

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
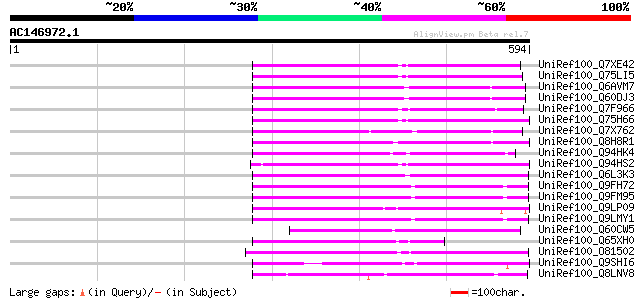
Query= AC146972.1 + phase: 0 /pseudo
(594 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XE42 Putative mutator-like transposase [Oryza sativa] 188 3e-46
UniRef100_Q75LI5 Putative transposon protein [Oryza sativa] 183 1e-44
UniRef100_Q6AVM7 Putative polyprotein [Oryza sativa] 177 1e-42
UniRef100_Q60DJ3 MuDR family transposase protein [Oryza sativa] 176 2e-42
UniRef100_Q7F966 OSJNBa0091C07.2 protein [Oryza sativa] 174 7e-42
UniRef100_Q75H66 Hypothetical protein OSJNBb0007E22.22 [Oryza sa... 172 2e-41
UniRef100_Q7X762 OSJNBb0118P14.3 protein [Oryza sativa] 168 5e-40
UniRef100_Q8H8R1 Putative mutator-like transposase [Oryza sativa] 157 6e-37
UniRef100_Q94HK4 Mutator-like transposase [Oryza sativa] 153 1e-35
UniRef100_Q94HS2 Putative transposon protein [Oryza sativa] 153 2e-35
UniRef100_Q6L3K3 Putative transposon MuDR mudrA-like protein [So... 148 4e-34
UniRef100_Q9FH72 Arabidopsis thaliana genomic DNA, chromosome 5,... 137 9e-31
UniRef100_Q9FM95 Similarity to mutator-like transposase [Arabido... 137 1e-30
UniRef100_Q9LP09 F9C16.9 [Arabidopsis thaliana] 137 1e-30
UniRef100_Q9LMY1 F21F23.10 protein [Arabidopsis thaliana] 136 2e-30
UniRef100_Q60CW5 Putative mutator transposable element-related p... 128 5e-28
UniRef100_Q65XH0 Hypothetical protein OJ1126_B11.9 [Oryza sativa] 125 3e-27
UniRef100_O81502 F9D12.2 protein [Arabidopsis thaliana] 120 1e-25
UniRef100_Q9SHI6 Similar to mudrA protein [Arabidopsis thaliana] 118 4e-25
UniRef100_Q8LNV8 Putative mutator protein [Oryza sativa] 117 1e-24
>UniRef100_Q7XE42 Putative mutator-like transposase [Oryza sativa]
Length = 1005
Score = 188 bits (478), Expect = 3e-46
Identities = 98/307 (31%), Positives = 161/307 (51%), Gaps = 4/307 (1%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+GL+ + + P HR C H+ N K+ + +E ++ + A+ FN ++
Sbjct: 552 KGLLSIVSTLFPFAEHRMCARHIYANWRKKHRLQEYQKRFWKIAKAPNEQLFNHYKRKLA 611
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
+ + W+ L K W +A+F ++ NN E FNS I+ R PI+TM E+IR
Sbjct: 612 AKTPRGWQDLEKTNPIHWSRAWFRLGSNCESVDNNMSESFNSWIIESRFKPIITMLEDIR 671
Query: 398 CYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRI 457
V R++ +N+ + +CP + K + + LWNGD + + + +
Sbjct: 672 IQVTRRIQENRSNSERWTMTVCPNIIRKFNKIRHRTQFCHVLWNGDAG---FEVRDK-KW 727
Query: 458 KVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQP 517
+ VD+ +TC+CR+WQ++ +PC HAC+AL + + H +L Y++TY + +QP
Sbjct: 728 RFTVDLTSKTCSCRYWQVSGIPCQHACAALFKMAQEPNNCIHECFSLERYKKTYQHVLQP 787
Query: 518 MNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGLM 577
+ W +P KP+PP+VK+ GRPKKNRRKD ++ G+ K I+C RCG
Sbjct: 788 VEHESAWPVSPNPKPLPPRVKKMPGRPKKNRRKDPSKPVKSGTKSSKVGTKIRCRRCGNY 847
Query: 578 GHNSRSC 584
GHNSR+C
Sbjct: 848 GHNSRTC 854
>UniRef100_Q75LI5 Putative transposon protein [Oryza sativa]
Length = 839
Score = 183 bits (464), Expect = 1e-44
Identities = 94/310 (30%), Positives = 156/310 (50%), Gaps = 4/310 (1%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+GL+ ++ + P HR C H+ N K+ + +E ++ + AR + FN ++
Sbjct: 437 KGLLSVIEHLFPKAEHRMCARHIYANWRKRHRLQEYQKRFWKIARSSNAVLFNHYKSKLA 496
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
+ WE L K W +A+F ++ NN CE FN+ I+ R PI+TM E+IR
Sbjct: 497 NKTPMGWEDLEKTNPIHWCRAWFKLGSNCDSVENNICESFNNWIIEARFKPIITMLEDIR 556
Query: 398 CYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRI 457
V R++ +NK + +CP ++ K + A+ LWNG + + + +
Sbjct: 557 MKVTRRIQENKTNSERWTMGICPNILKKINKIRHATQFCHVLWNGSSG---FEVREK-KW 612
Query: 458 KVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQP 517
+ VD+ TC+CR+WQ++ +PC HAC+A + +H + ++ YR TY +QP
Sbjct: 613 RFTVDLSANTCSCRYWQISGIPCQHACAAYFKMAEEPNNHVNMCFSIDQYRNTYQDVLQP 672
Query: 518 MNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGLM 577
+ W + +P+PP+VK+ G PK+ RRKD E + K ++CG C
Sbjct: 673 VEHESVWPLSTNPRPLPPRVKKMPGSPKRARRKDPTEAAGSSTKSSKRGGSVKCGFCHEK 732
Query: 578 GHNSRSCAKQ 587
GHNSR C K+
Sbjct: 733 GHNSRGCKKK 742
>UniRef100_Q6AVM7 Putative polyprotein [Oryza sativa]
Length = 1006
Score = 177 bits (448), Expect = 1e-42
Identities = 112/318 (35%), Positives = 166/318 (51%), Gaps = 10/318 (3%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+GLIPA+Q+V P HR+CV HL N Q+K + + + CAR ++ ++N+ M+ ++
Sbjct: 558 KGLIPAVQQVFPESEHRFCVRHLYSNFQLQFKGEVPKNQLWACARSSSVQEWNKNMDVMR 617
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
++N+ A+E+L K P W++A+F+E K ++NN CEVFN IL R PILTM E+I+
Sbjct: 618 NLNKSAYEWLEKLPPNTWVRAFFSEFPKCDILLNNNCEVFNKYILEARELPILTMLEKIK 677
Query: 398 CYVM-RKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCR 456
+M R ++ K D G +CP + ++ K A++ L G Q + E
Sbjct: 678 GQLMTRHFNKQKELADQFQGLICPKIRKKVLKNADAANTCYALPAGQGIFQVHERE---- 733
Query: 457 IKVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQ 516
+ VDI C CR W LT +PC HA S L E T ++ + Y + I
Sbjct: 734 YQYIVDINAMHCDCRRWDLTGIPCNHAISCLRHERINAESILPNCYTTDAFSKAYGFNIW 793
Query: 517 PMNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDI--QCGRC 574
P N WE + PP ++ AGRPKK+RRK E IG + K T + + C C
Sbjct: 794 PCNDKSKWENINGPEIKPPVYEKKAGRPKKSRRK-APYEVIGKNGPKLTKHGVMMHCKYC 852
Query: 575 GLMGHNSRSC--AKQGVA 590
G HNS C KQG++
Sbjct: 853 GEENHNSGGCKLKKQGIS 870
>UniRef100_Q60DJ3 MuDR family transposase protein [Oryza sativa]
Length = 1030
Score = 176 bits (446), Expect = 2e-42
Identities = 112/318 (35%), Positives = 166/318 (51%), Gaps = 10/318 (3%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+GLIPA+Q+V P HR+CV HL N Q+K + L+ + CAR ++ ++N+ M+ ++
Sbjct: 582 KGLIPAVQQVFPESEHRFCVRHLYSNFQLQFKGEVLKNQLWACARSSSVQEWNKNMDVMR 641
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
++N+ A+E+L K P W++A+F+E K ++NN CEVFN IL R PILTM E+I+
Sbjct: 642 NLNKSAYEWLEKLPPNTWVRAFFSEFPKCDILLNNNCEVFNKYILEARELPILTMLEKIK 701
Query: 398 CYVM-RKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCR 456
+M R ++ K D G +CP + ++ K A++ L G Q + E
Sbjct: 702 GQLMTRHFNKQKELADQFQGLICPKIRKKVLKNADAANTCYALPAGQGIFQVHERE---- 757
Query: 457 IKVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQ 516
+ VDI C CR W LT +PC HA S L E T ++ + Y + I
Sbjct: 758 YQYIVDINAMYCDCRRWDLTGIPCNHAISCLRHERINAESILPNCYTTDAFSKAYGFNIW 817
Query: 517 PMNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQC--GRC 574
P N WE + PP ++ AGRPKK+RRK E IG + K T + + C
Sbjct: 818 PCNDKSKWENVNGPEIKPPVYEKKAGRPKKSRRK-APYEVIGKNGPKLTKHGVMMHYKYC 876
Query: 575 GLMGHNSRSC--AKQGVA 590
G HNS C KQG++
Sbjct: 877 GEENHNSGGCKLKKQGIS 894
>UniRef100_Q7F966 OSJNBa0091C07.2 protein [Oryza sativa]
Length = 879
Score = 174 bits (441), Expect = 7e-42
Identities = 94/311 (30%), Positives = 160/311 (51%), Gaps = 5/311 (1%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+GL+ A++E +P + HR C H+ N K+++++ ++ +CA+ + FN ++
Sbjct: 513 KGLVSAVEEFLPQIEHRMCTRHIYANWRKKYRDQAFQKPFWKCAKASCRPFFNFCRAKLA 572
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
+ + W +A+F ++ NN CE FN+ I++ R HPI++M E IR
Sbjct: 573 QLTPAGAKXXXSTEPMHWSRAWFRIGSNCDSVDNNMCESFNNWIIDIRAHPIISMFEGIR 632
Query: 398 CYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRI 457
V ++ QN+ K G G +CP +L K S N +WNG + + + + +
Sbjct: 633 TKVYVRIQQNRSKAKGWLGRICPNILKKLNKYIDLSGNCEAIWNGKDG---FEVTDKDK- 688
Query: 458 KVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQP 517
+ VD+ K+TC+CR+WQL +PC HA +AL + + ED+ ++ Y + Y + + P
Sbjct: 689 RYTVDLEKRTCSCRYWQLAGIPCAHAITALFVSSKQPEDYIADCYSVEVYNKIYDHCMMP 748
Query: 518 MNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGLM 577
M + W T + KP PP + GRP+K RR+D E G + KT +C +C
Sbjct: 749 MEGMMQWPITGHPKPGPPGYVKMPGRPRKERRRDPLEAK-KGKKLSKTGTKGRCSQCKQT 807
Query: 578 GHNSRSCAKQG 588
HN R+C +G
Sbjct: 808 THNIRTCPMKG 818
>UniRef100_Q75H66 Hypothetical protein OSJNBb0007E22.22 [Oryza sativa]
Length = 981
Score = 172 bits (436), Expect = 2e-41
Identities = 99/318 (31%), Positives = 162/318 (50%), Gaps = 5/318 (1%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+GLIPA+Q++ P HR+CV HL +N + +K + L+ + CAR ++ ++N E +K
Sbjct: 547 KGLIPAVQQLFPDSEHRFCVRHLYQNFQQSFKGEILKNQLWACARSSSVQEWNTKFEEMK 606
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
++N+ A+ +L + W++A+F++ K ++NN+CEVFN IL R PILTM E+I+
Sbjct: 607 ALNEDAYNWLEQMAPNTWVRAFFSDFPKCDILLNNSCEVFNKYILEAREMPILTMLEKIK 666
Query: 398 CYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRI 457
+M + + + GP+CP + +L K ++ L G + +E
Sbjct: 667 GQLMTRFFNKQKEAQKWQGPICPKIRKKLLKIAEQANICYVLPAGKGV---FQVEER-GT 722
Query: 458 KVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQP 517
K VD+ + C CR W LT +PC HA + + ED ++ +++ Y+ I P
Sbjct: 723 KYIVDVVTKHCDCRRWDLTGIPCCHAIACIREDHLSEEDFLPHCYSINAFKAVYAENIIP 782
Query: 518 MNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRRKDGNE-EPIGGSSMKKTYNDIQCGRCGL 576
N WE + +PP ++ GRPKK+RRK E + G + K I C C
Sbjct: 783 CNDKANWEKMNGPQILPPVYEKKVGRPKKSRRKQPQEVQGRNGPKLTKHGVTIHCSYCHE 842
Query: 577 MGHNSRSCAKQGVARRPK 594
HN + C + RPK
Sbjct: 843 ANHNKKGCELRKKGIRPK 860
>UniRef100_Q7X762 OSJNBb0118P14.3 protein [Oryza sativa]
Length = 939
Score = 168 bits (425), Expect = 5e-40
Identities = 103/312 (33%), Positives = 164/312 (52%), Gaps = 8/312 (2%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+GL+PA++ R+CV HL +N K + L+ + AR +T ++N ME++K
Sbjct: 517 KGLVPAVRREFSDAEQRFCVRHLYQNFQVLHKGETLKNQLWAIARSSTVPEWNANMEKMK 576
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
+++ +A++YL + P W +A+F++ K ++NN EVFN IL+ R PIL+M E IR
Sbjct: 577 ALSSEAYKYLEEIPPNQWCRAFFSDFPKCDILLNNNSEVFNKYILDAREMPILSMLERIR 636
Query: 398 CYVMRKMSQNKMKLDGRAGP--LCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNC 455
+M ++ + +L+ R P +CP + ++EK ++ G A Q I +
Sbjct: 637 NQIMNRLYTKQKELE-RNWPCGICPKIKRKVEKNTEMANTCYVFPAGMGAFQVSDIGSQY 695
Query: 456 RIKVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFI 515
++++V + C CR WQLT +PC HA S L K ED + Y + YSY I
Sbjct: 696 IVELNV----KRCDCRRWQLTGIPCNHAISCLRHERIKPEDVVSFCYSTRCYEQAYSYNI 751
Query: 516 QPMNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCG 575
P+ I+WE + PP ++ GRPKK RRK +E GG+ + K ++ C C
Sbjct: 752 MPLRDSIHWEKMQGIEVKPPVYEKKVGRPKKTRRKQ-PQELEGGTKISKHGVEMHCSYCK 810
Query: 576 LMGHNSRSCAKQ 587
GHN SC K+
Sbjct: 811 NGGHNKTSCKKR 822
>UniRef100_Q8H8R1 Putative mutator-like transposase [Oryza sativa]
Length = 746
Score = 157 bits (398), Expect = 6e-37
Identities = 98/317 (30%), Positives = 155/317 (47%), Gaps = 5/317 (1%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+GLIPA+++ P HR+CV HL +N +K + L+ + AR TT ++N E++K
Sbjct: 393 EGLIPAIKDEFPDSEHRFCVRHLYQNFAVLYKGEALKNQLWAIARSTTVPEWNVNTEKMK 452
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
++N+ A+ YL + P W +A+F + K ++NN E IL R IL+M E+IR
Sbjct: 453 AVNKDAYGYLEEIPPNQWCRAFFRDFSKCDILLNNNLECHVRYILEARELTILSMLEKIR 512
Query: 398 CYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRI 457
+M ++ + + +CP + ++EK S+ L +R +
Sbjct: 513 SKLMNRIYTKQEECKKWVFDICPKIKQKVEKNIEMSNTCYAL----PSRMGVFQVTDRDK 568
Query: 458 KVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQP 517
+ VDI + C CR WQL +PC HA S L K ED T+ S+++ Y + I P
Sbjct: 569 QFVVDIKNKQCDCRRWQLIGIPCNHAISCLRHERIKPEDEVSFCYTIQSFKQAYMFNIMP 628
Query: 518 MNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGLM 577
+ +WE PP ++ GRPK RRK +E G+ + K I C C +
Sbjct: 629 VRDKTHWEKMNGVPVNPPVYEKKVGRPKTTRRKQ-PQELDAGTKISKHGVQIHCSYCKNV 687
Query: 578 GHNSRSCAKQGVARRPK 594
GHN + C K+ A+ K
Sbjct: 688 GHNKKGCKKRKAAQNGK 704
>UniRef100_Q94HK4 Mutator-like transposase [Oryza sativa]
Length = 725
Score = 153 bits (387), Expect = 1e-35
Identities = 103/303 (33%), Positives = 145/303 (46%), Gaps = 9/303 (2%)
Query: 279 GLIPAMQEVMPGVPHRYCVMHL*KNC-TKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
GL+ A+ V P HRYC MHL +N K W+ ++ + V TT +++ ME +K
Sbjct: 411 GLMNAIPIVFPDSEHRYCKMHLLQNMGNKGWRGEKYKGFVDAAIYATTVWDYDKAMEDLK 470
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
+N KAWE+L KE + + F+ K+ +VNN EVFN IL+ R PI+TM E IR
Sbjct: 471 KLNLKAWEWLIAIGKEHFSRHAFSPKAKSDLVVNNLSEVFNKYILDARDKPIVTMVEHIR 530
Query: 398 CYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEK-IASHNWTPLWNGDNARQRYHIENNCR 456
VM ++S + D + P +LE EK A + W + + H +
Sbjct: 531 RKVMVRLSLKRQGGDAAQWEITPIVAGKLEMEKNHARYCW--CYQSNLTTWEVHCLDR-- 586
Query: 457 IKVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQ 516
VDI +TC C WQLT +PC H AL G ED+ + +Y TY+ I
Sbjct: 587 -SFAVDISARTCACHKWQLTGIPCKHDVCALYKAGHTPEDYVADYFRKDAYMRTYTAVIY 645
Query: 517 PMNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGL 576
P+ W T PPK + GRPKK+RR+ +E P +KT + C C
Sbjct: 646 PVPDEHRWTKTDSPYIDPPKFDKHVGRPKKSRRRGPDEGPRVQGPARKTTS--TCSNCRR 703
Query: 577 MGH 579
GH
Sbjct: 704 DGH 706
>UniRef100_Q94HS2 Putative transposon protein [Oryza sativa]
Length = 841
Score = 153 bits (386), Expect = 2e-35
Identities = 98/321 (30%), Positives = 157/321 (48%), Gaps = 7/321 (2%)
Query: 276 NWQGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMER 335
+W+ + ++E + G+ + Y + +K + L+ + CAR ++ ++N ME
Sbjct: 400 SWRWFLQTLKEDL-GIDNTYPWTIMTDKQKVHFKGENLKNQLWACARSSSEVEWNANMEE 458
Query: 336 VKSINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEE 395
+KS+NQ A+E+L K P + W+KAYF+E K ++NN CEVFN IL R PIL+M E+
Sbjct: 459 MKSLNQDAYEWLQKMPPKTWVKAYFSEFPKCDILLNNNCEVFNKYILEARELPILSMFEK 518
Query: 396 IRC-YVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENN 454
I+ + R S+ K + GP+CP + ++ K ++ L G + +E+
Sbjct: 519 IKSQLISRHYSKQKEVAEQWHGPICPKIRKKVLKNADMANTCYVLPAGKGI---FQVEDR 575
Query: 455 CRIKVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYF 514
K VD+ + C CR W LT +PC HA S L E +L ++ Y++
Sbjct: 576 -NFKYIVDLSAKHCDCRRWDLTGIPCNHAISCLRSERISAESILPPCYSLEAFSRAYAFN 634
Query: 515 IQPMNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRRKDGNE-EPIGGSSMKKTYNDIQCGR 573
I P N W + PP ++ GRP K+RRK +E + G M K ++ C
Sbjct: 635 IWPYNDMTKWVQVNGPEVKPPIYEKKVGRPPKSRRKAPHEVQGKNGPKMSKHGVEMHCSF 694
Query: 574 CGLMGHNSRSCAKQGVARRPK 594
C HN + C + RPK
Sbjct: 695 CKEPRHNKKGCPLRKARLRPK 715
>UniRef100_Q6L3K3 Putative transposon MuDR mudrA-like protein [Solanum demissum]
Length = 873
Score = 148 bits (374), Expect = 4e-34
Identities = 89/318 (27%), Positives = 152/318 (46%), Gaps = 6/318 (1%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+GL+ A+ +V P HR+C H+ N +K WK ++R+++ A T +F+ ++ +
Sbjct: 409 KGLLDAVSQVFPKAHHRWCARHIEANWSKAWKGVQMRKLLWWSAWSTYEEEFHDQLKVMG 468
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
+++++A + L +P + W +AYF+ CK ++ NN E FN IL R PI+ M E IR
Sbjct: 469 AVSKQAAKDLVWYPAQNWCRAYFDTVCKNHSCENNFTESFNKWILEARAKPIIKMLENIR 528
Query: 398 CYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRI 457
+M ++ + + + G P+ I + NGD + E+
Sbjct: 529 IKIMNRLQKLEEEGKNWKGDFSPYAMELYNDFNIIAQCCQVQSNGDQGYEVVEGED---- 584
Query: 458 KVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQP 517
+ V++ ++ CTCR W LT + C HA A + D W + +Y Y + IQP
Sbjct: 585 RHVVNLNRKKCTCRTWDLTGILCPHAIKAYLHDKQEPLDQLSWWYSREAYMLVYMHKIQP 644
Query: 518 MNSHIYWEPTPYEKPVPPKVKRSAGRP--KKNRRKDGNEEPIGGSSMKKTYNDIQCGRCG 575
+ +W+ P PP++ + GRP K+ R KD + G S + + CG C
Sbjct: 645 VRGEKFWKVDPSHAMEPPEIHKLVGRPKLKRKREKDEARKREGVWSASRKGLKMTCGHCS 704
Query: 576 LMGHNSRSCAKQGVARRP 593
GHN R C +++P
Sbjct: 705 ATGHNQRRCPMLQRSKQP 722
>UniRef100_Q9FH72 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MRD20
[Arabidopsis thaliana]
Length = 733
Score = 137 bits (345), Expect = 9e-31
Identities = 93/321 (28%), Positives = 160/321 (48%), Gaps = 11/321 (3%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNK-ELRRVV*ECARETTPTQFNRIMERV 336
+GLI A++ V+P HR C H+ N K++K L +V +CAR T F + +E++
Sbjct: 365 KGLIYAIKNVLPYAEHRMCARHIFANLQKRYKQMGPLHKVFWKCARAYNETVFWKQLEKM 424
Query: 337 KSINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEI 396
K+I +A++ + + W +A+F++ K+ + NN E +N+ + + R P++ + E+I
Sbjct: 425 KTIKFEAYDEVKRSVGSNWSRAFFSDITKSAAVENNISESYNAVLKDAREKPVVALLEDI 484
Query: 397 RCYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCR 456
R ++M ++ G + P + +EK K + P NG + H +N
Sbjct: 485 RRHIMASNLVKIKEMQNVTGLITPKAIAIMEKRKKSLKWCYPFSNGRGIYEVDHGKNKYV 544
Query: 457 IKVDVDIFKQTCTCRFWQLT*MPCMHACSAL--ALRGFKL-EDHCHAWLTLGSYRETYSY 513
+ V K +CTCR + ++ +PC H SA+ + KL E W ++ ++ Y+
Sbjct: 545 VHVR---DKTSCTCREYDVSGIPCCHIMSAMWAEYKETKLPETAILDWYSVEKWKLCYNS 601
Query: 514 FIQPMNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRR-KDGNEEPIGGSSMKKTYNDIQCG 572
+ P+N WE +PP + GRPKKN R KD +EE +S K + C
Sbjct: 602 LLFPVNGMELWETHSDVVVMPPPDRIMPGRPKKNDRIKDPSEEASSENSQKAL---VTCS 658
Query: 573 RCGLMGHNSRSCAKQGVARRP 593
CG +GHN R+C + V + P
Sbjct: 659 NCGQIGHNKRTCQIELVPKPP 679
>UniRef100_Q9FM95 Similarity to mutator-like transposase [Arabidopsis thaliana]
Length = 733
Score = 137 bits (344), Expect = 1e-30
Identities = 93/321 (28%), Positives = 159/321 (48%), Gaps = 11/321 (3%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNK-ELRRVV*ECARETTPTQFNRIMERV 336
+GLI A++ V+P HR C H+ N K++K L +V +CAR T F + +E++
Sbjct: 365 KGLIYAIKNVLPYAEHRMCARHIFANLQKRYKQMGPLHKVFWKCARAYNETVFWKQLEKM 424
Query: 337 KSINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEI 396
K+I +A++ + + W +A+F++ K+ + NN E +N+ + + R P++ + E+I
Sbjct: 425 KTIKFEAYDEVKRSVGSNWSRAFFSDITKSAAVENNISESYNAVLKDAREKPVVALLEDI 484
Query: 397 RCYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCR 456
R ++M ++ G + P + +EK K + P NG + H +N
Sbjct: 485 RRHIMASNLVKLKEMQNVTGLITPKAIAIMEKRKKSLKWCYPFSNGRGIYEVDHGKNKYV 544
Query: 457 IKVDVDIFKQTCTCRFWQLT*MPCMHACSAL--ALRGFKL-EDHCHAWLTLGSYRETYSY 513
+ V K +CTCR + ++ +PC H SA+ + KL E W ++ ++ Y+
Sbjct: 545 VHVR---DKTSCTCREYDVSGIPCCHIMSAMWAEYKETKLPETAILDWYSVEKWKLCYNS 601
Query: 514 FIQPMNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRR-KDGNEEPIGGSSMKKTYNDIQCG 572
+ P+N WE +PP + GRPKKN R KD +EE +S K + C
Sbjct: 602 LLFPVNGMELWETHSDVVVMPPPDRIMPGRPKKNDRIKDPSEEASSENSQKAL---VTCS 658
Query: 573 RCGLMGHNSRSCAKQGVARRP 593
CG GHN R+C + V + P
Sbjct: 659 NCGQTGHNKRTCQIELVPKPP 679
>UniRef100_Q9LP09 F9C16.9 [Arabidopsis thaliana]
Length = 946
Score = 137 bits (344), Expect = 1e-30
Identities = 93/325 (28%), Positives = 147/325 (44%), Gaps = 12/325 (3%)
Query: 279 GLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVKS 338
GL+ A+ V+P HR C H+ +N + + EL+R+ + R T +F M +KS
Sbjct: 508 GLVKAIHSVIPQAEHRQCARHIMENWKRNSHDMELQRLFWKIVRSYTEGEFGAHMRALKS 567
Query: 339 INQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIRC 398
N A+E L K W +A+F + +NN E FN I R P+L M E+IR
Sbjct: 568 YNASAFELLLKTLPVTWSRAFFKIGSCCNDNLNNLSESFNRTIREARRKPLLDMLEDIRR 627
Query: 399 YVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRIK 458
M + + + D + +EK IA W ++ I+
Sbjct: 628 QCMVRNEKRYVIADRLRTRFTKRAHAEIEK-MIAGSQVCQRWMA--RHNKHEIKVGPVDS 684
Query: 459 VDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQPM 518
VD+ TC C WQ+T +PC+HA S + + K+ED+ W T +++TY+ I P+
Sbjct: 685 FTVDMNNNTCGCMKWQMTGIPCIHAASVIIGKRQKVEDYVSDWYTTSMWKQTYNDGIGPV 744
Query: 519 NSHIYWEPTPYEKPVPPKVKR-SAGRPKKNRRKDGNEEPIGGSS-----MKKTYNDIQCG 572
+ W +PP +R + GRPK + R+ G E SS + + + + C
Sbjct: 745 QGKLLWPTVNKVGVLPPPWRRGNPGRPKNHARRKGVFESSTASSSSTTELSRLHRVMTCS 804
Query: 573 RCGLMGHNSRSCAKQGV---ARRPK 594
C GHN + C + V A+RP+
Sbjct: 805 NCQGEGHNKQGCKNETVAPPAKRPR 829
>UniRef100_Q9LMY1 F21F23.10 protein [Arabidopsis thaliana]
Length = 753
Score = 136 bits (343), Expect = 2e-30
Identities = 93/321 (28%), Positives = 159/321 (48%), Gaps = 11/321 (3%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNK-ELRRVV*ECARETTPTQFNRIMERV 336
+GLI A++ V+P HR C H+ N K++K L +V +CAR T F + +E++
Sbjct: 385 KGLIYAIKNVLPYAEHRMCARHIFANLQKRYKQMGPLHKVFWKCARAYNETVFWKQLEKM 444
Query: 337 KSINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEI 396
K+I +A++ + + W +A+F++ K+ + NN E +N+ + + R P++ + E+I
Sbjct: 445 KTIKFEAYDEVKRSVGSNWSRAFFSDITKSAAVENNISESYNAVLKDAREKPVVALLEDI 504
Query: 397 RCYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCR 456
R ++M ++ G + P + +EK K + P NG + H +N
Sbjct: 505 RRHIMASNLVKIKEMQNVTGLITPKAIAIMEKRKKSLKWCYPFSNGRGIYEVDHGKNKYV 564
Query: 457 IKVDVDIFKQTCTCRFWQLT*MPCMHACSAL--ALRGFKL-EDHCHAWLTLGSYRETYSY 513
+ V K +CTCR + ++ +PC H SA+ + KL E W ++ ++ Y+
Sbjct: 565 VHVR---DKTSCTCREYDVSGIPCCHIMSAMWAEYKETKLPETAILDWYSVEKWKLCYNS 621
Query: 514 FIQPMNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRR-KDGNEEPIGGSSMKKTYNDIQCG 572
+ P+N WE +PP + GRPKKN R KD +EE +S K + C
Sbjct: 622 LLFPVNGMELWETHSDVVVMPPPDRIMPGRPKKNDRIKDPSEEASSENSQKAL---VTCS 678
Query: 573 RCGLMGHNSRSCAKQGVARRP 593
CG GHN R+C + V + P
Sbjct: 679 NCGQTGHNKRTCQIELVPKPP 699
>UniRef100_Q60CW5 Putative mutator transposable element-related protein [Solanum
tuberosum]
Length = 493
Score = 128 bits (321), Expect = 5e-28
Identities = 72/267 (26%), Positives = 130/267 (47%), Gaps = 5/267 (1%)
Query: 321 ARETTPTQFNRIMERVKSINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSK 380
A+ TT +F+ M R++ ++ A ++LN W +++ + + K ++NN CEVFNS
Sbjct: 170 AKATTVKEFDACMVRIRELDPNAVDWLNDKEPSQWSRSHLSSDAKCDILLNNICEVFNSM 229
Query: 381 ILNYRGHPILTMAEEIRCYVMRKMSQNKMKL-DGRAGPLCPWQQSRLEKEKIASHNWTPL 439
I + R PI+T+ E++R +M +M N+ K + +CP + L K + A+ + P
Sbjct: 230 IFDARDKPIVTLLEKLRYLLMARMLANREKAHKWSSNDVCPKIKDILHKNQTAAGEYIP- 288
Query: 440 WNGDNARQRYHIENNCRIKVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCH 499
N R+ I VD+ + C+C W + +PC HA +A+ + + D+
Sbjct: 289 -RKSNQRKYEIIGATIHDSWAVDLENRICSCTKWSIMGIPCKHAIAAIRAKKDNILDYVD 347
Query: 500 AWLTLGSYRETYSYFIQPMNSHIYWEPTPYEKPVPPKV--KRSAGRPKKNRRKDGNEEPI 557
+ +YR Y + I +N W + P+P + + GR +K RRK+ +E
Sbjct: 348 DCYKVETYRRIYEHAILSINGPQMWPKSTKVPPLPLTIVGNKKTGRKQKARRKEADEVGA 407
Query: 558 GGSSMKKTYNDIQCGRCGLMGHNSRSC 584
+ +K+ + C C GHN ++C
Sbjct: 408 SRTKIKRKQQSLDCSTCNKPGHNKKTC 434
>UniRef100_Q65XH0 Hypothetical protein OJ1126_B11.9 [Oryza sativa]
Length = 792
Score = 125 bits (315), Expect = 3e-27
Identities = 66/220 (30%), Positives = 118/220 (53%), Gaps = 4/220 (1%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+GL+ A++E +P + HR C H+ N K+++++ ++ +CA+ + FN ++
Sbjct: 513 KGLVSAVEEFLPQIEHRMCTRHIYANWRKKYRDQAFQKPFWKCAKASCRPFFNFCRAKLA 572
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
+ + + W +A+F ++ NN CE FN+ I++ R HPI++M E IR
Sbjct: 573 QLTPAGAKDMMSTEPMHWSRAWFRIGSNCDSVDNNMCESFNNWIIDIRAHPIISMFEGIR 632
Query: 398 CYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRI 457
V ++ QN+ K G G +CP +L K S N +WNG + + + + +
Sbjct: 633 TKVYVRIQQNRSKAKGWLGRICPNILKKLNKYIDLSGNCEAIWNG---KDGFEVTDKDK- 688
Query: 458 KVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDH 497
+ VD+ K+TC+CR+WQL +PC HA +AL + + ED+
Sbjct: 689 RYTVDLEKRTCSCRYWQLAGIPCAHAITALFVSSKQPEDY 728
>UniRef100_O81502 F9D12.2 protein [Arabidopsis thaliana]
Length = 940
Score = 120 bits (300), Expect = 1e-25
Identities = 87/325 (26%), Positives = 140/325 (42%), Gaps = 5/325 (1%)
Query: 270 FVVYCLNWQGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQF 329
F V +GLI A+ +++P HR+C H+ N K + + A T +
Sbjct: 499 FTVISDKQKGLINAVADLLPQAEHRHCARHVYANWKKVYGDYCHESYFWAIAYSATEGDY 558
Query: 330 NRIMERVKSINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPI 389
+ M+ ++S + A + L K W +A+F+ + ++ NN E FN I R P+
Sbjct: 559 SYNMDALRSYDPDACDDLLKTDPTTWCRAFFSTHSSCEDVSNNLSESFNRTIREARKLPV 618
Query: 390 LTMAEEIRCYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRY 449
+ M EE+R M+++S+ K P LE + +S L +G+N ++
Sbjct: 619 VNMLEEVRRISMKRISKLCDKTAKCETRFPPKIMQILEGNRKSSKYCQVLKSGEN---KF 675
Query: 450 HIENNCRIKVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRE 509
I VD+ +TC CR W+LT +PC HA + ED+ L ++
Sbjct: 676 EILEGSGY-YSVDLLTRTCGCRQWELTGIPCPHAIYVITEHNRDPEDYVDRLLETQVWKA 734
Query: 510 TYSYFIQPMNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRR-KDGNEEPIGGSSMKKTYND 568
TY I+PMN W+ + P + GRPKK R K+ E + + +
Sbjct: 735 TYKDNIEPMNGERLWKRRGKGRIEVPDKRGKRGRPKKFARIKEPGESSTNPTKLTREGKT 794
Query: 569 IQCGRCGLMGHNSRSCAKQGVARRP 593
+ C C +GHN SC V P
Sbjct: 795 VTCSNCKQIGHNKGSCKNPTVQLAP 819
>UniRef100_Q9SHI6 Similar to mudrA protein [Arabidopsis thaliana]
Length = 622
Score = 118 bits (296), Expect = 4e-25
Identities = 87/320 (27%), Positives = 137/320 (42%), Gaps = 27/320 (8%)
Query: 279 GLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVKS 338
GL+ A+ ++P HR C H+ N + + EL+R+ + AR T +FN +K
Sbjct: 190 GLVKAIHTILPQAEHRQCSKHIMDNWKRDSHDIELQRLFWKIARSYTVEEFNNYQADLK- 248
Query: 339 INQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIRC 398
+A+F + +NN E FN I R P+L + E+IR
Sbjct: 249 ------------------RAFFRTGTCCNDNLNNLSESFNRTIRQARRKPLLDLLEDIRR 290
Query: 399 YVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRIK 458
M + ++ + D P + +EK N + DN + Y N
Sbjct: 291 QCMVRTAKRFIIADRCKTKYTPRAHAEIEKMIAGFQNTQRYMSRDNLHEIY--VNGVGYF 348
Query: 459 VDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQPM 518
VD+D+ +TC CR WQ+ +PC+HA + + K+E + + + T +RETY I+P+
Sbjct: 349 VDMDL--KTCGCRKWQMVGIPCVHATCVIIGKKEKVESYVNDYYTRNRWRETYFRGIRPV 406
Query: 519 NSHIYWEPTPYEKPVPPKVKR-SAGRPKKNRRKDGNEEPIGGSSMKKTYND---IQCGRC 574
W +PP +R +AGRP R+ G E S+ K + + C C
Sbjct: 407 QGMPLWGRLNRLPVLPPPWRRGNAGRPSNYARRKGRNEVASSSNPNKMSREKRIMTCSNC 466
Query: 575 GLMGHNSRSCAKQGVARRPK 594
GHN +SC V PK
Sbjct: 467 LQEGHNKKSCKNATVLSPPK 486
>UniRef100_Q8LNV8 Putative mutator protein [Oryza sativa]
Length = 896
Score = 117 bits (293), Expect = 1e-24
Identities = 89/322 (27%), Positives = 145/322 (44%), Gaps = 13/322 (4%)
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+GL A++ V HR C HL +N K+++ + AR P +F+ M ++
Sbjct: 581 KGLENAVKNVFQRAEHRECFWHLMQNFIKKFQGPVFGNMY-PAARSYMPERFDHYMNKIY 639
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
N YL + K W+++ F+E K I NN E +N I + + PI+ +A+ IR
Sbjct: 640 EANSDVKPYLETYHKLLWMRSKFSEEIKCDFITNNLAESWNKWIKDMKHLPIVELADGIR 699
Query: 398 CYVMRKMSQNKM---KLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENN 454
M +++ + KLDG P+ Q + + +E H + D A +
Sbjct: 700 SKTMNLLARRRKIGEKLDGVMLPIVVRQLNAMTRE--LGHLKVVQGDRDQAEVTEITAEH 757
Query: 455 CRIKVDVDIFKQTCTCRFWQLT*MPCMHACS-ALALRGFKLEDHCHAWLTLGSYRETYSY 513
I+ V++ TCTCR WQ++ PC HA + ++ R + D+ ++ Y+ Y+
Sbjct: 758 EIIRHAVNLVNHTCTCREWQVSGKPCPHALALIISYRNPNMADYLDPCYSVERYKLAYAG 817
Query: 514 FIQPMNSHIYWEPTPYE-KPVPPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCG 572
I P+ W K +PP KR GR +KNR ++ G + K +QC
Sbjct: 818 VILPLPDKSQWPKVNIGFKLLPPLTKREVGRQRKNRIVGCLDK---GKARSKGKWQVQCK 874
Query: 573 RCGLMGHNSRS--CAKQGVARR 592
RC MGH S S C G ++
Sbjct: 875 RCLAMGHRSTSPKCPLNGTKKK 896
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.350 0.154 0.574
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 927,137,968
Number of Sequences: 2790947
Number of extensions: 37013638
Number of successful extensions: 170535
Number of sequences better than 10.0: 280
Number of HSP's better than 10.0 without gapping: 166
Number of HSP's successfully gapped in prelim test: 114
Number of HSP's that attempted gapping in prelim test: 169722
Number of HSP's gapped (non-prelim): 496
length of query: 594
length of database: 848,049,833
effective HSP length: 133
effective length of query: 461
effective length of database: 476,853,882
effective search space: 219829639602
effective search space used: 219829639602
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 78 (34.7 bits)
Medicago: description of AC146972.1


