (81%), Positives = 121/143 (83%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+K AGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP GTCARV IGQVL
Sbjct: 76 MTKSAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGTCARVDIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N HA EALRRAKFKFP RQKII SRKWGFTK S A+YLK KSE RIVPDGV
Sbjct: 136 LSVRCKDNNAAHASEALRRAKFKFPARQKIIESRKWGFTKFSRADYLKYKSEGRIVPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK+L HG L R PG+AFL A
Sbjct: 196 NAKLLANHGRLEKRAPGKAFLDA 218
>UniRef100_Q40650 Wilms' tumor-related protein QM [Oryza sativa]
Length = 173
Score = 234 bits (598), Expect = 2e-61
Identities = 114/141 (80%), Positives = 123/141 (86%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+K AGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP GTCARV IGQVL
Sbjct: 25 MTKNAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGTCARVDIGQVL 84
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N HA+EALRRAKFKFPGRQKII SRKWGFTK + EY+KLK+E RI+ DGV
Sbjct: 85 LSVRCKESNAKHAEEALRRAKFKFPGRQKIIHSRKWGFTKFTREEYVKLKAEGRIMSDGV 144
Query: 121 NAKVLGCHGPLANRQPGRAFL 141
NA++LG HG LA R PG+AFL
Sbjct: 145 NAQLLGSHGRLAKRAPGKAFL 165
>UniRef100_P45635 60S ribosomal protein L10-1 [Oryza sativa]
Length = 224
Score = 231 bits (589), Expect = 3e-60
Identities = 114/141 (80%), Positives = 120/141 (84%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+K AGKD FHLRVRVHP HVLRINKMLSCAGADRLQTGMRGAFGKP GTCARV IGQVL
Sbjct: 76 MTKNAGKDAFHLRVRVHPVHVLRINKMLSCAGADRLQTGMRGAFGKPQGTCARVDIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N HA+EALRRAKFKFPGRQKII SRKWGFTK S LKLK+E RI+ DGV
Sbjct: 136 LSVRCKESNAKHAEEALRRAKFKFPGRQKIIHSRKWGFTKFSRVRVLKLKAEGRIMSDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFL 141
NAK+LG HG LA R PG+AFL
Sbjct: 196 NAKLLGSHGRLAKRAPGKAFL 216
>UniRef100_Q93VL0 At1g14320/F14L17_28 [Arabidopsis thaliana]
Length = 119
Score = 206 bits (523), Expect = 1e-52
Identities = 98/117 (83%), Positives = 105/117 (88%)
Query: 27 MLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVLLSVRCKSGNGDHAQEALRRAKFKFPG 86
MLSCAGADRLQTGMRGAFGK LGTCARV+IGQVLLSVRCK +G HAQEALRRAKFKFPG
Sbjct: 1 MLSCAGADRLQTGMRGAFGKALGTCARVAIGQVLLSVRCKDAHGHHAQEALRRAKFKFPG 60
Query: 87 RQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGVNAKVLGCHGPLANRQPGRAFLPA 143
RQKIIVSRKWGFTK + A++ KL+ E R+VPDGVNAK L CHGPLANRQPG AFLPA
Sbjct: 61 RQKIIVSRKWGFTKFNRADFTKLRQEKRVVPDGVNAKFLSCHGPLANRQPGSAFLPA 117
>UniRef100_Q7YYM1 Ribsomal protein L10, probable [Cryptosporidium parvum]
Length = 222
Score = 202 bits (515), Expect = 1e-51
Identities = 97/139 (69%), Positives = 110/139 (78%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M KF GKD FHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGA+GKP GT ARV+IGQVL
Sbjct: 76 MIKFCGKDNFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAYGKPTGTAARVNIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
+S+RCK A ALRRAK+KFPGRQK++VS KWGFTK + EYLK ++E RIVPDGV
Sbjct: 136 MSIRCKEDKTQTAVMALRRAKYKFPGRQKVVVSDKWGFTKFTKEEYLKYQAEGRIVPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRA 139
N K + C G L+ P A
Sbjct: 196 NCKYISCRGSLSRIFPEAA 214
>UniRef100_Q86QS5 Ribosomal protein L10 [Branchiostoma belcheri tsingtaunese]
Length = 216
Score = 191 bits (486), Expect = 2e-48
Identities = 92/130 (70%), Positives = 102/130 (77%)
Query: 3 KFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVLLS 62
K GKD FH+R+R HPFHV+RINKMLSCAGADRLQTGMRGAFGKP GT ARV IGQ L+S
Sbjct: 78 KHCGKDAFHIRIRAHPFHVIRINKMLSCAGADRLQTGMRGAFGKPQGTVARVHIGQPLIS 137
Query: 63 VRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGVNA 122
R + N H EALRRAKFKFPGRQKI VS+KWGFTK +YL+LK+ +R VPDGV
Sbjct: 138 CRARDANKAHVIEALRRAKFKFPGRQKIYVSKKWGFTKFDQPDYLELKNADRFVPDGVGV 197
Query: 123 KVLGCHGPLA 132
K L HGPLA
Sbjct: 198 KYLPEHGPLA 207
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.140 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 230,178,481
Number of Sequences: 2790947
Number of extensions: 8471699
Number of successful extensions: 19559
Number of sequences better than 10.0: 166
Number of HSP's better than 10.0 without gapping: 164
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 19389
Number of HSP's gapped (non-prelim): 168
length of query: 143
length of database: 848,049,833
effective HSP length: 119
effective length of query: 24
effective length of database: 515,927,140
effective search space: 12382251360
effective search space used: 12382251360
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147002.5


