

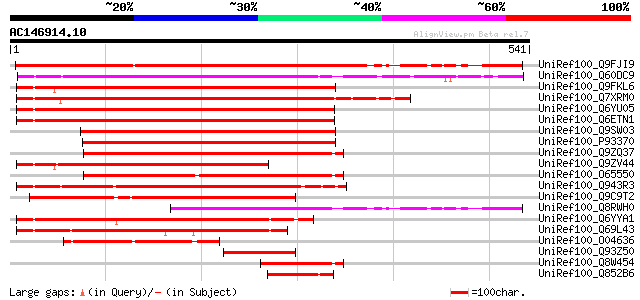
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146914.10 + phase: 0
(541 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FJI9 Similarity to calmodulin-binding protein [Arabi... 461 e-128
UniRef100_Q60DC9 Hypothetical protein OSJNBb0013C14.20 [Oryza sa... 393 e-108
UniRef100_Q9FKL6 Calmodulin-binding protein [Arabidopsis thaliana] 349 1e-94
UniRef100_Q7XRM0 OSJNBa0058G03.4 protein [Oryza sativa] 345 2e-93
UniRef100_Q6YU05 Putative calmodulin-binding protein [Oryza sativa] 344 4e-93
UniRef100_Q6ETN1 Putative calmodulin-binding protein [Oryza sativa] 342 2e-92
UniRef100_Q9SW03 Putative calmodulin-binding protein [Arabidopsi... 316 1e-84
UniRef100_P93370 Calmodulin-binding protein [Nicotiana tabacum] 304 5e-81
UniRef100_Q9ZQ37 Putative calmodulin-binding protein [Arabidopsi... 279 1e-73
UniRef100_Q9ZV44 Putative calmodulin-binding protein [Arabidopsi... 276 1e-72
UniRef100_O65550 Putative calmodulin-binding protein [Arabidopsi... 273 1e-71
UniRef100_Q943R3 Calmodulin-binding protein-like [Oryza sativa] 263 1e-68
UniRef100_Q9C9T2 Putative calmodulin-binding protein; 77122-7370... 261 4e-68
UniRef100_Q8RWH0 Hypothetical protein At5g62570 [Arabidopsis tha... 258 4e-67
UniRef100_Q6YYA1 Putative calmodulin-binding protein [Oryza sativa] 219 2e-55
UniRef100_Q69L43 Calmodulin-binding protein-like [Oryza sativa] 214 7e-54
UniRef100_O04636 F2P16.9 protein [Arabidopsis thaliana] 143 1e-32
UniRef100_Q93Z50 At1g73800/F25P22_22 [Arabidopsis thaliana] 95 5e-18
UniRef100_Q8W454 Putative calmodulin-binding protein [Arabidopsi... 72 4e-11
UniRef100_Q852B6 Putative calmodulin-binding protein [Oryza sativa] 72 4e-11
>UniRef100_Q9FJI9 Similarity to calmodulin-binding protein [Arabidopsis thaliana]
Length = 505
Score = 461 bits (1187), Expect = e-128
Identities = 259/530 (48%), Positives = 349/530 (64%), Gaps = 45/530 (8%)
Query: 7 FSVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTL 66
FSVV+EVM+LQ+V++ +EP+LEPL+R+VV+EEVELAL KHL+ IK C KE + ESR L
Sbjct: 12 FSVVQEVMRLQTVKHFLEPVLEPLIRKVVKEEVELALGKHLAGIKWICEKETHPLESRNL 71
Query: 67 QLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEE 126
QL+F N++SLPVFT ARIEG++G +R+ L+D TG++ +GP SSAK+E+ V+EGDF
Sbjct: 72 QLKFLNNLSLPVFTSARIEGDEGQAIRVGLIDPSTGQIFSSGPASSAKLEVFVVEGDFNS 131
Query: 127 ESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLG 186
SD W ED +NNIVRER+GKKPLL G+V L DG+ ++ EIS+TDNSSWTRSR+FRLG
Sbjct: 132 VSD-WTDEDIRNNIVREREGKKPLLNGNVFAVLNDGIGVMDEISFTDNSSWTRSRKFRLG 190
Query: 187 VRVVDNFDGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSR 246
VR+VD FD ++IREA T+SF+VRDHRGELYKKHHPPSL DEVWRLEKIGKDGAFHRRL+
Sbjct: 191 VRIVDQFDYVKIREAITESFVVRDHRGELYKKHHPPSLFDEVWRLEKIGKDGAFHRRLNL 250
Query: 247 EKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFA-SHSQQ 305
I TVKDFLT +L+ +KLR +LGTGMS+KMWE+T++HAR+CVLD++ HV A ++
Sbjct: 251 SNINTVKDFLTHFHLNSSKLRQVLGTGMSSKMWEITLDHARSCVLDSSVHVYQAPGFQKK 310
Query: 306 PHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFISDHPLVSLADAQISVISALNQC-DFA 364
VVFN V +V GLL + +Y+ +KLSE EKA I + LA A++ VI AL+ +
Sbjct: 311 TAVVFNVVAQVLGLLVDFQYIPAEKLSEIEKALILSVYIFDLAQAEVMVIDALSHLNEVI 370
Query: 365 SFEDEVSLMDGYSHLTNVHYSPSSPRTEGSSANKLLALQKTGGFNYTQESASSTDIMPSI 424
S++DEVS+M NV +P+S +GS A G +Y+ S +S D +
Sbjct: 371 SYDDEVSMM------RNVLNAPAS---QGSVA----------GIDYSGLSLTSLDGYGFV 411
Query: 425 YSVGGTSSLDDYGLPNFESLGLRYDQHLGFPVQVSNSLICDMDSIVHAFGDEDHLQFFDA 484
S+ T+ + + + + H G N C H G E+
Sbjct: 412 SSLHNTAECSG---KHSDDVDMEVTPH-GLYEDYDNLWNCS-----HILGLEEP------ 456
Query: 485 DLQSQCHIEADLHSAVDSFMPVSSTSMTKGKAQRRWRKVVNVLKWFMVKK 534
+++L SA+D FM + S+ +RW K+ +V +W V K
Sbjct: 457 --------QSELQSALDDFMSQKNASVGGKAHSKRWTKLFSVSRWLSVFK 498
>UniRef100_Q60DC9 Hypothetical protein OSJNBb0013C14.20 [Oryza sativa]
Length = 548
Score = 393 bits (1009), Expect = e-108
Identities = 237/541 (43%), Positives = 327/541 (59%), Gaps = 44/541 (8%)
Query: 9 VVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTLQL 68
V+ EVM+ S+ L I EPL+RRVV+EE+E A H + + +T ++ S S+ QL
Sbjct: 27 VITEVMRKSSIEKLFTAI-EPLIRRVVKEEIESAFANHATMMARTV-MDVVPSSSKNFQL 84
Query: 69 QFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEE-E 127
QF +SLP+FTG++IEGE ++ I LVD +T +VV +G ES KVEIVVLEGDFE E
Sbjct: 85 QFMTKLSLPIFTGSKIEGESSLSITIALVDTVTREVVASGDESLMKVEIVVLEGDFEGGE 144
Query: 128 SDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGV 187
D W ++F NNI+R R+GK+PLL+GD+ + L G+ VGE+S+TDNSSWTRSR+FRLG
Sbjct: 145 GDDWTAQEFNNNIIRAREGKRPLLSGDIFVGLIKGIGAVGELSFTDNSSWTRSRKFRLGA 204
Query: 188 RVVD-NFDGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSR 246
+ D +++G+R+REAK++SF+V+DHRGELYKKHHPP L DEVWRLEKIGK+GAFH+RL+R
Sbjct: 205 KTEDGSYNGVRVREAKSESFVVKDHRGELYKKHHPPILDDEVWRLEKIGKEGAFHKRLNR 264
Query: 247 EKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQP 306
EKI TVK+FLTLL+LD +LR ILG+GMS KMWEVTVEH++TC+L H+ + +
Sbjct: 265 EKIVTVKEFLTLLHLDAPRLRKILGSGMSTKMWEVTVEHSKTCILPDKVHLYYPDSLSKT 324
Query: 307 HVVFNAVGEVTGLLAESEYVAVDKLSETEKAFISDHPLVSLADAQISVISALNQCDFASF 366
VVFN VGEV GL++E ++V D L E EK A+A +V A
Sbjct: 325 AVVFNVVGEVRGLISE-KFVCADDLREKEK-----------AEAYAAVKQAYENWKNVFT 372
Query: 367 EDEVSLMDGYSHLTNVHYSPSSPRTEGSSANKLLALQKTGGFNYTQESASSTDIMPSIYS 426
D +L+ S L +V + + ++ T GF +Q S S P I+S
Sbjct: 373 CDNETLLANPSQLLDVR----TTSLHENDYDQFPTQVSTDGFGLSQSSIPS----PDIFS 424
Query: 427 VGGTSSLDDYGLPNFESLGLRYDQHL-----GFPVQV-------SNSLICDMDSIVHAFG 474
+ +S+LD L E+ +Y L P QV SNSL+ + D H
Sbjct: 425 IDPSSALDPCPLETAENNENQYQSELPPLGGHAPPQVSQTVDKFSNSLVYE-DCTSHPSF 483
Query: 475 DEDHLQFFDADLQSQCHIEADLHSAVDSFMPVSSTSMTKGKAQRRWRKVVNVLKWFMVKK 534
ED+ + D + DL +A+ F+ +++K KA R WR + V+ W K
Sbjct: 484 SEDYYRCSDPSVSFDTQ---DLGAALKGFI----ATISKPKAYRGWRTLSYVIGWIFYTK 536
Query: 535 R 535
+
Sbjct: 537 K 537
>UniRef100_Q9FKL6 Calmodulin-binding protein [Arabidopsis thaliana]
Length = 647
Score = 349 bits (896), Expect = 1e-94
Identities = 173/338 (51%), Positives = 251/338 (74%), Gaps = 7/338 (2%)
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKK----HLSSIKQTCGKEMNTSES 63
SV+ E +K+ S++ L LEP++RRVV EE+E AL K L+ + K + +
Sbjct: 33 SVIVEALKVDSLQKLCSS-LEPILRRVVSEELERALAKLGPARLTGSSGSSPKRIEGPDG 91
Query: 64 RTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGD 123
R LQL F++ +SLP+FTG ++EGE G+ + + L+DA TG+ V GPE+SAK+ IVVLEGD
Sbjct: 92 RKLQLHFKSRLSLPLFTGGKVEGEQGAVIHVVLIDANTGRAVVYGPEASAKLHIVVLEGD 151
Query: 124 FEEESDI-WMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRR 182
F E D W E+F++++V+ER GK+PLLTG+V + LK+G+ +GE+ +TDNSSW RSR+
Sbjct: 152 FNTEDDEDWTQEEFESHVVKERSGKRPLLTGEVYVTLKEGVGTLGELVFTDNSSWIRSRK 211
Query: 183 FRLGVRVVDNF-DGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFH 241
FRLG+RVV DG+RIREAKT++F+V+DHRGELYKKH+PP+L+D+VWRL+KIGKDGAFH
Sbjct: 212 FRLGLRVVSGCCDGMRIREAKTEAFVVKDHRGELYKKHYPPALNDDVWRLDKIGKDGAFH 271
Query: 242 RRLSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFAS 301
++L+ E I TV+DFL ++ D KLRTILG+GMS KMW+ VEHA+TCV + ++ +A
Sbjct: 272 KKLTAEGINTVEDFLRVMVKDSPKLRTILGSGMSNKMWDALVEHAKTCVQSSKLYIYYAE 331
Query: 302 HSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFI 339
S+ VVFN + E++GL++ +Y + D L++++K ++
Sbjct: 332 DSRNVGVVFNNIYELSGLISGDQYFSADSLTDSQKVYV 369
>UniRef100_Q7XRM0 OSJNBa0058G03.4 protein [Oryza sativa]
Length = 652
Score = 345 bits (885), Expect = 2e-93
Identities = 193/419 (46%), Positives = 279/419 (66%), Gaps = 17/419 (4%)
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIK------QTCGKEMNTS 61
SV+ E +K+ S++ L LEP++RRVV EEVE AL K + ++ K +
Sbjct: 41 SVIVEALKVDSLQKLCSS-LEPILRRVVSEEVERALAKLGPAATPARIQGRSSPKRIEGP 99
Query: 62 ESRTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLE 121
LQLQF + +SLP+FTG ++EGE G+ + + L+DA TG+VV +GPES AK++++VLE
Sbjct: 100 SGINLQLQFRSRLSLPLFTGGKVEGEQGAAIHVVLLDANTGRVVTSGPESFAKLDVLVLE 159
Query: 122 GDFEEESDI-WMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRS 180
GDF +E D W E+F+N+IV+ER+GK+PLLTGD+ + LK+G+ +GE+ +TDNSSW RS
Sbjct: 160 GDFNKEQDEDWTEEEFENHIVKEREGKRPLLTGDLQVTLKEGVGTIGELIFTDNSSWIRS 219
Query: 181 RRFRLGVRVVDNF-DGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGA 239
R+FRLG+RV F +G+R++EAKT++F V+DHRGELYKKH+PP+L D+VWRLEKIGKDGA
Sbjct: 220 RKFRLGLRVSSGFCEGVRVKEAKTEAFTVKDHRGELYKKHYPPALKDDVWRLEKIGKDGA 279
Query: 240 FHRRLSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSF 299
FH++L+ I TV+ FL LL D KLRTILG+ MS KMWE VEHA+TCVL ++ +
Sbjct: 280 FHKKLNSNGIYTVEHFLQLLVRDQQKLRTILGSNMSNKMWESLVEHAKTCVLSGKHYIYY 339
Query: 300 ASHSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFISDHPLVSLADAQISVISALN 359
+S ++ +FN + E TGL+A+ +Y++ + LSE ++ F +D + D I+V+ +
Sbjct: 340 SSDARSVGAIFNNIYEFTGLIADDQYISAENLSENQRLF-ADTLVKQAYDDWINVVE-YD 397
Query: 360 QCDFASFEDEVSLMDGYSHLTNVHYSPSSPRTEGSSANKLLALQKTGG-FNYTQESASS 417
+ F+ + + S S S P + GS+ + Q TGG N Q S SS
Sbjct: 398 GKELLRFKQKKKSVTTRSDTAKA--STSYPSSYGSTHSH---KQLTGGPVNIEQSSMSS 451
>UniRef100_Q6YU05 Putative calmodulin-binding protein [Oryza sativa]
Length = 652
Score = 344 bits (882), Expect = 4e-93
Identities = 173/335 (51%), Positives = 248/335 (73%), Gaps = 5/335 (1%)
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQ--TCGKEMNTSESRT 65
SV+ E +K+ S++ L LEP++RRVV EEVE AL K + Q + K + + R
Sbjct: 42 SVIVEALKVDSLQKLCSS-LEPILRRVVSEEVERALAKLGPARIQGRSSPKRIEGPDGRN 100
Query: 66 LQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFE 125
LQL+F +SLP+FTG ++EGE G+ + + L+DA TG V +GPES AK++++VLEGDF
Sbjct: 101 LQLKFTTRLSLPLFTGGKVEGEQGAAIHVVLLDANTGVAVTSGPESCAKLDVLVLEGDFN 160
Query: 126 -EESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFR 184
EE + W E+F+++IV+ER+GK+PLLTGD+ + LK+G+ +GE+ +TDNSSW RSR+FR
Sbjct: 161 NEEDEDWTEEEFESHIVKEREGKRPLLTGDLQVTLKEGVGTIGELIFTDNSSWIRSRKFR 220
Query: 185 LGVRVVD-NFDGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRR 243
LG+RV +F+GIR+REAKT++F V+DHRGELYKKH+PP+L D+VWRLEKIGKDGAFH++
Sbjct: 221 LGLRVAPGSFEGIRVREAKTEAFTVKDHRGELYKKHYPPALKDDVWRLEKIGKDGAFHKK 280
Query: 244 LSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHS 303
L+ I TV+DFL LL D +LR+ILG+GMS KMWE VEHA+TCVL +V +A S
Sbjct: 281 LNASGIYTVEDFLQLLVKDQQRLRSILGSGMSNKMWESLVEHAKTCVLSGKHYVYYAIDS 340
Query: 304 QQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAF 338
+ +FN + E TGL+A+ ++++ + L++ +K +
Sbjct: 341 RNVGAIFNNIYEFTGLIADDQFISAENLTDNQKIY 375
>UniRef100_Q6ETN1 Putative calmodulin-binding protein [Oryza sativa]
Length = 624
Score = 342 bits (876), Expect = 2e-92
Identities = 170/335 (50%), Positives = 249/335 (73%), Gaps = 5/335 (1%)
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKK--HLSSIKQTCGKEMNTSESRT 65
SV+ E +K+ S++ L LEP++RRVV EEVE AL + + ++ K + + R
Sbjct: 30 SVIVEALKVDSLQRLCSS-LEPILRRVVSEEVERALGRLGPATITGRSSPKRIEGPDGRN 88
Query: 66 LQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFE 125
LQLQF +SLP+FTG ++EGE G+ + + L+DA TG VV +GPES AK++IVVLEGDF
Sbjct: 89 LQLQFRTRLSLPLFTGGKVEGEQGAAIHVVLLDAGTGCVVSSGPESCAKLDIVVLEGDFN 148
Query: 126 -EESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFR 184
E+ + W E+F++++V+ER+GK+PLLTGDV + LK+G+ VGE+++TDNSSW RSR+FR
Sbjct: 149 NEDEEGWSGEEFESHVVKEREGKRPLLTGDVQVTLKEGVGTVGELTFTDNSSWIRSRKFR 208
Query: 185 LGVRVVDNF-DGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRR 243
LG+++ F +GIRIREAKT++F+V+DHRGELYKKH+PP+L DEVWRLEKIGKDG+FH+R
Sbjct: 209 LGLKISSGFCEGIRIREAKTEAFMVKDHRGELYKKHYPPALKDEVWRLEKIGKDGSFHKR 268
Query: 244 LSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHS 303
L++ I TV+DFL L+ DP KLR+ILG+GMS KMW++ VEHA+TCVL ++ ++ +
Sbjct: 269 LNKAGISTVEDFLRLVVRDPQKLRSILGSGMSNKMWDILVEHAKTCVLSGKYYIYYSDEN 328
Query: 304 QQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAF 338
+ +FN + GL++ ++ + + L +++K F
Sbjct: 329 RSIGAIFNNIYAFCGLISGEQFYSSESLDDSQKLF 363
>UniRef100_Q9SW03 Putative calmodulin-binding protein [Arabidopsis thaliana]
Length = 536
Score = 316 bits (809), Expect = 1e-84
Identities = 151/268 (56%), Positives = 207/268 (76%), Gaps = 2/268 (0%)
Query: 74 ISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEESDI-WM 132
+SLP+FTG R+EGE G+ + + L+DA TG+ V GPE+S K+E+VVL GDF E D W
Sbjct: 10 LSLPLFTGGRVEGEQGATIHVVLIDANTGRPVTVGPEASLKLEVVVLGGDFNNEDDEDWT 69
Query: 133 PEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVVDN 192
E+F++++V+ER+GK+PLLTGD+ + LK+G+ +GEI +TDNSSW RSR+FRLG+RV
Sbjct: 70 QEEFESHVVKEREGKRPLLTGDLFVVLKEGVGTLGEIVFTDNSSWIRSRKFRLGLRVPSG 129
Query: 193 F-DGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRT 251
+ DGIRIREAKT++F V+DHRGELYKKH+PP+L+DEVWRLEKIGKDGAFH+RL+ I T
Sbjct: 130 YCDGIRIREAKTEAFSVKDHRGELYKKHYPPALNDEVWRLEKIGKDGAFHKRLTAAGIVT 189
Query: 252 VKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFN 311
V+ FL L D KLR ILG+GMS KMW++ VEHA+TCVL ++ + S+ VVFN
Sbjct: 190 VEGFLRQLVRDSTKLRAILGSGMSNKMWDLLVEHAKTCVLSGKLYIYYTEDSRSVGVVFN 249
Query: 312 AVGEVTGLLAESEYVAVDKLSETEKAFI 339
+ E++GL+ E +Y++ D LSE++K ++
Sbjct: 250 NIYELSGLITEDQYLSADSLSESQKVYV 277
>UniRef100_P93370 Calmodulin-binding protein [Nicotiana tabacum]
Length = 551
Score = 304 bits (778), Expect = 5e-81
Identities = 142/265 (53%), Positives = 205/265 (76%), Gaps = 2/265 (0%)
Query: 77 PVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEESDI-WMPED 135
P+FTGA++EGE G+ + + L+DA TG +V TG ES K+++VVLEGDF E D W E+
Sbjct: 16 PLFTGAKVEGEHGAAIHVVLIDADTGHLVTTGAESCIKLDVVVLEGDFNTEDDEGWTQEE 75
Query: 136 FKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVVDNF-D 194
F +++V+ER+GK+PLLTG++ + LK G+ +G++++TDNSSW RSR+FRLG++V + +
Sbjct: 76 FDSHVVKEREGKRPLLTGELQVTLKGGVGTLGDLTFTDNSSWIRSRKFRLGMKVASGYCE 135
Query: 195 GIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKD 254
G+RIREAKT++F V DHRGELYKKH+PP+L+D+VWRLEKIGKDG+FH+RL++ I TV+D
Sbjct: 136 GVRIREAKTEAFTVEDHRGELYKKHYPPALNDDVWRLEKIGKDGSFHKRLNKAAIFTVED 195
Query: 255 FLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVG 314
FL L+ DP KLR ILG+GMS KMW+ +EHA+TCVL +V ++ S+ VVFN +
Sbjct: 196 FLRLVVRDPQKLRNILGSGMSNKMWDALIEHAKTCVLSGKLYVYYSDDSRNVGVVFNNIY 255
Query: 315 EVTGLLAESEYVAVDKLSETEKAFI 339
E+ GL+A +Y + D LS+++K ++
Sbjct: 256 ELNGLIAGEQYYSADSLSDSQKVYV 280
>UniRef100_Q9ZQ37 Putative calmodulin-binding protein [Arabidopsis thaliana]
Length = 503
Score = 279 bits (714), Expect = 1e-73
Identities = 140/273 (51%), Positives = 195/273 (71%), Gaps = 4/273 (1%)
Query: 78 VFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEESDI-WMPEDF 136
+FTG ++EGE GS + + L+DA TG VV TG ES++K+ +VVLEGDF +E D W E F
Sbjct: 5 LFTGGKVEGERGSAIHVVLIDANTGNVVQTGEESASKLNVVVLEGDFNDEDDEDWTREHF 64
Query: 137 KNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVVDNF-DG 195
++ V+ER+GK+P+LTGD + LK+G+ +GE+++TDNSSW RSR+FRLGV+ + D
Sbjct: 65 ESFEVKEREGKRPILTGDTQIVLKEGVGTLGELTFTDNSSWIRSRKFRLGVKPASGYGDS 124
Query: 196 IRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDF 255
IREAKT+ F V+DHRGELYKKH+PP++ DEVWRL++I KDG H++L + I TV+DF
Sbjct: 125 FCIREAKTEPFAVKDHRGELYKKHYPPAVHDEVWRLDRIAKDGVLHKKLLKANIVTVEDF 184
Query: 256 LTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGE 315
L LL DP KLR +LG+GMS +MWE TVEHA+TCVL +V + + VVFN + E
Sbjct: 185 LRLLVKDPQKLRNLLGSGMSNRMWENTVEHAKTCVLGGKLYVFYTDQTHATGVVFNHIYE 244
Query: 316 VTGLLAESEYVAVDKLSETEKAFISDHPLVSLA 348
GL+ ++++++ L+ +K IS LV LA
Sbjct: 245 FRGLITNGQFLSLESLNHDQK--ISADILVKLA 275
>UniRef100_Q9ZV44 Putative calmodulin-binding protein [Arabidopsis thaliana]
Length = 353
Score = 276 bits (706), Expect = 1e-72
Identities = 142/268 (52%), Positives = 199/268 (73%), Gaps = 9/268 (3%)
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKK----HLSSIKQTCGKEMNTSES 63
SV+ E +K+ S++ L LEP++RRVV EEVE AL K LS +++ K +
Sbjct: 39 SVIVEALKMDSLQRLCSS-LEPILRRVVSEEVERALAKLGPARLS--ERSSPKRIEGIGG 95
Query: 64 RTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGD 123
R LQLQF + +S+P+FTG +IEGE G+ + + L+D TG V+ GPE+SAK+++VVL+GD
Sbjct: 96 RNLQLQFRSRLSVPLFTGGKIEGEQGAAIHVVLLDMTTGHVLTVGPEASAKLDVVVLDGD 155
Query: 124 FE-EESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRR 182
F E+ D W E+F+ ++V+ER GK+PLLTGDV + LK+G+ +GE+ +TDNSSW R R+
Sbjct: 156 FNTEDDDGWSGEEFEGHLVKERQGKRPLLTGDVQVTLKEGVGTLGELIFTDNSSWIRCRK 215
Query: 183 FRLGVRVVDNF-DGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFH 241
FRLG+RV + +G+R+REAKT++F V+DHRGELYKKH+PP+L DEVWRLEKIGKDGAFH
Sbjct: 216 FRLGLRVSSGYCEGMRVREAKTEAFTVKDHRGELYKKHYPPALDDEVWRLEKIGKDGAFH 275
Query: 242 RRLSREKIRTVKDFLTLLNLDPAKLRTI 269
++L++ I VK+FL L+ D KLRT+
Sbjct: 276 KKLNKAGIYNVKEFLRLMVKDSQKLRTV 303
>UniRef100_O65550 Putative calmodulin-binding protein [Arabidopsis thaliana]
Length = 467
Score = 273 bits (697), Expect = 1e-71
Identities = 137/272 (50%), Positives = 191/272 (69%), Gaps = 6/272 (2%)
Query: 78 VFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEESDI-WMPEDF 136
+FTG ++EGE GS + + L+DA TG V+ TG ES K+ IVVL+GDF +E D W E F
Sbjct: 10 LFTGGKVEGEQGSAIHVVLIDANTGNVIQTGEESMTKLNIVVLDGDFNDEDDKDWTREHF 69
Query: 137 KNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVVDNFDGI 196
++ V+ER+GK+P+LTGD + +K+G+ +G++++TDNSSW RSR+FRLGV+ F
Sbjct: 70 ESFEVKEREGKRPILTGDRHVIIKEGVGTLGKLTFTDNSSWIRSRKFRLGVKPATGF--- 126
Query: 197 RIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFL 256
IREAKT+ F V+DHRGE+YKKH+PP L DEVWRL+KI KDGA H++L + I TV+DFL
Sbjct: 127 HIREAKTEPFAVKDHRGEVYKKHYPPVLHDEVWRLDKIAKDGALHKKLLKSNIVTVEDFL 186
Query: 257 TLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGEV 316
+L DP KLR++LG+GMS +MW+ TVEHA+TCVL + + + Q VVFN + E
Sbjct: 187 QILMKDPQKLRSLLGSGMSNRMWDNTVEHAKTCVLGGKLYAYYTDQTHQTAVVFNHIYEF 246
Query: 317 TGLLAESEYVAVDKLSETEKAFISDHPLVSLA 348
GL+A +++ + L+ +K IS LV A
Sbjct: 247 QGLIANGHFLSSESLNHDQK--ISADTLVKTA 276
>UniRef100_Q943R3 Calmodulin-binding protein-like [Oryza sativa]
Length = 438
Score = 263 bits (671), Expect = 1e-68
Identities = 153/348 (43%), Positives = 223/348 (63%), Gaps = 14/348 (4%)
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTLQ 67
+V+RE + ++ ++ L LEPL+RRVV+EE++ L + I++ E ++ +
Sbjct: 29 TVIREALMVKQMQTLFVA-LEPLLRRVVQEELQAGLVRSPRYIERM-SPETPPAQPPMWK 86
Query: 68 LQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFE-E 126
L F LP+FTG++IE +G+ L I LVD TG +VE+V + GDF +
Sbjct: 87 LAFRFKPQLPIFTGSKIEDVNGNPLEIILVDVDTGAPATIS--QPLRVEVVPVLGDFPPD 144
Query: 127 ESDIWMPEDFKNN-IVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRL 185
+ + W E+F+ IV+ER GK+PLLTGDV L ++DG +V E+ +TDNSSW R RRFR+
Sbjct: 145 DREHWTAEEFQQRGIVKERSGKRPLLTGDVSLTMRDGCVVVNELQFTDNSSWVRCRRFRI 204
Query: 186 GVRVVD-NFDGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRL 244
GVRVV ++DG RI EA T+ F+VRDHRGELY+KH+PP L D+VWRLEKIGK+GAFHR+L
Sbjct: 205 GVRVVPGSYDGPRIGEAMTEPFVVRDHRGELYRKHYPPVLGDDVWRLEKIGKEGAFHRKL 264
Query: 245 SREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQ 304
++ +R V++FL LL + P +LR I+G GM+ +MWEVT HA+ CV ++ H
Sbjct: 265 TQHNVRNVQEFLRLLTVKPDELRAIMGDGMTDRMWEVTTSHAKKCVPGDKVYMYSTQHGT 324
Query: 305 QPHVVFNAVGEVTGL-LAESEYVAVDKLSETEKAFISDHPLVSLADAQ 351
V N++ E+ + LA EY + +L+ +KAF+ H L+ A Q
Sbjct: 325 ---VYVNSIFELVKVELAGVEY-QLHQLNRGQKAFV--HQLLLAAYEQ 366
>UniRef100_Q9C9T2 Putative calmodulin-binding protein; 77122-73705 [Arabidopsis
thaliana]
Length = 451
Score = 261 bits (667), Expect = 4e-68
Identities = 133/281 (47%), Positives = 189/281 (66%), Gaps = 7/281 (2%)
Query: 21 NLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGK-EMNTSESRTLQLQFENSISLPVF 79
N + +LEP++R+VVR+EVE + K + + + E + + TL+L F ++ P+F
Sbjct: 44 NTLRSVLEPVIRKVVRQEVEYGISKRFRLSRSSSFRIEAPEATTPTLKLIFRKNLMTPIF 103
Query: 80 TGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEESDIWMPEDFKNN 139
TG++I D + L I LVD V P K++IV L GDF D W ++F++N
Sbjct: 104 TGSKISDVDNNPLEIILVDDSNKPVNLNRP---IKLDIVALHGDFPS-GDKWTSDEFESN 159
Query: 140 IVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVV--DNFDGIR 197
I++ERDGK+PLL G+V + +++G+ +GEI +TDNSSW RSR+FR+G +V + G+
Sbjct: 160 IIKERDGKRPLLAGEVSVTVRNGVATIGEIVFTDNSSWIRSRKFRIGAKVAKGSSGQGVV 219
Query: 198 IREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLT 257
+ EA T++ +VRDHRGELYKKHHPP L DEVWRLEKIGKDGAFH++LS I TV+DFL
Sbjct: 220 VCEAMTEAIVVRDHRGELYKKHHPPMLEDEVWRLEKIGKDGAFHKKLSSRHINTVQDFLK 279
Query: 258 LLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVS 298
L +D +LR ILG GMS + WEVT++HAR C+L ++S
Sbjct: 280 LSVVDVDELRQILGPGMSDRKWEVTLKHARECILGNKLYIS 320
>UniRef100_Q8RWH0 Hypothetical protein At5g62570 [Arabidopsis thaliana]
Length = 325
Score = 258 bits (658), Expect = 4e-67
Identities = 160/369 (43%), Positives = 217/369 (58%), Gaps = 55/369 (14%)
Query: 168 EISYTDNSSWTRSRRFRLGVRVVDNFDGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDE 227
EIS+TDNSSWTRSR+FRLGVR+VD FD ++IREA T+SF+VRDHRGELYKKHHPPSL DE
Sbjct: 3 EISFTDNSSWTRSRKFRLGVRIVDQFDYVKIREAITESFVVRDHRGELYKKHHPPSLFDE 62
Query: 228 VWRLEKIGKDGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHAR 287
VWRLEKI KDGAFHR L+ I TVKDFLT +L+ +KLR +LGTGMS+KMWE+T++HAR
Sbjct: 63 VWRLEKIVKDGAFHRLLNLSNINTVKDFLTHFHLNSSKLRQVLGTGMSSKMWEITLDHAR 122
Query: 288 TCVLDTTRHVSFA-SHSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFISDHPLVS 346
+CVLD++ HV A ++ VVFN V +V GLL + +Y+ +KLSE EK
Sbjct: 123 SCVLDSSVHVYQAPGFQKKTAVVFNVVAQVLGLLVDFQYIPAEKLSEIEK---------- 172
Query: 347 LADAQISVISALNQC-DFASFEDEVSLMDGYSHLTNVHYSPSSPRTEGSSANKLLALQKT 405
A A++ VI AL+ + S++DEVS+M NV +P+S +GS A
Sbjct: 173 -AQAEVMVIDALSHLNEVISYDDEVSMM------RNVLNAPAS---QGSVA--------- 213
Query: 406 GGFNYTQESASSTDIMPSIYSVGGTSSLDDYGLPNFESLGLRYDQHLGFPVQVSNSLICD 465
G +Y+ S +S D + S+ T+ + + + + H G N C
Sbjct: 214 -GIDYSGLSLTSLDGYGFVSSLHNTAECSG---KHSDDVDMEVTPH-GLYEDYDNLWNCS 268
Query: 466 MDSIVHAFGDEDHLQFFDADLQSQCHIEADLHSAVDSFMPVSSTSMTKGKAQRRWRKVVN 525
H G E+ +++L SA+D FM + S+ +RW K+ +
Sbjct: 269 -----HILGLEEP--------------QSELQSALDDFMSQKNASVGGKAHSKRWTKLFS 309
Query: 526 VLKWFMVKK 534
V +W V K
Sbjct: 310 VSRWLSVFK 318
>UniRef100_Q6YYA1 Putative calmodulin-binding protein [Oryza sativa]
Length = 520
Score = 219 bits (557), Expect = 2e-55
Identities = 125/320 (39%), Positives = 198/320 (61%), Gaps = 17/320 (5%)
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSI--KQTCGKEMNTSESRT 65
SVVR + ++++ ++ LEP++RRVVREE+ ++ + + + + S
Sbjct: 45 SVVRRAVAAETIQQIVLN-LEPVIRRVVREEIRNIFPQYGHDLPHRSLPLQIQDVGVSPP 103
Query: 66 LQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPE----SSAKVEIVVLE 121
L+L F + LP+FT ++ D + + I+LVD T +V S+ K+E++VL+
Sbjct: 104 LKLVFTKQLKLPIFTNNKLVDIDNNPIEIQLVDTRTNLIVTPSNTHLGYSAIKLEVLVLD 163
Query: 122 GDF--EEESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKD-GLCMVGEISYTDNSSWT 178
GDF +E+ W + F IV+ R+G++PLL G V + + + G+ ++ ++S+TDNSSW
Sbjct: 164 GDFRYDEDGARWTDDQFSTAIVKAREGRRPLLVGTVSVTMSNHGVAVIDDVSFTDNSSWI 223
Query: 179 RSRRFRLGVRVVDNFD--GIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGK 236
RSR+FR+GVRVV D G+RI+EA ++SF V+DHRGELYKKH PP L+D VWRL IGK
Sbjct: 224 RSRKFRIGVRVVMLTDSCGLRIQEAVSESFTVKDHRGELYKKHFPPLLTDNVWRLRNIGK 283
Query: 237 DGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRH 296
DG +RL E I+ V+DFL L ++P KL++++ GMS + W T++ A++C + +
Sbjct: 284 DGPIDKRLEAEGIKNVQDFLKLNTMNPNKLKSLV--GMSDRQWSATLKQAKSCDMGGKCY 341
Query: 297 VSFASHSQQPHVVFNAVGEV 316
V S+ + FN VGE+
Sbjct: 342 V---FKSEGCEIKFNPVGEI 358
>UniRef100_Q69L43 Calmodulin-binding protein-like [Oryza sativa]
Length = 429
Score = 214 bits (544), Expect = 7e-54
Identities = 118/290 (40%), Positives = 182/290 (62%), Gaps = 12/290 (4%)
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTLQ 67
SV+R M + ++ L LEP R+ V+EE+E +L KH + ++ +N+ +S +L+
Sbjct: 35 SVMRGAMAAEKIQKLGLD-LEPFFRKAVQEELERSLSKHGHLLYRSPPMLVNSVDS-SLK 92
Query: 68 LQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEE 127
L F + LP+FT ++ D + L++ L+ + K+E++VL+GDF
Sbjct: 93 LAFAKRLQLPIFTNNKLVDVDNNPLQVHLLHMSSTTTSHHHLPMIKKLEVLVLDGDFSHG 152
Query: 128 SDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKD---GLCMVGEISYTDNSSWTRSRRFR 184
+ W ++F IVRER+G++PLL G + + + D G+ + ++++TDNSSWTRSRRFR
Sbjct: 153 DEGWSSDEFSGAIVREREGRRPLLVGTLNVAMADDHLGVAFIDDVAFTDNSSWTRSRRFR 212
Query: 185 LGVRVV---DNFDG--IRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGA 239
+GVR V + DG +RIREA ++SF+V+DHRGE YKKH PP DEVWRL+ I KDG
Sbjct: 213 IGVRAVAVAGSGDGGELRIREAVSESFMVKDHRGESYKKHFPPRPDDEVWRLKNIRKDGP 272
Query: 240 FHRRLSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTC 289
H+RL E++R V+ FL L +P KLR ++ MS ++W+ T+ HA+TC
Sbjct: 273 IHKRLESERVRNVQGFLNLHATNPEKLRKLV--VMSDRLWKATLHHAKTC 320
>UniRef100_O04636 F2P16.9 protein [Arabidopsis thaliana]
Length = 550
Score = 143 bits (360), Expect = 1e-32
Identities = 82/163 (50%), Positives = 112/163 (68%), Gaps = 8/163 (4%)
Query: 57 EMNTSESRTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVE 116
E +S SR L+L F NS +FTG++IE EDGS L I LVDA T +V TGP SS++VE
Sbjct: 7 ETPSSRSR-LKLCFINSPPSSIFTGSKIEAEDGSPLVIELVDATTNTLVSTGPFSSSRVE 65
Query: 117 IVVLEGDFEEESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMV-GEISYTDNS 175
+V L DF EES W E F NI+ +R+GK+PLLTGD+ + LK+G+ ++ G+I+++DNS
Sbjct: 66 LVPLNADFTEES--WTVEGFNRNILTQREGKRPLLTGDLTVMLKNGVGVITGDIAFSDNS 123
Query: 176 SWTRSRRFRLGVRVVDNFDGIRIREAKTDSFIVRDHRGELYKK 218
SWTRSR+FRLG ++ G EA++++F RD RGE K
Sbjct: 124 SWTRSRKFRLGAKLT----GDGAVEARSEAFGCRDQRGEWVSK 162
>UniRef100_Q93Z50 At1g73800/F25P22_22 [Arabidopsis thaliana]
Length = 207
Score = 95.1 bits (235), Expect = 5e-18
Identities = 45/75 (60%), Positives = 56/75 (74%)
Query: 224 LSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTV 283
L DEVWRLEKIGKDGAFH++LS I TV+DFL L +D +LR ILG GMS + WEVT+
Sbjct: 2 LEDEVWRLEKIGKDGAFHKKLSSRHINTVQDFLKLSVVDVDELRQILGPGMSDRKWEVTL 61
Query: 284 EHARTCVLDTTRHVS 298
+HAR C+L ++S
Sbjct: 62 KHARECILGNKLYIS 76
>UniRef100_Q8W454 Putative calmodulin-binding protein [Arabidopsis thaliana]
Length = 278
Score = 72.0 bits (175), Expect = 4e-11
Identities = 37/87 (42%), Positives = 54/87 (61%), Gaps = 2/87 (2%)
Query: 262 DPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGEVTGLLA 321
DP KLR++LG+GMS +MW+ TVEHA+TCVL + + + Q VVFN + E GL+A
Sbjct: 3 DPQKLRSLLGSGMSNRMWDNTVEHAKTCVLGGKLYAYYTDQTHQTAVVFNHIYEFQGLIA 62
Query: 322 ESEYVAVDKLSETEKAFISDHPLVSLA 348
+++ + L+ +K IS LV A
Sbjct: 63 NGHFLSSESLNHDQK--ISADTLVKTA 87
>UniRef100_Q852B6 Putative calmodulin-binding protein [Oryza sativa]
Length = 166
Score = 72.0 bits (175), Expect = 4e-11
Identities = 36/69 (52%), Positives = 48/69 (69%), Gaps = 1/69 (1%)
Query: 269 ILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGEVTGLLAESEYVAV 328
ILG+GMS KMWEVTVEH++TC+L H+ + + VVFN VGEV GL++E ++V
Sbjct: 55 ILGSGMSTKMWEVTVEHSKTCILTDKVHLYYPHSLIKTAVVFNVVGEVRGLISE-KFVCA 113
Query: 329 DKLSETEKA 337
D L E +KA
Sbjct: 114 DDLMEKDKA 122
Score = 36.6 bits (83), Expect = 1.9
Identities = 15/31 (48%), Positives = 23/31 (73%)
Query: 150 LLTGDVILYLKDGLCMVGEISYTDNSSWTRS 180
L +GD+ + L +G+ +GE+S+T NSSWT S
Sbjct: 3 LASGDIFVGLTNGISTMGELSFTHNSSWTCS 33
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.134 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 877,489,081
Number of Sequences: 2790947
Number of extensions: 36801757
Number of successful extensions: 98740
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 98654
Number of HSP's gapped (non-prelim): 38
length of query: 541
length of database: 848,049,833
effective HSP length: 132
effective length of query: 409
effective length of database: 479,644,829
effective search space: 196174735061
effective search space used: 196174735061
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146914.10


