

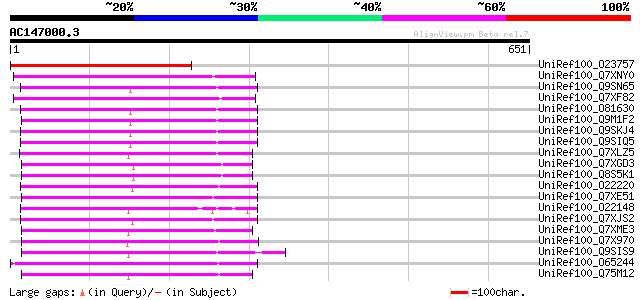
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.3 + phase: 0 /pseudo
(651 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O23757 Reverse transcriptase [Beta vulgaris subsp. vul... 251 5e-65
UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa] 216 1e-54
UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis tha... 215 3e-54
UniRef100_Q7XF82 Putative retroelement [Oryza sativa] 215 3e-54
UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana] 215 3e-54
UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis tha... 214 5e-54
UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcrip... 213 2e-53
UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcrip... 213 2e-53
UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa] 206 2e-51
UniRef100_Q7XGD3 Putative retroelement [Oryza sativa] 206 2e-51
UniRef100_Q8S5K1 Putative retroelement [Oryza sativa] 206 2e-51
UniRef100_O22220 Putative non-LTR retroelement reverse transcrip... 202 2e-50
UniRef100_Q7XE51 Putative non-LTR retroelement reverse transcrip... 201 7e-50
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 201 7e-50
UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcrip... 200 1e-49
UniRef100_Q7XME3 OSJNBa0061G20.16 protein [Oryza sativa] 200 1e-49
UniRef100_Q7X970 BZIP-like protein [Oryza sativa] 199 2e-49
UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcrip... 199 2e-49
UniRef100_O65244 F21E10.5 protein [Arabidopsis thaliana] 199 2e-49
UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa] 196 1e-48
>UniRef100_O23757 Reverse transcriptase [Beta vulgaris subsp. vulgaris]
Length = 507
Score = 251 bits (641), Expect = 5e-65
Identities = 115/227 (50%), Positives = 161/227 (70%)
Query: 2 LPFRKLSLREAGNLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQF 61
L F KLS E +L PF+HEE+ +AV CD+ K+PGP+G +F FIK W+++K D +
Sbjct: 281 LNFNKLSESECSSLIAPFTHEEIDEAVDSCDAQKAPGPDGFNFKFIKSSWEVIKKDIYKI 340
Query: 62 LVDFHRNGKLIKGVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIG 121
+ DF +G L G NS FIALIPK +P+ N++ PIS+VGCLYK++AK+LA RL+ V+
Sbjct: 341 VEDFWASGSLPHGCNSAFIALIPKTQHPRGFNEFRPISMVGCLYKIIAKILARRLQRVMD 400
Query: 122 SVVSDSQSAFIKGN*ILDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVM 181
++ QS+FIKG ILDG+LIA+E++D RRM E ++ ++DF KA+DS+ YL+ V+
Sbjct: 401 HLIGPYQSSFIKGRQILDGVLIASELIDSCRRMKNEAVVLKLDFHKAFDSISWNYLEWVL 460
Query: 182 VTMNFPTLWQKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLS 228
MNFP +W+ WI C+ +A AAVL+NG P F ++RGLRQGDPLS
Sbjct: 461 CQMNFPLVWRSWIRSCVMSASAAVLINGSPSSVFKLQRGLRQGDPLS 507
>UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa]
Length = 1026
Score = 216 bits (551), Expect = 1e-54
Identities = 113/303 (37%), Positives = 178/303 (58%), Gaps = 2/303 (0%)
Query: 6 KLSLREAGNLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDF 65
K++ +A L P + EE++ V+D KSPGP+G+ F FWDL+K D + + DF
Sbjct: 226 KVTEEDAIGLITPVTMEELRNVVFDLKKNKSPGPDGMPGEFYIHFWDLIKHDLLDLINDF 285
Query: 66 HRNGKLIKGVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVS 125
RN I +N I LIPK + ++ + PI L+ +K++ KVL NRL + ++S
Sbjct: 286 QRNSVNIDRLNFGVITLIPKTKDASQIQKFRPICLLNVSFKIITKVLMNRLNGTMAYIIS 345
Query: 126 DSQSAFIKGN*ILDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMN 185
Q+AF+K I++G++I +EV++ + + +LF+VDFEKAYD V+ ++ ++
Sbjct: 346 KQQTAFLKNRFIMEGVVILHEVLNSIHQKKQSGILFKVDFEKAYDKVNWVFIYRMLKAKG 405
Query: 186 FPTLWQKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQ 245
FP W WI + + K AV VN F +GLRQGDPLSP LF LAAE +L+++
Sbjct: 406 FPDQWCDWIMKVVMGGKVAVKVNDQIGSFFKTHKGLRQGDPLSPLLFNLAAEALTLLVQR 465
Query: 246 MAGAQLFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNF 305
L G +G G+ + LQ+ADDT+ + + ++++ +L LFE++SGLK+NF
Sbjct: 466 AEENSLIEG--LGTNGDNKIAILQYADDTIFLINDKLDHAKNLKYILCLFEQLSGLKINF 523
Query: 306 HKS 308
+KS
Sbjct: 524 NKS 526
>UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis thaliana]
Length = 1294
Score = 215 bits (548), Expect = 3e-54
Identities = 112/301 (37%), Positives = 176/301 (58%), Gaps = 4/301 (1%)
Query: 14 NLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIK 73
+LTK S E+ A+ K+PGP+G++ F K W++V D ++ + F R + +
Sbjct: 785 DLTKDLSDLEIYNAICHIGDDKAPGPDGLTARFYKSCWEIVGPDVIKEVKIFFRTSYMKQ 844
Query: 74 GVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIK 133
+N T I +IPK+ NP+ L+DY PI+L LYK+++K L RL+ + ++VSDSQ+AFI
Sbjct: 845 SINHTNICMIPKITNPETLSDYRPIALCNVLYKIISKCLVERLKGHLDAIVSDSQAAFIP 904
Query: 134 GN*ILDGILIANEVVDD---ARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLW 190
G + D ++IA+E++ +R+ + + + D KAYD V+ +L+ M F W
Sbjct: 905 GRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGFSETW 964
Query: 191 QKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQ 250
KWI + + +VLVNG P +RG+RQGDPLSP+LF+L A+ N L+K
Sbjct: 965 IKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHLIKNRVAEG 1024
Query: 251 LFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSML 310
G +G G +THLQFADD+L + + N ++++ V ++E SG K+N KSM+
Sbjct: 1025 DIRGIRIGN-GVPGVTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSGQKINMSKSMI 1083
Query: 311 T 311
T
Sbjct: 1084 T 1084
>UniRef100_Q7XF82 Putative retroelement [Oryza sativa]
Length = 586
Score = 215 bits (548), Expect = 3e-54
Identities = 108/303 (35%), Positives = 182/303 (59%), Gaps = 2/303 (0%)
Query: 6 KLSLREAGNLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDF 65
++S + LTKPFS EEVK+ V++ ++PGP+G F +FW ++K + + + DF
Sbjct: 6 QVSKEDKVELTKPFSLEEVKKVVFEMRQNRAPGPDGFPAEFYVKFWGIIKFELLDLINDF 65
Query: 66 HRNGKLIKGVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVS 125
H ++ +N + L+PK + +++ + PI L+ +K++ KVL NRL ++ ++S
Sbjct: 66 HNGFLEVERLNYGIVTLVPKTKDAEQIQKFRPICLLNVSFKIITKVLMNRLDRIMTYIIS 125
Query: 126 DSQSAFIKGN*ILDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMN 185
+Q+AF+K I++GI+I +E+++ +LF+VDFEKAYD VD ++ ++ +
Sbjct: 126 KNQTAFLKNRFIMEGIVILHELLNSLHTKKSSGILFKVDFEKAYDMVDWGFIYRILHSKG 185
Query: 186 FPTLWQKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQ 245
FP W W+ + + K A+ VN F+ +GLRQGDPLSP LF LAAE +L+++
Sbjct: 186 FPDQWCDWVMKVVRGGKVAIKVNDEVGPYFNTHKGLRQGDPLSPLLFNLAAEALTLLVQR 245
Query: 246 MAGAQLFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNF 305
L G + +V T LQ+ADDT+ + + R+++ +L LFE++SGLK+NF
Sbjct: 246 AENNSLVEGLPINENNKV--TILQYADDTIFMIKDDPEQARNLKFILCLFEQLSGLKINF 303
Query: 306 HKS 308
+KS
Sbjct: 304 NKS 306
>UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana]
Length = 1662
Score = 215 bits (548), Expect = 3e-54
Identities = 112/301 (37%), Positives = 176/301 (58%), Gaps = 4/301 (1%)
Query: 14 NLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIK 73
+LTK S E+ A+ K+PGP+G++ F K W++V D ++ + F R + +
Sbjct: 805 DLTKDLSDLEIYNAICHIGDDKAPGPDGLTARFYKSCWEIVGPDVIKEVKIFFRTSYMKQ 864
Query: 74 GVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIK 133
+N T I +IPK+ NP+ L+DY PI+L LYK+++K L RL+ + ++VSDSQ+AFI
Sbjct: 865 SINHTNICMIPKITNPETLSDYRPIALCNVLYKIISKCLVERLKGHLDAIVSDSQAAFIP 924
Query: 134 GN*ILDGILIANEVVDD---ARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLW 190
G + D ++IA+E++ +R+ + + + D KAYD V+ +L+ M F W
Sbjct: 925 GRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGFSETW 984
Query: 191 QKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQ 250
KWI + + +VLVNG P +RG+RQGDPLSP+LF+L A+ N L+K
Sbjct: 985 IKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHLIKNRVAEG 1044
Query: 251 LFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSML 310
G +G G +THLQFADD+L + + N ++++ V ++E SG K+N KSM+
Sbjct: 1045 DIRGIRIGN-GVPGVTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSGQKINMSKSMI 1103
Query: 311 T 311
T
Sbjct: 1104 T 1104
>UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis thaliana]
Length = 851
Score = 214 bits (546), Expect = 5e-54
Identities = 114/300 (38%), Positives = 178/300 (59%), Gaps = 4/300 (1%)
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
LT+ F E+ +A+ K+PGP+G++ F K+ WD+V +D ++ + F + +
Sbjct: 45 LTQDFRDSEIFEAICQIGDDKAPGPDGLTARFYKQCWDIVGNDVIKEVKLFFESSHMKTS 104
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
VN T I +IPK+ NPQ L+DY PI+L LYKV++K + NRL+ + S+VSDSQ+AFI G
Sbjct: 105 VNHTNICMIPKIQNPQTLSDYRPIALCNVLYKVISKCMVNRLKAHLNSIVSDSQAAFIPG 164
Query: 135 N*ILDGILIANEVVDD---ARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQ 191
I D ++IA+E++ +R+ K + + D KAYD V+ +L+ M F W
Sbjct: 165 RIINDNVMIAHEIMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCDKWI 224
Query: 192 KWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQL 251
WI + + +VL+NG P S RG+RQGDPLSP+LF+L + + L+K A +
Sbjct: 225 GWIMAAVKSVHYSVLINGSPHGYISPTRGIRQGDPLSPYLFILCGDILSHLIKVKASSGD 284
Query: 252 FNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSMLT 311
G +G G +THLQFADD+L + + N ++++ V ++E SG K+N KS++T
Sbjct: 285 IRGVRIGN-GAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSLIT 343
>UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1750
Score = 213 bits (541), Expect = 2e-53
Identities = 115/301 (38%), Positives = 174/301 (57%), Gaps = 4/301 (1%)
Query: 14 NLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIK 73
+LTK FS E+ A+ K+PGP+G++ F K WD+V D + + F +
Sbjct: 805 DLTKEFSDTEIYDAICQIGDDKAPGPDGLTARFYKNCWDIVGYDVILEVKKFFETSFMKP 864
Query: 74 GVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIK 133
+N T I +IPK+ NP L+DY PI+L LYKV++K L NRL++ + S+VSDSQ+AFI
Sbjct: 865 SINHTNICMIPKITNPTTLSDYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIP 924
Query: 134 GN*ILDGILIANEVVDD---ARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLW 190
G I D ++IA+EV+ +R+ K + + D KAYD V+ +L+ M F W
Sbjct: 925 GRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKW 984
Query: 191 QKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQ 250
WI + + +VL+NG P + RG+RQGDPLSP+LF+L + + L+ A +
Sbjct: 985 IGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASSG 1044
Query: 251 LFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSML 310
G +G G +THLQFADD+L + + N ++++ V ++E SG K+N KSM+
Sbjct: 1045 DLRGVRIGN-GAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMI 1103
Query: 311 T 311
T
Sbjct: 1104 T 1104
>UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1524
Score = 213 bits (541), Expect = 2e-53
Identities = 115/301 (38%), Positives = 174/301 (57%), Gaps = 4/301 (1%)
Query: 14 NLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIK 73
+LTK FS E+ A+ K+PGP+G++ F K WD+V D + + F +
Sbjct: 579 DLTKEFSDTEIYDAICQIGDDKAPGPDGLTARFYKNCWDIVGYDVILEVKKFFETSFMKP 638
Query: 74 GVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIK 133
+N T I +IPK+ NP L+DY PI+L LYKV++K L NRL++ + S+VSDSQ+AFI
Sbjct: 639 SINHTNICMIPKITNPTTLSDYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIP 698
Query: 134 GN*ILDGILIANEVVDD---ARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLW 190
G I D ++IA+EV+ +R+ K + + D KAYD V+ +L+ M F W
Sbjct: 699 GRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKW 758
Query: 191 QKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQ 250
WI + + +VL+NG P + RG+RQGDPLSP+LF+L + + L+ A +
Sbjct: 759 IGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASSG 818
Query: 251 LFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSML 310
G +G G +THLQFADD+L + + N ++++ V ++E SG K+N KSM+
Sbjct: 819 DLRGVRIGN-GAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMI 877
Query: 311 T 311
T
Sbjct: 878 T 878
>UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa]
Length = 1205
Score = 206 bits (524), Expect = 2e-51
Identities = 104/295 (35%), Positives = 171/295 (57%), Gaps = 4/295 (1%)
Query: 13 GNLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLI 72
G LTKPF+ EE+ A++ K+PGP+G F +R W ++K D ++ + +F G+
Sbjct: 410 GMLTKPFTDEEISDALFQIGPLKAPGPDGFPARFFQRNWGVLKRDVIEGVREFFETGEWK 469
Query: 73 KGVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFI 132
+G+N T I +IPK N P + D+ P+SL +YKV+AK L NRLR ++ ++S++QSAF+
Sbjct: 470 EGMNDTVIVMIPKTNAPVEMKDFRPVSLCNVIYKVVAKCLVNRLRPLLQEIISETQSAFV 529
Query: 133 KGN*ILDGILIANEV---VDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTL 189
G I D L+A E + R ++ ++D KAYD VD +LD + + F +
Sbjct: 530 PGRMITDNALVAFECFHSIHKCTRESQDFCALKLDLSKAYDRVDWGFLDGALQKLGFGNI 589
Query: 190 WQKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGA 249
W+KWI C+ + + +V +NG +E F RGLR+GDPL+P+LFL A+G + ++++
Sbjct: 590 WRKWIMSCVTSVRYSVRLNGNMLEPFYPTRGLREGDPLNPYLFLFIADGLSNILQRRRDE 649
Query: 250 QLFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVN 304
+ V R ++HL FADD+L+ + + ++ L L+E +G +N
Sbjct: 650 RQIQPLKVCR-SAPGVSHLLFADDSLLFFKAEVIQATRIKEALDLYERCTGQLIN 703
>UniRef100_Q7XGD3 Putative retroelement [Oryza sativa]
Length = 1694
Score = 206 bits (523), Expect = 2e-51
Identities = 114/293 (38%), Positives = 163/293 (54%), Gaps = 4/293 (1%)
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
L K FS EE+ A++ K+PGP+G F +R W ++KDD ++ + +F G + G
Sbjct: 1038 LCKTFSEEEIANALFQIGPLKAPGPDGFPGRFYQRNWAILKDDIVRAVQEFFSLGTMPSG 1097
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
VN T I LIPK PQ L D+ PISL +YK+++K L NRLR ++ +VS +QSAF+ G
Sbjct: 1098 VNETAIVLIPKTEQPQELKDFRPISLCNVVYKIVSKCLVNRLRPILDDLVSQNQSAFVPG 1157
Query: 135 N*ILDGILIANEVVDDARR---MDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQ 191
I D LIA E +R + +++D KAYD VD ++L+ MV + F W
Sbjct: 1158 RLITDNALIAFEYFHHIQRNKNPENAYSAYKLDLSKAYDRVDWEFLEQAMVKLGFAHCWV 1217
Query: 192 KWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQL 251
KWI CI + AV +NG + F+ RGLRQGDPLSPFLFL A+G + L+++
Sbjct: 1218 KWIMACITLVRYAVKLNGTLLNTFAPSRGLRQGDPLSPFLFLFVADGLSFLLEEKVDQGA 1277
Query: 252 FNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVN 304
N + R L HL FADDT + + + M+ VL + +G +N
Sbjct: 1278 MNPIHICRHAPAVL-HLLFADDTPLFFKAANDQAVVMKEVLETYASCTGQLIN 1329
>UniRef100_Q8S5K1 Putative retroelement [Oryza sativa]
Length = 1888
Score = 206 bits (523), Expect = 2e-51
Identities = 114/293 (38%), Positives = 163/293 (54%), Gaps = 4/293 (1%)
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
L K FS EE+ A++ K+PGP+G F +R W ++KDD ++ + +F G + G
Sbjct: 995 LCKTFSEEEIANALFQIGPLKAPGPDGFPGRFYQRNWAILKDDIVRAVQEFFSLGTMPSG 1054
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
VN T I LIPK PQ L D+ PISL +YK+++K L NRLR ++ +VS +QSAF+ G
Sbjct: 1055 VNETAIVLIPKTEQPQELKDFRPISLCNVVYKIVSKCLVNRLRPILDDLVSQNQSAFVPG 1114
Query: 135 N*ILDGILIANEVVDDARR---MDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQ 191
I D LIA E +R + +++D KAYD VD ++L+ MV + F W
Sbjct: 1115 RLITDNALIAFEYFHHIQRNKNPENAYSAYKLDLSKAYDRVDWEFLEQAMVKLGFAHCWV 1174
Query: 192 KWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQL 251
KWI CI + AV +NG + F+ RGLRQGDPLSPFLFL A+G + L+++
Sbjct: 1175 KWIMACITLVRYAVKLNGTLLNTFAPSRGLRQGDPLSPFLFLFVADGLSFLLEEKVDQGA 1234
Query: 252 FNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVN 304
N + R L HL FADDT + + + M+ VL + +G +N
Sbjct: 1235 MNPIHICRHAPAVL-HLLFADDTPLFFKAANDQAVVMKEVLETYASCTGQLIN 1286
>UniRef100_O22220 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1094
Score = 202 bits (515), Expect = 2e-50
Identities = 105/301 (34%), Positives = 174/301 (56%), Gaps = 4/301 (1%)
Query: 14 NLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIK 73
+LTK + +E+ +AV+ ++ +PGP+G + F +R W LVK+ + + F + G L +
Sbjct: 152 DLTKKVNEQEIYKAVFSINAESAPGPDGFTALFFQRQWPLVKNQIISDIELFFQTGILPE 211
Query: 74 GVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIK 133
N T + LIPK+ P R+ D PISL +YK+++K+L+ RL+ + +VS +QSAF+
Sbjct: 212 DWNHTHLCLIPKITKPARMADIRPISLCSVMYKIISKILSARLKKYLPVIVSPTQSAFVA 271
Query: 134 GN*ILDGILIANEVVDDAR---RMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLW 190
+ D I++A+E+V + R ++ K+ ++F+ D KAYD V+ +L +++ + F + W
Sbjct: 272 ERLVSDNIILAHEIVHNLRTNEKISKDFMVFKTDMSKAYDRVEWPFLKGILLALGFNSTW 331
Query: 191 QKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQ 250
W+ C+ + +VL+NG P + RGLRQGDPLSPFLF+L E ++ Q
Sbjct: 332 INWMMACVSSVSYSVLINGQPFGHITPHRGLRQGDPLSPFLFVLCTEALIHILNQAEKIG 391
Query: 251 LFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSML 310
+G G ++ HL FADDTL+I + S L + L + +SG +N KS +
Sbjct: 392 KISGIQFNGTGP-SVNHLLFADDTLLICKASQLECAEIMHCLSQYGHISGQMINSEKSAI 450
Query: 311 T 311
T
Sbjct: 451 T 451
>UniRef100_Q7XE51 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1652
Score = 201 bits (510), Expect = 7e-50
Identities = 105/296 (35%), Positives = 165/296 (55%), Gaps = 1/296 (0%)
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
L++ F EVK+A++ + K+PGP+G F + FWD++KDD M DFH +
Sbjct: 1143 LSESFKELEVKEAIFQMEHNKAPGPDGFPAEFYQVFWDVIKDDLMAVFCDFHEGTLPLHR 1202
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
+ I L+PK + R+ Y PI L+ +K+ KV+ANR+ V V+ SQ+ F+ G
Sbjct: 1203 LKFGIITLLPKQKDASRIQQYRPICLLNVSFKIFTKVMANRIALVAQKVIKPSQTTFLSG 1262
Query: 135 N*ILDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQKWI 194
I++G++I +E + + + K ++ ++DFEKAYD VD K+L + F + W WI
Sbjct: 1263 RNIMEGVVILHETLHELHKKKKNGVILKLDFEKAYDKVDWKFLQQSLRMKGFSSKWCDWI 1322
Query: 195 SECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFNG 254
+ AV VN F +GLRQGDPLSP LF L + +L+++ F G
Sbjct: 1323 DSIVRGGSVAVKVNDEIGSYFQTRKGLRQGDPLSPILFNLVVDMLAILIQRAKDQGRFKG 1382
Query: 255 FLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSML 310
+V + + L+ LQ+ADDT++ + R ++ VL FE++S LK+NF+KS L
Sbjct: 1383 -VVPHLVDNGLSILQYADDTILFMDHDLDEARDLKLVLSTFEKLSSLKINFYKSEL 1437
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 201 bits (510), Expect = 7e-50
Identities = 110/306 (35%), Positives = 183/306 (58%), Gaps = 16/306 (5%)
Query: 14 NLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIK 73
NL P + EEV++A + + K PGP+G++ ++FW+ + D + + F R+G + +
Sbjct: 423 NLLAPITKEEVQRATFSINPHKCPGPDGMNGFLYQQFWETMGDQITEMVQAFFRSGSIEE 482
Query: 74 GVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIK 133
G+N T I LIPK+ +++ D+ PISL +YKV+ K++ANRL+ ++ S++S++Q+AF+K
Sbjct: 483 GMNKTNICLIPKILKAEKMTDFRPISLCNVIYKVIGKLMANRLKKILPSLISETQAAFVK 542
Query: 134 GN*ILDGILIANEV---VDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLW 190
G I D ILIA+E+ + + +E + + D KAYD V+ +L+ M + F W
Sbjct: 543 GRLISDNILIAHELLHALSSNNKCSEEFIAIKTDISKAYDRVEWPFLEKAMRGLGFADHW 602
Query: 191 QKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQ 250
+ I EC+ + + VL+NG P E RGLRQGDPLSP+LF++ E +L+K + A+
Sbjct: 603 IRLIMECVKSVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLFVICTE---MLVKMLQSAE 659
Query: 251 LFN---GFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEE---VSGLKVN 304
N G V R G ++HL FADD++ + +N ++ ++ + EE SG +VN
Sbjct: 660 QKNQITGLKVAR-GAPPISHLLFADDSMFYCK---VNDEALGQIIRIIEEYSLASGQRVN 715
Query: 305 FHKSML 310
+ KS +
Sbjct: 716 YLKSSI 721
>UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1344
Score = 200 bits (509), Expect = 1e-49
Identities = 108/301 (35%), Positives = 171/301 (55%), Gaps = 4/301 (1%)
Query: 14 NLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIK 73
NL + + EV AV+ + +PGP+G + F ++ WDLVK + + F G L +
Sbjct: 401 NLIQEVTELEVYNAVFSINKESAPGPDGFTALFFQQHWDLVKHQILTEIFGFFETGVLPQ 460
Query: 74 GVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIK 133
N T I LIPK+ +PQR++D PISL LYK+++K+L RL+ + ++VS +QSAF+
Sbjct: 461 DWNHTHICLIPKITSPQRMSDLRPISLCSVLYKIISKILTQRLKKHLPAIVSTTQSAFVP 520
Query: 134 GN*ILDGILIANEVVDDAR---RMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLW 190
I D IL+A+E++ R R+ KE + F+ D KAYD V+ +L+ +M + F W
Sbjct: 521 QRLISDNILVAHEMIHSLRTNDRISKEHMAFKTDMSKAYDRVEWPFLETMMTALGFNNKW 580
Query: 191 QKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQ 250
WI C+ + +VL+NG P RG+RQGDPLSP LF+L E ++ + A
Sbjct: 581 ISWIMNCVTSVSYSVLINGQPYGHIIPTRGIRQGDPLSPALFVLCTEALIHILNKAEQAG 640
Query: 251 LFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSML 310
G + + +V++ HL FADDTL++ + + + L + ++SG +N +KS +
Sbjct: 641 KITG-IQFQDKKVSVNHLLFADDTLLMCKATKQECEELMQCLSQYGQLSGQMINLNKSAI 699
Query: 311 T 311
T
Sbjct: 700 T 700
>UniRef100_Q7XME3 OSJNBa0061G20.16 protein [Oryza sativa]
Length = 1285
Score = 200 bits (508), Expect = 1e-49
Identities = 107/293 (36%), Positives = 164/293 (55%), Gaps = 4/293 (1%)
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
L + F EE+ QA++ KSPGP+G F +R W +K D + + +F ++G + +G
Sbjct: 976 LCQEFKEEEIAQAIFQIGPLKSPGPDGFPARFYQRNWGTLKSDIILAVRNFFQSGLMPEG 1035
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
VN T I LIPK + P L DY PISL +YKV++K L NRLR ++ +VS QSAFI+G
Sbjct: 1036 VNDTAIVLIPKKDQPIDLKDYRPISLCNVVYKVVSKCLVNRLRPILDDLVSKEQSAFIQG 1095
Query: 135 N*ILDGILIANEV---VDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQ 191
I D L+A E + ++ + +++D KAYD VD ++L++ + + F W
Sbjct: 1096 RMITDNALLAFECFHSIQKNKKANSAACAYKLDLSKAYDRVDWRFLELALNKLGFAHRWV 1155
Query: 192 KWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQL 251
WI C+ T + +V NG + F+ RGLRQGDPLSPFLFL A+G ++L+K+
Sbjct: 1156 SWIMSCVTTVRYSVKFNGTLLRSFAPTRGLRQGDPLSPFLFLFVADGLSLLLKEKVAQNS 1215
Query: 252 FNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVN 304
+ ++HL FADDTL+ + ++ VL + + +G +N
Sbjct: 1216 LTPLKI-CCQAPGISHLLFADDTLLFFKAEKKEAEVVKEVLTNYAQGTGQLIN 1267
>UniRef100_Q7X970 BZIP-like protein [Oryza sativa]
Length = 2367
Score = 199 bits (507), Expect = 2e-49
Identities = 111/302 (36%), Positives = 168/302 (54%), Gaps = 5/302 (1%)
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
L + F+ +E+ QA++ KSPGP+G F +R W +K D + + F + G + +G
Sbjct: 1158 LCEDFTEDEISQAIFQIGPLKSPGPDGFPARFYQRNWGTIKADIIGAVRRFFQTGLMPEG 1217
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
VN T I LIPK P L D+ PISL +YKV++K L NRLR ++ +VS QSAF++G
Sbjct: 1218 VNDTAIVLIPKKEQPVDLRDFRPISLCNVVYKVVSKCLVNRLRPILDDLVSVEQSAFVQG 1277
Query: 135 N*ILDGILIANE---VVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQ 191
I D L+A E + ++ + +++D KAYD VD ++L+M M + F W
Sbjct: 1278 RMITDNALLAFECFHAMQKNKKANHAACAYKLDLSKAYDRVDWRFLEMAMNKLGFARRWV 1337
Query: 192 KWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQL 251
WI +C+ + + V NG ++ F+ RGLRQGDPL PFLFL A+G ++L+K+
Sbjct: 1338 NWIMKCVTSVRYMVKFNGTLLQSFAPTRGLRQGDPLLPFLFLFVADGLSLLLKEKVAQNS 1397
Query: 252 FNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHK-SML 310
F V R ++HL FADDTL+ + ++ VL + +G +N K S+L
Sbjct: 1398 LTPFKVCRAAP-GISHLLFADDTLLFFKAHQREAEVVKEVLSSYAMGTGQLINPAKCSIL 1456
Query: 311 TG 312
G
Sbjct: 1457 MG 1458
>UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1715
Score = 199 bits (506), Expect = 2e-49
Identities = 111/335 (33%), Positives = 182/335 (54%), Gaps = 12/335 (3%)
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
L + + +EV+ AV+ + ++PG +G + F WDL+ +D + F + +
Sbjct: 786 LLQSVTDQEVRDAVFAIGADRAPGFDGFTAAFYHHLWDLIGNDVCLMVRHFFESDVMDNQ 845
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
+N T I LIPK+ +P+ ++DY PISL YK+++K+L RL+ +G V+SDSQ+AF+ G
Sbjct: 846 INQTQICLIPKIIDPKHMSDYRPISLCTASYKIISKILIKRLKQCLGDVISDSQAAFVPG 905
Query: 135 N*ILDGILIANEV---VDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQ 191
I D +L+A+E+ + R + + D KAYD V+ +L+ VM+ + F W
Sbjct: 906 QNISDNVLVAHELLHSLKSRRECQSGYVAVKTDISKAYDRVEWNFLEKVMIQLGFAPRWV 965
Query: 192 KWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQL 251
KWI C+ + VL+NG P + RG+RQGDPLSP+LFL AE + ++++ +
Sbjct: 966 KWIMTCVTSVSYEVLINGSPYGKIFPSRGIRQGDPLSPYLFLFCAEVLSNMLRKAEVNKQ 1025
Query: 252 FNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSMLT 311
+G + + + ++HL FADD+L S N+ + + +EE SG K+N+ KS
Sbjct: 1026 IHGMKITK-DCLAISHLLFADDSLFFCRASNQNIEQLALIFKKYEEASGQKINYAKS--- 1081
Query: 312 G*IYLSLGCGICIELSKREASFCLFGLTYWRGFSK 346
S+ G I +R+ L G+ RG K
Sbjct: 1082 -----SIIFGQKIPTMRRQRLHRLLGIDNVRGGGK 1111
>UniRef100_O65244 F21E10.5 protein [Arabidopsis thaliana]
Length = 928
Score = 199 bits (506), Expect = 2e-49
Identities = 115/310 (37%), Positives = 178/310 (57%), Gaps = 4/310 (1%)
Query: 2 LPFRKLSLREAGNLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQF 61
LP+R S + LT S EE+ + V+ + KSPGP+G + F K W+++ +F+
Sbjct: 325 LPYR-CSDSDKEMLTNHVSAEEIHKVVFSMPNDKSPGPDGYTAEFYKGAWNIIGAEFILA 383
Query: 62 LVDFHRNGKLIKGVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIG 121
+ F G L KG+NST +ALIPK + + DY PIS LYKV++K++ANRL+ V+
Sbjct: 384 IQSFFAKGFLPKGINSTILALIPKKKEAKEMKDYRPISCCNVLYKVISKIIANRLKLVLP 443
Query: 122 SVVSDSQSAFIKGN*ILDGILIANEVVDDARRMD-KELLLFRVDFEKAYDSVDLKYLDMV 180
+ +QSAF+K +++ +L+A E+V D + ++D KA+DSV K+L V
Sbjct: 444 KFIVGNQSAFVKDRLLIENVLLATEIVKDYHKDSVSSRCALKIDISKAFDSVQWKFLINV 503
Query: 181 MVTMNFPTLWQKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFN 240
+ MNFP + WI+ CI TA +V VNG FS R LRQG LSP+LF+++ + +
Sbjct: 504 LEAMNFPPEFTHWITLCITTASFSVQVNGELAGVFSSARELRQGCSLSPYLFVISMDVLS 563
Query: 241 VLMKQMAGAQLFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSG 300
++ + GA+ F R + LTHL FADD +I+ + ++ + VL F + SG
Sbjct: 564 KMLDKAVGARQFGYHPKCRA--IGLTHLSFADDLMILSDGKVRSIDGIVKVLYEFAKWSG 621
Query: 301 LKVNFHKSML 310
LK++ KS +
Sbjct: 622 LKISMEKSTM 631
>UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa]
Length = 1936
Score = 196 bits (499), Expect = 1e-48
Identities = 106/293 (36%), Positives = 164/293 (55%), Gaps = 4/293 (1%)
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
L + F EE+ QA++ KSP P+G F +R W +K D + + +F ++G + KG
Sbjct: 1064 LCQEFKEEEIAQAIFQIGPLKSPRPDGFPARFYQRNWGTLKSDIILAVRNFFQSGLMPKG 1123
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
VN T I LIPK + P L DY PISL +YKV++K L NRLR ++ +VS QSAFI+G
Sbjct: 1124 VNDTAIVLIPKKDQPIDLKDYRPISLCNVVYKVVSKCLVNRLRPILDDLVSKEQSAFIQG 1183
Query: 135 N*ILDGILIANEV---VDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQ 191
I D L+A E + ++ + +++D KAYD VD ++L++ + + F W
Sbjct: 1184 RMITDNALLAFECFHSIQKNKKANSAACAYKLDLSKAYDRVDWRFLELALNKLGFAHRWV 1243
Query: 192 KWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQL 251
WI C+ T + +V NG + F+ RGLRQG+PLSPFLFL A+G ++L+K+
Sbjct: 1244 SWIMLCVTTVRYSVKFNGTLLRSFAPTRGLRQGEPLSPFLFLFVADGLSLLLKEKVAQNS 1303
Query: 252 FNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVN 304
+ R +++L FADDTL+ + ++ VL + + +G +N
Sbjct: 1304 LTPLKICRQAP-GISYLLFADDTLLFFKAEKKEAEVVKEVLTNYAQGTGQLIN 1355
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.344 0.156 0.540
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,027,038,503
Number of Sequences: 2790947
Number of extensions: 42633013
Number of successful extensions: 181566
Number of sequences better than 10.0: 945
Number of HSP's better than 10.0 without gapping: 608
Number of HSP's successfully gapped in prelim test: 337
Number of HSP's that attempted gapping in prelim test: 179594
Number of HSP's gapped (non-prelim): 1209
length of query: 651
length of database: 848,049,833
effective HSP length: 134
effective length of query: 517
effective length of database: 474,062,935
effective search space: 245090537395
effective search space used: 245090537395
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC147000.3


