

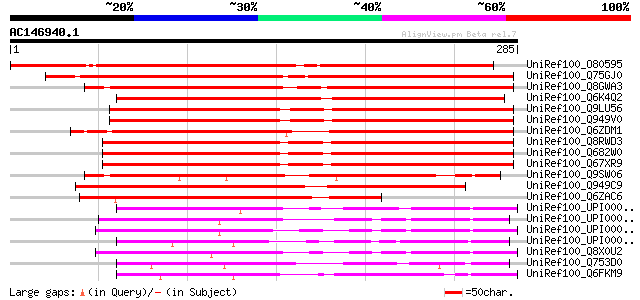
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146940.1 - phase: 0
(285 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O80595 T27I1.6 protein [Arabidopsis thaliana] 293 5e-78
UniRef100_Q75GJ0 Expressed protein [Oryza sativa] 280 3e-74
UniRef100_Q8GWA3 Hypothetical protein At4g25770/F14M19_50 [Arabi... 253 4e-66
UniRef100_Q6K4Q2 Hypothetical protein OSJNBa0054K20.7 [Oryza sat... 246 5e-64
UniRef100_Q9LU56 Arabidopsis thaliana genomic DNA, chromosome 5,... 233 4e-60
UniRef100_Q949V0 Hypothetical protein At5g51180 [Arabidopsis tha... 232 1e-59
UniRef100_Q6ZDM1 Hypothetical protein P0025F03.21 [Oryza sativa] 224 2e-57
UniRef100_Q8RWD3 Hypothetical protein At1g29120:At1g29130 [Arabi... 208 1e-52
UniRef100_Q682W0 MRNA, complete cds, clone: RAFL21-69-E05 [Arabi... 208 1e-52
UniRef100_Q67XR9 MRNA, complete cds, clone: RAFL25-28-O12 [Arabi... 208 1e-52
UniRef100_Q9SW06 Hypothetical protein AT4g25770 [Arabidopsis tha... 206 7e-52
UniRef100_Q949C9 Hypothetical protein C780ERIPDK [Oryza sativa] 189 7e-47
UniRef100_Q6ZAC6 Hypothetical protein P0429B05.29 [Oryza sativa] 173 5e-42
UniRef100_UPI00002350F7 UPI00002350F7 UniRef100 entry 105 1e-21
UniRef100_UPI000023E046 UPI000023E046 UniRef100 entry 100 6e-20
UniRef100_UPI0000219C2A UPI0000219C2A UniRef100 entry 97 4e-19
UniRef100_UPI000042CA0C UPI000042CA0C UniRef100 entry 92 2e-17
UniRef100_Q8X0U2 Hypothetical protein 18A7.150 [Neurospora crassa] 89 2e-16
UniRef100_Q753D0 AFR386Cp [Ashbya gossypii] 86 8e-16
UniRef100_Q6FKM9 Similar to tr|Q08448 Saccharomyces cerevisiae Y... 85 2e-15
>UniRef100_O80595 T27I1.6 protein [Arabidopsis thaliana]
Length = 402
Score = 293 bits (749), Expect = 5e-78
Identities = 154/273 (56%), Positives = 199/273 (72%), Gaps = 8/273 (2%)
Query: 1 MESQQIPKVQHSVVDVQINNKNIKKLKFPKF-GCFRIQHDATGDGFDIEVVDASGHRSNP 59
+E + PK + + ++K KK K + GC R + D +G+ D+ VD G R+ P
Sbjct: 22 LEGIRKPKKMKKMRSRKSDDKETKKKKKKYWMGCLRAESDESGN-VDL-TVDFPGERTEP 79
Query: 60 THLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEV 119
THL++MVNGLIGSA NW++AAKQ LK+YP D++VHCS+ N ST TFDGVDV G RLAEEV
Sbjct: 80 THLVVMVNGLIGSAQNWRFAAKQMLKKYPQDLLVHCSKRNHSTQTFDGVDVMGERLAEEV 139
Query: 120 ISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQISNQECHVR 179
SVIKRHPS++KISF+ HSLGGLIARYAI +LYE++ +EL H I + +C +
Sbjct: 140 RSVIKRHPSLQKISFVGHSLGGLIARYAIGRLYEQESREELP----HNSDDIGD-KCSIE 194
Query: 180 KYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSRFLGKTGKHLFLT 239
+ + +IAGLEP+ FITSATPHLG RGHKQVPL G ++LE+ A+R+S LGKTGKHLFL
Sbjct: 195 EPKARIAGLEPVYFITSATPHLGSRGHKQVPLFSGSYTLERLATRMSGCLGKTGKHLFLA 254
Query: 240 DGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRV 272
D KPPLLL+MV+DS D+KF+SAL+ FKR +
Sbjct: 255 DSDGGKPPLLLRMVKDSRDLKFISALQCFKRHL 287
>UniRef100_Q75GJ0 Expressed protein [Oryza sativa]
Length = 385
Score = 280 bits (716), Expect = 3e-74
Identities = 147/263 (55%), Positives = 186/263 (69%), Gaps = 5/263 (1%)
Query: 21 KNIKKLKFPKFGCFRIQHDATGDGFDIEVVDASGHRSNPTHLIIMVNGLIGSAHNWKYAA 80
+ ++ P+ GCF + A +G G R PTHL++ VNG++GSA NW+YAA
Sbjct: 16 RRLRAACLPRPGCFTVS--AADEGPSGSGGGGGGSRPAPTHLVVTVNGIVGSAENWRYAA 73
Query: 81 KQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEVISVIKRHPSVRKISFIAHSLG 140
K F+K++P DV+VHCS CN + TFDGVDV G RLAEEV+S+++R P ++KISF+AHSLG
Sbjct: 74 KHFIKKHPEDVVVHCSGCNGAVRTFDGVDVMGTRLAEEVLSLVQRRPELQKISFVAHSLG 133
Query: 141 GLIARYAIAKLYERDISKELSQGNVHCEGQISNQECHVRKYEGKIAGLEPINFITSATPH 200
GLIARYAIA LY+ + E+ H E QI++ + GKIAGLEPINFIT ATPH
Sbjct: 134 GLIARYAIALLYKS--ATEIDSHEEH-EKQITDVSSNQLIDRGKIAGLEPINFITFATPH 190
Query: 201 LGCRGHKQVPLLCGFHSLEKTASRLSRFLGKTGKHLFLTDGKNEKPPLLLQMVRDSEDIK 260
LG R HKQ+PLL G + LEK A R+S G++GKHLFL D ++ KPPLLLQMV D D+
Sbjct: 191 LGTRSHKQIPLLRGSYKLEKMAYRISWIAGRSGKHLFLKDIEDGKPPLLLQMVTDYGDLH 250
Query: 261 FMSALRSFKRRVAYANIRYDRIL 283
FMSALRSFKRRVAY+NI D I+
Sbjct: 251 FMSALRSFKRRVAYSNICNDFIV 273
>UniRef100_Q8GWA3 Hypothetical protein At4g25770/F14M19_50 [Arabidopsis thaliana]
Length = 418
Score = 253 bits (646), Expect = 4e-66
Identities = 130/242 (53%), Positives = 173/242 (70%), Gaps = 14/242 (5%)
Query: 43 DGFDIEVVDASGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSST 102
D FD +V++++ P HL++MVNG++GSA +WKYAA+QF+K++P V+VH SE NS+T
Sbjct: 78 DFFDADVMESA---EKPDHLVVMVNGIVGSAADWKYAAEQFVKKFPDKVLVHRSESNSAT 134
Query: 103 LTFDGVDVTGNRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQ 162
LTF GVD G RLA EV+ V+K V+KISF+AHSLGGL+ARYAI KLYE+
Sbjct: 135 LTFGGVDKMGERLANEVLGVVKHRSGVKKISFVAHSLGGLVARYAIGKLYEQ-------P 187
Query: 163 GNVHCEGQISNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTA 222
G V S ++ G+IAGLEP+NFIT ATPHLG RGH+Q P+LCG LE+TA
Sbjct: 188 GEVDSLDSPSKEK---SARGGEIAGLEPMNFITFATPHLGSRGHRQFPILCGLPFLERTA 244
Query: 223 SRLSRF-LGKTGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDR 281
S+ + G+TGKHLFL D + PLL++M DS+D+KF+SAL +FKRRVAYAN+ +D
Sbjct: 245 SQTAHLAAGRTGKHLFLVDNDDGNAPLLIRMATDSDDLKFISALNAFKRRVAYANVNFDS 304
Query: 282 IL 283
++
Sbjct: 305 MV 306
>UniRef100_Q6K4Q2 Hypothetical protein OSJNBa0054K20.7 [Oryza sativa]
Length = 432
Score = 246 bits (628), Expect = 5e-64
Identities = 126/219 (57%), Positives = 162/219 (73%), Gaps = 7/219 (3%)
Query: 61 HLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEVI 120
HL++MVNGL GS+ +WK+AA+QF+KR P V VH S+CN S LT+DGVD+ G RLAEEV
Sbjct: 136 HLVVMVNGLYGSSADWKFAAEQFVKRLPGKVFVHRSQCNHSKLTYDGVDLMGERLAEEVR 195
Query: 121 SVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQISNQECHVRK 180
V++R +++KISF+AHSLGGL+ RYAI KLY+ I++E S + ++ +
Sbjct: 196 QVVQRRSNLQKISFVAHSLGGLVTRYAIGKLYDPSINEEASLDKENFSNELRTSD----- 250
Query: 181 YEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSRFL-GKTGKHLFLT 239
GKIAGLEPINFI ATPHLG R +KQ+P L G LE+TA+ + F+ G+TGKHLFLT
Sbjct: 251 -GGKIAGLEPINFIAVATPHLGSRWNKQLPFLFGVPLLERTAAVTAHFIVGRTGKHLFLT 309
Query: 240 DGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIR 278
D + KPPLLL+M D +D KFMSALRSFKRRVAYAN++
Sbjct: 310 DSDDGKPPLLLRMAEDCDDGKFMSALRSFKRRVAYANLQ 348
>UniRef100_Q9LU56 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MWD22
[Arabidopsis thaliana]
Length = 360
Score = 233 bits (594), Expect = 4e-60
Identities = 125/228 (54%), Positives = 161/228 (69%), Gaps = 10/228 (4%)
Query: 57 SNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLA 116
S+ HL++MV+G++GS +WK+ A+QF+K+ P V VHCSE N S LT DGVDV G RLA
Sbjct: 31 SSADHLVVMVHGILGSTDDWKFGAEQFVKKMPDKVFVHCSEKNVSALTLDGVDVMGERLA 90
Query: 117 EEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQISNQEC 176
EV +I+R P++ KISF+AHSLGGL ARYAI KLY K +Q +V S+QE
Sbjct: 91 AEVRHIIQRKPNICKISFVAHSLGGLAARYAIGKLY-----KPANQEDVKDSVADSSQET 145
Query: 177 HVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSRFLGK-TGKH 235
+G I GLE +NFIT ATPHLG G+KQVP L GF S+EK A + ++ K TG+H
Sbjct: 146 P----KGTICGLEAMNFITVATPHLGSMGNKQVPFLFGFSSIEKVAGLIIHWIFKRTGRH 201
Query: 236 LFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
LFL D + KPPLL +MV D++D F+SALR+FKRRVAY+N+ +DRIL
Sbjct: 202 LFLKDEEEGKPPLLRRMVEDTDDCHFISALRAFKRRVAYSNVGHDRIL 249
>UniRef100_Q949V0 Hypothetical protein At5g51180 [Arabidopsis thaliana]
Length = 357
Score = 232 bits (591), Expect = 1e-59
Identities = 122/228 (53%), Positives = 161/228 (70%), Gaps = 10/228 (4%)
Query: 57 SNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLA 116
S+ HL++MV+G++GS +WK+ A+QF+K+ P V VHCSE N S LT DGVDV G RLA
Sbjct: 31 SSADHLVVMVHGILGSTDDWKFGAEQFVKKMPDKVFVHCSEKNVSALTLDGVDVMGERLA 90
Query: 117 EEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQISNQEC 176
EV+ +I+R P++ KISF+AHSLGGL ARYAI KLY K +Q +V S+QE
Sbjct: 91 AEVLDIIQRKPNICKISFVAHSLGGLAARYAIGKLY-----KPANQEDVKDSVADSSQET 145
Query: 177 HVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSRFLGK-TGKH 235
+G I GLE +NFIT ATPHLG G+KQVP L GF S+EK A + ++ K TG+H
Sbjct: 146 P----KGTICGLEAMNFITVATPHLGSMGNKQVPFLFGFSSIEKVAGLIIHWIFKRTGRH 201
Query: 236 LFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
LFL D + KPPLL +MV D++D F+SALR+FKRRVAY+N+ +D ++
Sbjct: 202 LFLKDEEEGKPPLLRRMVEDTDDCHFISALRAFKRRVAYSNVGHDHVV 249
>UniRef100_Q6ZDM1 Hypothetical protein P0025F03.21 [Oryza sativa]
Length = 350
Score = 224 bits (571), Expect = 2e-57
Identities = 128/252 (50%), Positives = 165/252 (64%), Gaps = 26/252 (10%)
Query: 35 RIQHDATGDGFDIEVVDASGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVH 94
R + A+G G D+ S H +P HL++MV+G++GS +W+YAA +F+K+ P DVIVH
Sbjct: 13 RAEESASG-GVDVWSDAVSSH--DPDHLLVMVHGILGSNADWQYAANEFVKQLPDDVIVH 69
Query: 95 CSECNSSTLTFDGVDVTGNRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYER 154
CSE N +TLT +GVDV G RLA+EVI VI R P + KISF+AHS+GGL ARYAIAKLY
Sbjct: 70 CSEKNINTLTLEGVDVMGERLADEVIDVIIRKPELTKISFLAHSVGGLAARYAIAKLYRH 129
Query: 155 --DISKELSQGNVHCEGQISNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLL 212
D SK + +G I GLE +NFIT ATPHLG RG+ QVPLL
Sbjct: 130 PSDTSKS--------------------ETKGTIGGLEAMNFITVATPHLGSRGNNQVPLL 169
Query: 213 CGFHSLEKTASRLSRFL-GKTGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRR 271
G ++E ASR+ ++ +TGKHLFLTD +PPLL +M D D+ F+SAL +F+RR
Sbjct: 170 FGSIAMENFASRVVHWIFRRTGKHLFLTDDDEGEPPLLQRMAEDYGDLYFISALHAFRRR 229
Query: 272 VAYANIRYDRIL 283
VAYAN D I+
Sbjct: 230 VAYANADCDHIV 241
>UniRef100_Q8RWD3 Hypothetical protein At1g29120:At1g29130 [Arabidopsis thaliana]
Length = 455
Score = 208 bits (529), Expect = 1e-52
Identities = 108/232 (46%), Positives = 153/232 (65%), Gaps = 9/232 (3%)
Query: 53 SGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTG 112
S ++ P HL+++V+G++ S +W Y + +R +++ S N+ T TF G+D G
Sbjct: 94 SDDKNEPDHLLVLVHGILASPSDWLYVEAELKRRLGRRFLIYASSSNTFTKTFGGIDGAG 153
Query: 113 NRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQIS 172
RLAEEV V+++ S++KISF+AHSLGGL +R+A+A LY S ++Q + Q
Sbjct: 154 KRLAEEVRQVVQKSKSLKKISFLAHSLGGLFSRHAVAVLY----SAAMAQVSDVAVSQSG 209
Query: 173 NQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSR-FLGK 231
N G+IAGLEPINFIT ATPHLG RG KQ+P L G LEK A+ ++ F+G+
Sbjct: 210 NSNL----LRGRIAGLEPINFITLATPHLGVRGRKQLPFLLGVPILEKLAAPIAPFFVGR 265
Query: 232 TGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
TG LFLTDGK +KPPLLL+M D ED+KF+SAL +F+ R+ YAN+ YD ++
Sbjct: 266 TGSQLFLTDGKADKPPLLLRMASDGEDLKFLSALGAFRSRIIYANVSYDHMV 317
>UniRef100_Q682W0 MRNA, complete cds, clone: RAFL21-69-E05 [Arabidopsis thaliana]
Length = 398
Score = 208 bits (529), Expect = 1e-52
Identities = 108/232 (46%), Positives = 153/232 (65%), Gaps = 9/232 (3%)
Query: 53 SGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTG 112
S ++ P HL+++V+G++ S +W Y + +R +++ S N+ T TF G+D G
Sbjct: 95 SDDKNEPDHLLVLVHGILASPSDWLYVEAELKRRLGRRFLIYASSSNTFTKTFGGIDGAG 154
Query: 113 NRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQIS 172
RLAEEV V+++ S++KISF+AHSLGGL +R+A+A LY S ++Q + Q
Sbjct: 155 KRLAEEVRQVVQKSKSLKKISFLAHSLGGLFSRHAVAVLY----SAAMAQVSDVAVSQSG 210
Query: 173 NQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSR-FLGK 231
N G+IAGLEPINFIT ATPHLG RG KQ+P L G LEK A+ ++ F+G+
Sbjct: 211 NSNL----LRGRIAGLEPINFITLATPHLGVRGRKQLPFLLGVPILEKLAAPIAPFFVGR 266
Query: 232 TGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
TG LFLTDGK +KPPLLL+M D ED+KF+SAL +F+ R+ YAN+ YD ++
Sbjct: 267 TGSQLFLTDGKADKPPLLLRMASDGEDLKFLSALGAFRSRIIYANVSYDHMV 318
>UniRef100_Q67XR9 MRNA, complete cds, clone: RAFL25-28-O12 [Arabidopsis thaliana]
Length = 455
Score = 208 bits (529), Expect = 1e-52
Identities = 108/232 (46%), Positives = 153/232 (65%), Gaps = 9/232 (3%)
Query: 53 SGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTG 112
S ++ P HL+++V+G++ S +W Y + +R +++ S N+ T TF G+D G
Sbjct: 94 SDDKNEPDHLLVLVHGILASPSDWLYVEAELKRRLGRRFLIYASSSNTFTKTFGGIDGAG 153
Query: 113 NRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQIS 172
RLAEEV V+++ S++KISF+AHSLGGL +R+A+A LY S ++Q + Q
Sbjct: 154 KRLAEEVRQVVQKSKSLKKISFLAHSLGGLFSRHAVAVLY----SAAMAQVSDVAVSQSG 209
Query: 173 NQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSR-FLGK 231
N G+IAGLEPINFIT ATPHLG RG KQ+P L G LEK A+ ++ F+G+
Sbjct: 210 NSNL----LRGRIAGLEPINFITLATPHLGVRGRKQLPFLLGVPILEKLAAPIAPFFVGR 265
Query: 232 TGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
TG LFLTDGK +KPPLLL+M D ED+KF+SAL +F+ R+ YAN+ YD ++
Sbjct: 266 TGSQLFLTDGKADKPPLLLRMASDGEDLKFLSALGAFRSRIIYANVSYDHMV 317
>UniRef100_Q9SW06 Hypothetical protein AT4g25770 [Arabidopsis thaliana]
Length = 395
Score = 206 bits (523), Expect = 7e-52
Identities = 121/258 (46%), Positives = 162/258 (61%), Gaps = 51/258 (19%)
Query: 43 DGFDIEVVDASGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVH-------- 94
D FD +V++++ P HL++MVNG++GSA +WKYAA+QF+K++P V+VH
Sbjct: 78 DFFDADVMESA---EKPDHLVVMVNGIVGSAADWKYAAEQFVKKFPDKVLVHRACFLTYR 134
Query: 95 -C-SECNSSTLTFDGVDVTGNRLAEEVI----------SVIKRHPSVRKISFIAHSLGGL 142
C SE NS+TLTFDGVD G RLA EV+ V+K ++KISF+AHSLGGL
Sbjct: 135 YCGSESNSATLTFDGVDKMGERLANEVVFGCIVVSSVLGVVKHRSGLKKISFVAHSLGGL 194
Query: 143 IARYAIAKLYERDISKELSQGNVHCEGQISNQECHVRKYE---GKIAGLEPINFITSATP 199
+ARYAI KLYE+ G++ + + ++ G+IAGLEP+NFIT ATP
Sbjct: 195 VARYAIGKLYEQP-------------GEVDSLDSPSKEKSARGGEIAGLEPMNFITFATP 241
Query: 200 HLGCRGHKQVPLLCGFHSLEKTASRLSRF-LGKTGKHLFLTDGKNEKPPLLLQMVRDSED 258
HLG RGH+Q P+LCG LE+TAS+ + G+TGKHLFL D ++ S D
Sbjct: 242 HLGSRGHRQFPILCGLPFLERTASQTAHLAAGRTGKHLFLID----------MLIISSYD 291
Query: 259 IKFMSALRSFKRRVAYAN 276
+ SAL +FKRRVAYAN
Sbjct: 292 LN-RSALNAFKRRVAYAN 308
>UniRef100_Q949C9 Hypothetical protein C780ERIPDK [Oryza sativa]
Length = 346
Score = 189 bits (480), Expect = 7e-47
Identities = 104/220 (47%), Positives = 134/220 (60%), Gaps = 13/220 (5%)
Query: 38 HDATGDGFDIEVVDASGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSE 97
H+A + G P HL++MV+G++GSA +WK+ A+QF K VIVH S
Sbjct: 9 HEAAAAEEEEAQAPGGGVGPEPDHLVVMVHGIVGSAADWKFGAEQFEKLLSDKVIVHRSN 68
Query: 98 CNSSTLTFDGVDVTGNRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDIS 157
N LT DGVDV G RLA+EV+ + P +RKISF+AHS+GGL+ARYAI +LY
Sbjct: 69 RNMYKLTLDGVDVMGERLAQEVVEETNKRPQIRKISFVAHSVGGLVARYAIGRLYRPPKQ 128
Query: 158 KELSQGNVHCEGQISNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHS 217
S N++ +G I GLE +NFIT A+PHLG RG+KQVP L GF +
Sbjct: 129 TSQSSQNLN------------NTNKGTIHGLEAVNFITVASPHLGSRGNKQVPFLFGFTA 176
Query: 218 LEKTASRLSRFL-GKTGKHLFLTDGKNEKPPLLLQMVRDS 256
+E AS + + GKTGKHLFLTD + KPPLLL+M DS
Sbjct: 177 IETFASYIIHLIFGKTGKHLFLTDNDDGKPPLLLRMWTDS 216
>UniRef100_Q6ZAC6 Hypothetical protein P0429B05.29 [Oryza sativa]
Length = 338
Score = 173 bits (438), Expect = 5e-42
Identities = 90/174 (51%), Positives = 119/174 (67%), Gaps = 13/174 (7%)
Query: 40 ATGDGFDIEVVDASGHRSN----PTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHC 95
A G G D+ A + P HL++MVNGL+GSA +WK+AA+QF++R P VIVH
Sbjct: 67 ARGGGEDVWSAQAEAEVAQGGGFPEHLVVMVNGLVGSADDWKFAAEQFVRRMPEKVIVHR 126
Query: 96 SECNSSTLTFDGVDVTGNRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERD 155
S+CNS+T TFDGVD+ G RLA EV+SV+++ V+KISF+AHSLGGL+ARYAI +LYE +
Sbjct: 127 SQCNSATQTFDGVDLMGERLANEVLSVVEQRRGVKKISFVAHSLGGLVARYAIGRLYEPN 186
Query: 156 ISKELSQGNVHCEGQISNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQV 209
+ S EG+ + EG IAGLEP+NFIT A+PHLG G+KQ+
Sbjct: 187 NKTKSSSEKSRDEGE---------RLEGFIAGLEPMNFITFASPHLGSSGNKQI 231
>UniRef100_UPI00002350F7 UPI00002350F7 UniRef100 entry
Length = 465
Score = 105 bits (263), Expect = 1e-21
Identities = 75/230 (32%), Positives = 112/230 (48%), Gaps = 38/230 (16%)
Query: 61 HLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVI-VHCSECNSSTLTFDGVDVTGNRLAEEV 119
HL ++V+GL G+ + A KRY + + +ECN+ LT+DG++V G RLA E+
Sbjct: 16 HLCVLVHGLWGNPSHMGSIASALRKRYDEKQLHILVAECNTGNLTYDGIEVCGERLAHEI 75
Query: 120 ISVIKRHPS----VRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQISNQE 175
+ + + + +RK+S + +SLGGLI+RYAI LY R G + + +
Sbjct: 76 EATLSKLEADGHKMRKLSVVGYSLGGLISRYAIGLLYAR--------------GWLDDDK 121
Query: 176 CHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSRFLGKTGKH 235
LEP+NF T A+PHLG R + F+ L SR + +GK
Sbjct: 122 ------------LEPVNFTTFASPHLGARAPVRGVQNLLFNGLG------SRTISTSGKQ 163
Query: 236 LFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRILLF 285
+FL D + LL + D I F+ L+ FK R Y N+ DR F
Sbjct: 164 MFLADTFQDTGKPLLSALADPNSI-FIEGLKRFKNRCVYGNVVNDRTTAF 212
>UniRef100_UPI000023E046 UPI000023E046 UniRef100 entry
Length = 444
Score = 100 bits (248), Expect = 6e-20
Identities = 74/236 (31%), Positives = 110/236 (46%), Gaps = 40/236 (16%)
Query: 51 DASGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVI-VHCSECNSSTLTFDGVD 109
D +G N THL ++V+GL G+ + + AK Y D + + ++ NS T+DG++
Sbjct: 9 DVTGGSVNATHLCVLVHGLWGNPSHLRNVAKSLRDNYSEDELYILLAKQNSGNHTYDGIE 68
Query: 110 VTGNRLA----EEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNV 165
G R+ +E+ + K+ + K+S +SLGGL++RYA+ LY +
Sbjct: 69 TGGERVCAEIEQELKDIEKKGGKITKLSIAGYSLGGLVSRYAVGLLYAK----------- 117
Query: 166 HCEGQISNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRL 225
G + LE +NF T ATPHLG R PL ++
Sbjct: 118 -----------------GVLDDLECMNFTTFATPHLGVR----TPLKGWLSNIYNVLG-- 154
Query: 226 SRFLGKTGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDR 281
+R L +G+ LF D LL ++ D I FMS L+ FKRR YANI DR
Sbjct: 155 ARTLSMSGRQLFTIDSFRNTNRPLLAVLADPNSI-FMSGLKKFKRRTLYANIVNDR 209
>UniRef100_UPI0000219C2A UPI0000219C2A UniRef100 entry
Length = 445
Score = 97.4 bits (241), Expect = 4e-19
Identities = 76/242 (31%), Positives = 116/242 (47%), Gaps = 40/242 (16%)
Query: 49 VVDASGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVI-VHCSECNSSTLTFDG 107
V+ +SG + HL ++V+GL G+ + AK RY D + + ++ NS + T+DG
Sbjct: 4 VLASSGGTARADHLCVLVHGLWGNPAHMAQVAKALRDRYSRDQLYIIVAKRNSGSFTYDG 63
Query: 108 VDVTGNRLA----EEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQG 163
+++ G R+ EE+ + K + KISFI +S+GGL+ARYAI G
Sbjct: 64 IELGGQRVCREIEEELEKIKKTGGKITKISFIGYSMGGLVARYAI--------------G 109
Query: 164 NVHCEGQISNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTAS 223
+ +G + +C INF A+P LGCR PL + L
Sbjct: 110 LLEAKGVLEKLQC--------------INFTAFASPFLGCR----TPLKGWNNHLFNVLG 151
Query: 224 RLSRFLGKTGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
+R L +G+ LF D + L+ ++ D E I FMS LR FKR Y+NI DR
Sbjct: 152 --ARTLSLSGRQLFGIDKFRDTGRPLIAVMTDQESI-FMSGLRRFKRHTLYSNIVNDRAA 208
Query: 284 LF 285
++
Sbjct: 209 VY 210
>UniRef100_UPI000042CA0C UPI000042CA0C UniRef100 entry
Length = 434
Score = 91.7 bits (226), Expect = 2e-17
Identities = 76/231 (32%), Positives = 109/231 (46%), Gaps = 45/231 (19%)
Query: 61 HLIIMVNGLIGSAHNWKYAAKQFLKR-YPYD---VIVHCSECNSSTLTFDGVDVTGNRLA 116
HLII+V+G+ G++ + Y KQ + + YD + H + +S LT+DG+DV G R++
Sbjct: 5 HLIILVHGVWGNSSHLAYVEKQIHENIHSYDGTILYTHKTGSHSGYLTYDGIDVNGKRIS 64
Query: 117 EEVISVIK-----RHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQI 171
+EV K + V K S + +SLGGLI+RY C G +
Sbjct: 65 DEVWEQTKLIEQEKGGKVTKFSVVGYSLGGLISRY--------------------CIGYL 104
Query: 172 SNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLS-RFLG 230
S+Q G +EPINF T TPH+G VP F + + +R++ FL
Sbjct: 105 SSQ--------GYFDNIEPINFTTFCTPHVGV----SVPQSHNFSA--RLYNRIAPLFLA 150
Query: 231 KTGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDR 281
TG FL D E LL + D KF AL FK + YAN+ D+
Sbjct: 151 DTGSQFFLRDKVGEFGKPLLVWMADPRS-KFYKALAKFKYKSLYANVVNDK 200
>UniRef100_Q8X0U2 Hypothetical protein 18A7.150 [Neurospora crassa]
Length = 436
Score = 88.6 bits (218), Expect = 2e-16
Identities = 72/242 (29%), Positives = 113/242 (45%), Gaps = 40/242 (16%)
Query: 49 VVDASGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVI-VHCSECNSSTLTFDG 107
+ D G HL ++V+GL G+ ++ AK +YP D + + ++ N+ + T+DG
Sbjct: 1 MADYIGGNIEADHLCVLVHGLWGNPNHMASVAKALRAQYPRDKLNILVAKRNAGSFTYDG 60
Query: 108 VDVTG----NRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQG 163
+++ G N + EE+ +V + ++KIS +SLGGL+ARYAI LY R
Sbjct: 61 IELGGERVCNEIEEELQAVESKGGKIKKISIAGYSLGGLVARYAIGLLYAR--------- 111
Query: 164 NVHCEGQISNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTAS 223
G + N EC + F A+P LG R PL + +
Sbjct: 112 -----GVLDNLEC--------------MTFTAFASPFLGVR----TPLRGWPNHVWNVLG 148
Query: 224 RLSRFLGKTGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
+R L +G+ LF D + LL ++ D + I FM L FKRR+ Y NI DR
Sbjct: 149 --ARTLCMSGRQLFGIDHFRDTGKPLLAVLADPKSI-FMCGLAKFKRRILYTNIVNDRSA 205
Query: 284 LF 285
++
Sbjct: 206 VY 207
>UniRef100_Q753D0 AFR386Cp [Ashbya gossypii]
Length = 504
Score = 86.3 bits (212), Expect = 8e-16
Identities = 66/233 (28%), Positives = 108/233 (46%), Gaps = 47/233 (20%)
Query: 61 HLIIMVNGLIGSAHNWKY---AAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAE 117
HL+ +V+GL G+ + +Y A Q +R P ++V+ ++ N T+DG+D+ G R+A+
Sbjct: 5 HLVFLVHGLWGNVTHMEYLGRAVSQLQERSPETLVVYAAKMNQGYRTYDGIDICGYRVAK 64
Query: 118 EV------ISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQI 171
E+ ++ + V K S + +S+GGLI+RYA+ LY K+
Sbjct: 65 EIQEQVATLNCPETGTVVTKFSIVGYSMGGLISRYAVGLLYSNQFFKKQD---------- 114
Query: 172 SNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSRFLGK 231
++ INF T +PH+G + + F+ + S LG
Sbjct: 115 ----------------IKLINFTTFCSPHVGVLAPGKNLAVRVFNFV------CSLILGN 152
Query: 232 TGKHLFLTD---GKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDR 281
+G+ +FL D N P ++L V DS F AL F+ R YANI D+
Sbjct: 153 SGRQMFLKDRIKAANGMPLIVLMSVGDS---IFYKALEQFQHRSLYANIVNDK 202
>UniRef100_Q6FKM9 Similar to tr|Q08448 Saccharomyces cerevisiae YOR059c [Candida
glabrata]
Length = 457
Score = 84.7 bits (208), Expect = 2e-15
Identities = 64/234 (27%), Positives = 107/234 (45%), Gaps = 42/234 (17%)
Query: 61 HLIIMVNGLIGSAHNWKYAAKQF-------LKRYPYDVIVHCSECNSSTLTFDGVDVTGN 113
HL ++++GL G+ + K K D + + N++ TFDG+++ G
Sbjct: 6 HLFVLIHGLWGNYKHMKSLEKVLDATLNGKKSGKDKDYVFFLPKQNATFKTFDGIEIIGY 65
Query: 114 RLAEEVISVIK--RHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQI 171
R E+ +K + ++ KISF+ +SLGGL+AR+ + K+Y
Sbjct: 66 RTLLELCEFMKEFKDGNITKISFVGYSLGGLVARFVVGKMY------------------- 106
Query: 172 SNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSRFLGK 231
EC+ +E F+T ATPHLG + + + L S LGK
Sbjct: 107 --SECN-----DIFGNIERCIFMTMATPHLGIQFYNPLGYLHRKLLFSTFTGLGSTILGK 159
Query: 232 TGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRILLF 285
+G+ LF+ + N+ +VR SE K++ AL F R+ +AN++ DR + F
Sbjct: 160 SGRELFIANSSND------ILVRLSEG-KYIEALEEFNHRILFANVKNDRTVAF 206
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 471,843,406
Number of Sequences: 2790947
Number of extensions: 19180995
Number of successful extensions: 45500
Number of sequences better than 10.0: 166
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 110
Number of HSP's that attempted gapping in prelim test: 45261
Number of HSP's gapped (non-prelim): 203
length of query: 285
length of database: 848,049,833
effective HSP length: 126
effective length of query: 159
effective length of database: 496,390,511
effective search space: 78926091249
effective search space used: 78926091249
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146940.1


