

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.5 - phase: 0
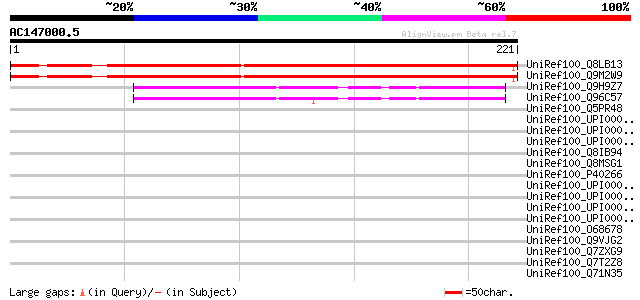
(221 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LB13 Hypothetical protein [Arabidopsis thaliana] 194 1e-48
UniRef100_Q9M2W9 Hypothetical protein T16K5.240 [Arabidopsis tha... 191 2e-47
UniRef100_Q9H9Z7 Hypothetical protein FLJ12448 [Homo sapiens] 47 4e-04
UniRef100_Q96C57 Hypothetical protein FLJ12448 [Homo sapiens] 46 8e-04
UniRef100_Q5PR48 Hypothetical protein [Brachydanio rerio] 44 0.002
UniRef100_UPI000046C1B5 UPI000046C1B5 UniRef100 entry 40 0.057
UniRef100_UPI0000465B0E UPI0000465B0E UniRef100 entry 39 0.074
UniRef100_UPI0000340513 UPI0000340513 UniRef100 entry 38 0.17
UniRef100_Q8IB94 Ubiquitin-protein ligase 1, putative [Plasmodiu... 38 0.17
UniRef100_Q8MSG1 GM13065p [Drosophila melanogaster] 38 0.17
UniRef100_P40266 Histone H1-I [Glyptotendipes salinus] 38 0.17
UniRef100_UPI000036A978 UPI000036A978 UniRef100 entry 37 0.37
UniRef100_UPI0000498AE9 UPI0000498AE9 UniRef100 entry 37 0.37
UniRef100_UPI00002F43FF UPI00002F43FF UniRef100 entry 37 0.37
UniRef100_UPI00002356AB UPI00002356AB UniRef100 entry 37 0.37
UniRef100_O68678 Gas vesicle protein GvpQ [Bacillus megaterium] 37 0.48
UniRef100_Q9VJG2 CG12288-PA [Drosophila melanogaster] 37 0.48
UniRef100_Q7ZXG9 Tnnt3a-prov protein [Xenopus laevis] 36 0.63
UniRef100_Q7T2Z8 Fast troponin T [Xenopus laevis] 36 0.63
UniRef100_Q71N35 Troponin T3 [Xenopus laevis] 36 0.63
>UniRef100_Q8LB13 Hypothetical protein [Arabidopsis thaliana]
Length = 220
Score = 194 bits (493), Expect = 1e-48
Identities = 102/222 (45%), Positives = 154/222 (68%), Gaps = 11/222 (4%)
Query: 1 MAEEELNNGYCSSSGDEDGDAAWKAAIDSVAGTSSYVTSFMNGFSATNNNDTKKKHNDNQ 60
MA+ EL+ G SS ED D W+AAI+S+A T+ Y G SAT T+ +N +
Sbjct: 1 MAKRELSGGDSSS---EDEDPKWRAAINSIATTTVY------GASATKPAATQSHNNGDF 51
Query: 61 NPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSKPGIVFD 120
K K+ H Q+K LL++++E T++ V++P+ + ++ P +DCG+RLF+ GIVFD
Sbjct: 52 RLKPKKLTHGQIKVKNLLNEMVEKTLDFVEDPVNIPEDKPE-NDCGVRLFKRCATGIVFD 110
Query: 121 HADEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAKD 180
H DE + P K+P L P + ++ SK+F++R++SIAVDG+D++ AA +A KK+ ARL+AK+
Sbjct: 111 HVDEIRGPKKKPNLRPDKGVEGSSKEFKKRVKSIAVDGSDILTAAMEAAKKASARLDAKE 170
Query: 181 AAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAKIK-IKH 221
AAK KAK+EEERI +LKK+RGE+WLPS+ + M+ ++K IKH
Sbjct: 171 VAAKDKAKKEEERIAELKKVRGEKWLPSIERAMKKEMKRIKH 212
>UniRef100_Q9M2W9 Hypothetical protein T16K5.240 [Arabidopsis thaliana]
Length = 220
Score = 191 bits (484), Expect = 2e-47
Identities = 101/222 (45%), Positives = 153/222 (68%), Gaps = 11/222 (4%)
Query: 1 MAEEELNNGYCSSSGDEDGDAAWKAAIDSVAGTSSYVTSFMNGFSATNNNDTKKKHNDNQ 60
MA+ EL+ G SS ED D W+AAI+S+A T+ Y G SAT T+ + +
Sbjct: 1 MAKRELSGGDSSS---EDEDPKWRAAINSIATTTVY------GASATKPAATQSHNYGDF 51
Query: 61 NPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSKPGIVFD 120
K K+ H Q+K LL++++E T++ V++P+ + ++ P +DCG+RLF+ GIVFD
Sbjct: 52 RLKPKKLTHGQIKVKNLLNEMVEKTLDFVEDPVNIPEDKPE-NDCGVRLFKRCATGIVFD 110
Query: 121 HADEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAKD 180
H DE + P K+P L P + ++ SK+F++R++SIAVDG+D++ AA +A KK+ ARL+AK+
Sbjct: 111 HVDEIRGPKKKPNLRPDKGVEGSSKEFKKRVKSIAVDGSDILTAAVEAAKKASARLDAKE 170
Query: 181 AAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAKIK-IKH 221
AAK KAK+EEERI +LKK+RGE+WLPS+ + M+ ++K IKH
Sbjct: 171 VAAKDKAKKEEERIAELKKVRGEKWLPSIERAMKKEMKRIKH 212
>UniRef100_Q9H9Z7 Hypothetical protein FLJ12448 [Homo sapiens]
Length = 262
Score = 47.0 bits (110), Expect = 4e-04
Identities = 44/162 (27%), Positives = 71/162 (43%), Gaps = 9/162 (5%)
Query: 55 KHNDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSK 114
+ + N+ +P+ + + K L L D E KEP + ++D G RLF S
Sbjct: 70 EQDGNELQTTPEFRAHVAKKLGALLDSFITISEAAKEPAKAKVQKVALEDDGFRLFFTSV 129
Query: 115 PGIVFDHADEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLA 174
PG + + PQP KR ED DE+ RR R AV +D++ ++ S
Sbjct: 130 PG-GREKEESPQPRRKRQPSSSSEDSDEE----WRRCREAAVSASDIL---QESAIHSPG 181
Query: 175 RLEAKDAAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAK 216
+E K+A K K K++ +++ + SMA + K
Sbjct: 182 TVE-KEAKKKRKLKKKAKKVASVDSAVAATTPTSMATVQKQK 222
>UniRef100_Q96C57 Hypothetical protein FLJ12448 [Homo sapiens]
Length = 263
Score = 45.8 bits (107), Expect = 8e-04
Identities = 45/163 (27%), Positives = 72/163 (43%), Gaps = 10/163 (6%)
Query: 55 KHNDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSK 114
+ + N+ +P+ + + K L L D E KEP + ++D G RLF S
Sbjct: 70 EQDGNELQTTPEFRAHVAKKLGALLDSFITISEAAKEPAKAKVQKVALEDDGFRLFFTSV 129
Query: 115 PGIVFDHADEPQPPMKR-PKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSL 173
PG + + PQP KR P ED DE+ RR R AV +D++ ++ S
Sbjct: 130 PG-GREKEESPQPRRKRQPSSSSSEDSDEE----WRRCREAAVSASDIL---QESAIHSP 181
Query: 174 ARLEAKDAAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAK 216
+E K+A K K K++ +++ + SMA + K
Sbjct: 182 GTVE-KEAKKKRKLKKKAKKVASVDSAVAATTPTSMATVQKQK 223
>UniRef100_Q5PR48 Hypothetical protein [Brachydanio rerio]
Length = 234
Score = 44.3 bits (103), Expect = 0.002
Identities = 48/223 (21%), Positives = 92/223 (40%), Gaps = 23/223 (10%)
Query: 13 SSGDEDGDAAWKAAIDSVAGTSSYVTSF-MNGFSATNNNDTKKKHNDNQNPKSPKIKHYQ 71
SS +++ A K A+ S + NG + + +K +H+ N+ +P+ + +
Sbjct: 4 SSSEDENTARLKEAVWSFKPEDVKINGKEKNGRQSHRADVSKHEHDGNELGTTPEFRSHV 63
Query: 72 LKAL-KLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSKPGIVFDHADEPQPPMK 130
K L LD + E+ + + + D+ RLF S PG + + P PP +
Sbjct: 64 AKKLGAYLDGCIS---EVCSDTVEPAQSENREDEEDFRLFSSSTPGKWMEQS-LPPPPKR 119
Query: 131 RPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAA----NDAYKKSLARLEAKDAAAKAK 186
RP VP + + RIR AV +D++ A ++ ++ + E +D K K
Sbjct: 120 RP--VPSS--SDSDSEMEMRIREAAVSLSDILGPAAQNLSEKTEEKSTKEETEDTVTKKK 175
Query: 187 AKRE---------EERIEKLKKIRGERWLPSMAKEMQAKIKIK 220
++ + EK + G + + + ++ K K K
Sbjct: 176 KRKTSSEESQDKVNHQTEKQSNVEGNQEQTTAGERLKKKKKKK 218
>UniRef100_UPI000046C1B5 UPI000046C1B5 UniRef100 entry
Length = 1757
Score = 39.7 bits (91), Expect = 0.057
Identities = 50/235 (21%), Positives = 95/235 (40%), Gaps = 52/235 (22%)
Query: 27 IDSVAGTSSYVTSFMNGFSATNNNDTKK---------------------------KHNDN 59
I+S+ T+S S+ + F T N TKK K N
Sbjct: 3 IESIKNTTSIGKSYTDKFLNTLTNKTKKLENIFKDASLNEHEQTNNELKKYFNNLKENLG 62
Query: 60 QNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDE----------------DPNVD 103
++PKS + + K KL +DI++ ++I K + E + N+
Sbjct: 63 KDPKSTLSQQFNEKE-KLFNDIIQKNVDINKNISNIEIEIYASIHNISDETENEIEENIK 121
Query: 104 DCGIRLFRHSKPGIVFDHADEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIA 163
++F K + + +E + +K+ E EK+KK+ I I D I
Sbjct: 122 SLNTQIFEKVKTNVA--NLNEIKEKLKQYNFDDFEK--EKNKKYNDEINKIKGD----IT 173
Query: 164 AANDAYKKSLARLEAKDAAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAKIK 218
++ K++A+L + ++++ A + +I+K +K+ + KE++ KIK
Sbjct: 174 TLDNKIDKNIAKLTEIEKSSESYASEIKIQIDKSEKVTDKTIYKEEPKEIEKKIK 228
>UniRef100_UPI0000465B0E UPI0000465B0E UniRef100 entry
Length = 836
Score = 39.3 bits (90), Expect = 0.074
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 8/76 (10%)
Query: 48 NNNDTKKKHNDNQNPKSPKIKHYQLKALKLLDDILE---NTIEIVKEPIPVLDEDPN--- 101
NN +T K+H DN+ KS + + LK K +DI + N I+ + + + +L PN
Sbjct: 504 NNPETMKQHEDNEQNKSNNVMYKHLKPPKNSNDISQLHFNEIKNMLKDLTILQSGPNWKT 563
Query: 102 --VDDCGIRLFRHSKP 115
+DD LF++ +P
Sbjct: 564 YKMDDLPKELFKNIRP 579
>UniRef100_UPI0000340513 UPI0000340513 UniRef100 entry
Length = 294
Score = 38.1 bits (87), Expect = 0.17
Identities = 37/136 (27%), Positives = 68/136 (49%), Gaps = 10/136 (7%)
Query: 67 IKHYQLKALKLLDDILENTIEIVKEPIPVLDE-DPNVDDCGIRLFRHSKPGIVFDHADEP 125
++ +Q K +LL+D E + + P+ DE + +DD RL R + + H E
Sbjct: 1 LEKHQTKLKRLLEDAREEMAGLNQ---PITDEVNRALDDENKRLRREIEDELEHKHRQE- 56
Query: 126 QPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAK--DAAA 183
+++ + +DI+++ + I+ I D A +A +++L + + + DA A
Sbjct: 57 ---IEQIRHANRQDIEQERFEKEHLIQEIERLKCDHQKALEEALEEALEKKQRQLEDAVA 113
Query: 184 KAKAKREEERIEKLKK 199
AKA E+ER+E LKK
Sbjct: 114 AAKASAEKERLELLKK 129
>UniRef100_Q8IB94 Ubiquitin-protein ligase 1, putative [Plasmodium falciparum]
Length = 8591
Score = 38.1 bits (87), Expect = 0.17
Identities = 18/63 (28%), Positives = 30/63 (47%)
Query: 7 NNGYCSSSGDEDGDAAWKAAIDSVAGTSSYVTSFMNGFSATNNNDTKKKHNDNQNPKSPK 66
NNG+ + GD D D+ + +S + + Y + N + NNND +NDN N
Sbjct: 6250 NNGFNPNEGDNDSDSDSNSDSNSDSDSDDYNDNSDNNNNNDNNNDNNDNNNDNNNENQNN 6309
Query: 67 IKH 69
+ +
Sbjct: 6310 VNN 6312
>UniRef100_Q8MSG1 GM13065p [Drosophila melanogaster]
Length = 435
Score = 38.1 bits (87), Expect = 0.17
Identities = 28/97 (28%), Positives = 55/97 (55%), Gaps = 6/97 (6%)
Query: 124 EPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAKDAAA 183
E +P K P +V + +KSK ++ +++ V +++ AA ++ KK L AKD A
Sbjct: 13 ELKPIKKEPGVVEDGKVTKKSKTKPKK-KTVKVPKDEVKAAVQESPKK----LSAKDLA- 66
Query: 184 KAKAKREEERIEKLKKIRGERWLPSMAKEMQAKIKIK 220
K K K+ +++ +KLKK + + + E +A++K++
Sbjct: 67 KIKKKKAKKQAQKLKKQQNKLAPKEIKTEPEAQVKVE 103
>UniRef100_P40266 Histone H1-I [Glyptotendipes salinus]
Length = 233
Score = 38.1 bits (87), Expect = 0.17
Identities = 30/96 (31%), Positives = 44/96 (45%), Gaps = 4/96 (4%)
Query: 123 DEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAKDAA 182
D+P+ K+ P +I++K KK + AV+ AA A KK++A+ AK AA
Sbjct: 131 DKPEAAPKKKAPRPKREIEKKEKKVVAKKPKPAVEKK----AAKPAAKKAVAKPAAKKAA 186
Query: 183 AKAKAKREEERIEKLKKIRGERWLPSMAKEMQAKIK 218
AK AK + K + P+ K AK K
Sbjct: 187 AKPAAKEPAAKASPKKAAAKPKAKPTPKKSTAAKPK 222
>UniRef100_UPI000036A978 UPI000036A978 UniRef100 entry
Length = 2460
Score = 37.0 bits (84), Expect = 0.37
Identities = 46/201 (22%), Positives = 79/201 (38%), Gaps = 45/201 (22%)
Query: 53 KKKHNDNQNPKSPKI----KHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIR 108
KK ND++ S KI K Y A+ +L + + I K +++ + D
Sbjct: 1396 KKDKNDSERQNSKKIEKELKPYGSSAINILKEKKKREIHREKWRD---EKERHRDRHADG 1452
Query: 109 LFRHSKPGIVFDHADEPQPPMKRPKLVPGEDIDEKSK---------KFRRRIRSIAV--- 156
L RH + ++ H DE Q P R K P + +KS+ K + + + A
Sbjct: 1453 LLRHHRDELLRHHRDE-QKPATRDKDSPPRVLKDKSRDDGPRLSDAKLKEKFKDSAEKEK 1511
Query: 157 -------DGNDLIAAANDAYKKSLA------------------RLEAKDAAAKAKAKREE 191
+GND +A D KK L KD + + KR +
Sbjct: 1512 GDPAKMSNGNDKVAPCKDPGKKDARPREKLLGDGDLMMTSFERMLSQKDLEIEERHKRHK 1571
Query: 192 ERIEKLKKIRGERWLPSMAKE 212
ER+++++K+R P + ++
Sbjct: 1572 ERMKQMEKLRHRSGDPKLKEK 1592
>UniRef100_UPI0000498AE9 UPI0000498AE9 UniRef100 entry
Length = 1226
Score = 37.0 bits (84), Expect = 0.37
Identities = 43/197 (21%), Positives = 81/197 (40%), Gaps = 20/197 (10%)
Query: 17 EDGDAAWKAAIDSVAGTSSYVTSFMNGFSATNNNDTKKKHNDNQNPKSPKIKHYQLKALK 76
++G+ ++ + GTS Y+ G S +K ND + K+ + K + +
Sbjct: 167 KNGEDGMLEFLEDIIGTSQYIEGIEKGTSEL------EKLNDEVHEKTNRFKIIERERNG 220
Query: 77 LLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSKPGIVFDHADEPQPPMKRPKLVP 136
+L + EN I +KE + N++ ++ + + +E + ++ K++
Sbjct: 221 ILKE-KENAISYLKEEYKMCGLKNNLNKLELKAVEEKE----VEGKEEREKLEQKRKIIQ 275
Query: 137 ---GEDIDEKSKKFR------RRIRSIAVDGNDLIAAANDAYKKSLARLEAKDAAAKAKA 187
E + EK+ K + R I+ I + N A KK E K KA
Sbjct: 276 DAKNEKLKEKTTKDKIIEEKERDIKKIEKEYEKQKGLINSAKKKKARAEEEKKQNEKAVL 335
Query: 188 KREEERIEKLKKIRGER 204
+ E+E E KKI+ E+
Sbjct: 336 RNEKEIKEMEKKIKDEK 352
>UniRef100_UPI00002F43FF UPI00002F43FF UniRef100 entry
Length = 90
Score = 37.0 bits (84), Expect = 0.37
Identities = 23/55 (41%), Positives = 32/55 (57%), Gaps = 1/55 (1%)
Query: 140 IDEKSKKFRRRIRSIAVDGNDLIAAA-NDAYKKSLARLEAKDAAAKAKAKREEER 193
ID +++ + S VDG L+AA ND K +LA L A AAA+A RE+E+
Sbjct: 26 IDRAELEYQLELNSTRVDGQMLLAALENDKLKVTLATLLAARAAARAARAREDEQ 80
>UniRef100_UPI00002356AB UPI00002356AB UniRef100 entry
Length = 384
Score = 37.0 bits (84), Expect = 0.37
Identities = 39/155 (25%), Positives = 64/155 (41%), Gaps = 19/155 (12%)
Query: 48 NNNDTKKKHNDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGI 107
NNN+ +N+N N + + Q D N+ + ++PI +D +VDD I
Sbjct: 86 NNNNNSHNNNNNNNNTNTNSNNSQDADSNADSDHNSNSDQNQQKPIIETTDDHSVDDQHI 145
Query: 108 RLFRHSKPGIVFDHADEPQPPMK----RPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIA 163
S DEP P+ RP P + + +++ +RR RS+ +D +A
Sbjct: 146 DTVSDS---------DEPSSPIMGIGLRPISKPVSVVVDNTRR-KRRARSLVASEDDDVA 195
Query: 164 AANDAYK-----KSLARLEAKDAAAKAKAKREEER 193
A + K K L R K A A+ + E +
Sbjct: 196 APSTRQKRARANKYLTRKTQKAIQAAARVRDPESK 230
>UniRef100_O68678 Gas vesicle protein GvpQ [Bacillus megaterium]
Length = 157
Score = 36.6 bits (83), Expect = 0.48
Identities = 27/132 (20%), Positives = 60/132 (45%), Gaps = 5/132 (3%)
Query: 68 KHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSKPGIVFDHADEPQP 127
K + ALK I+++T E VKE I + V + F G + + A+E
Sbjct: 4 KDVEKAALKAGKKIIDHTPEPVKEKI-----EEKVKEKAKEKFVEKTEGKLQEKANEASE 58
Query: 128 PMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAKDAAAKAKA 187
++ K + + K + + +++ + + D ++ +A + ++ + D K+K
Sbjct: 59 KLQETKEKNAQKVHGKGEDAKEKLQDVLLSVKDKLSDVKEAGENFQEKVSSSDDKEKSKN 118
Query: 188 KREEERIEKLKK 199
KR+ + + ++KK
Sbjct: 119 KRKIKGVNQIKK 130
>UniRef100_Q9VJG2 CG12288-PA [Drosophila melanogaster]
Length = 435
Score = 36.6 bits (83), Expect = 0.48
Identities = 27/97 (27%), Positives = 55/97 (55%), Gaps = 6/97 (6%)
Query: 124 EPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAKDAAA 183
E +P K P +V + +KSK ++ +++ V +++ AA ++ KK L AKD A
Sbjct: 13 ELKPIKKEPGVVEDGKVTKKSKTKPKK-KTVKVPKDEVKAAVQESPKK----LSAKDLA- 66
Query: 184 KAKAKREEERIEKLKKIRGERWLPSMAKEMQAKIKIK 220
K + K+ +++ +KLKK + + + E +A++K++
Sbjct: 67 KIEKKKAKKQAQKLKKQQNKLAPKEIKTEPEAQVKVE 103
>UniRef100_Q7ZXG9 Tnnt3a-prov protein [Xenopus laevis]
Length = 276
Score = 36.2 bits (82), Expect = 0.63
Identities = 32/114 (28%), Positives = 58/114 (50%), Gaps = 7/114 (6%)
Query: 90 KEPIPVLDEDPNVDDCGIRLFRHSKPGIVFDHADEPQPPMKRPKLVPGEDIDEKSKKFRR 149
+E PV++E P V++ + G +D ++P+P + PK+ GE +D + +R
Sbjct: 25 QEEEPVVEEAPQVEEA-YEEAEEEEHGEEYDE-EKPKPKLTAPKIPDGEKVDFDDIQKKR 82
Query: 150 RIRSIAVDGNDLIAAANDAYKKSLARLEA-KDAAAKAKAKREEE---RIEKLKK 199
+ + + ++ LI +A KK + A KD K +A+R E+ R EK K+
Sbjct: 83 QNKDL-IELQSLIDTHFEARKKEEEDIIALKDRIEKRRAERAEQLRIRTEKEKE 135
>UniRef100_Q7T2Z8 Fast troponin T [Xenopus laevis]
Length = 276
Score = 36.2 bits (82), Expect = 0.63
Identities = 32/114 (28%), Positives = 58/114 (50%), Gaps = 7/114 (6%)
Query: 90 KEPIPVLDEDPNVDDCGIRLFRHSKPGIVFDHADEPQPPMKRPKLVPGEDIDEKSKKFRR 149
+E PV++E P V++ + G +D ++P+P + PK+ GE +D + +R
Sbjct: 25 QEEEPVVEEAPQVEEA-YEEAEEEEHGEEYDE-EKPKPKLTAPKIPDGEKVDFDDIQKKR 82
Query: 150 RIRSIAVDGNDLIAAANDAYKKSLARLEA-KDAAAKAKAKREEE---RIEKLKK 199
+ + + ++ LI +A KK + A KD K +A+R E+ R EK K+
Sbjct: 83 QNKDL-IELQSLIDTHFEARKKEEEDIIALKDRIEKRRAERAEQLRIRTEKEKE 135
>UniRef100_Q71N35 Troponin T3 [Xenopus laevis]
Length = 273
Score = 36.2 bits (82), Expect = 0.63
Identities = 32/114 (28%), Positives = 58/114 (50%), Gaps = 7/114 (6%)
Query: 90 KEPIPVLDEDPNVDDCGIRLFRHSKPGIVFDHADEPQPPMKRPKLVPGEDIDEKSKKFRR 149
+E PV++E P V++ + G +D ++P+P + PK+ GE +D + +R
Sbjct: 22 QEEEPVVEEAPQVEEA-YEEAEEEEHGEEYDE-EKPKPKLTAPKIPDGEKVDFDDIQKKR 79
Query: 150 RIRSIAVDGNDLIAAANDAYKKSLARLEA-KDAAAKAKAKREEE---RIEKLKK 199
+ + + ++ LI +A KK + A KD K +A+R E+ R EK K+
Sbjct: 80 QNKDL-IELQSLIDTHFEARKKEEEDIIALKDRIEKRRAERAEQLRIRTEKEKE 132
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.311 0.131 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 392,108,974
Number of Sequences: 2790947
Number of extensions: 17304712
Number of successful extensions: 71182
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 126
Number of HSP's that attempted gapping in prelim test: 70952
Number of HSP's gapped (non-prelim): 324
length of query: 221
length of database: 848,049,833
effective HSP length: 123
effective length of query: 98
effective length of database: 504,763,352
effective search space: 49466808496
effective search space used: 49466808496
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC147000.5


