

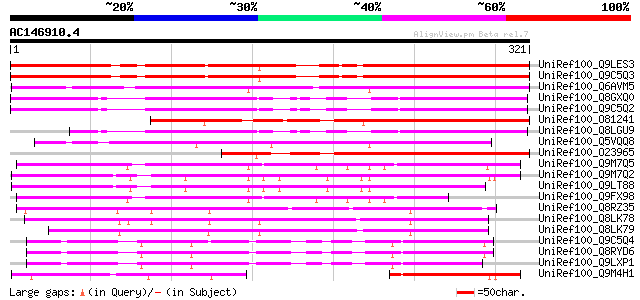
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146910.4 + phase: 0
(321 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LES3 Promoter-binding factor-like protein [Arabidops... 302 9e-81
UniRef100_Q9C5Q3 BZIP protein DPBF3 [Arabidopsis thaliana] 300 5e-80
UniRef100_Q6AVM5 Putative ABA-responsive element-binding protein... 247 3e-64
UniRef100_Q8GXQ0 Putative bZIP transcription factor AtbZIP12 / D... 241 1e-62
UniRef100_Q9C5Q2 BZIP protein DPBF4 [Arabidopsis thaliana] 241 2e-62
UniRef100_O81241 Dc3 promoter-binding factor-3 [Helianthus annuus] 226 8e-58
UniRef100_Q8LGU9 Basic leucine zipper transcription factor [Arab... 209 6e-53
UniRef100_Q5VQQ8 Putative bZIP protein DPBF3 [Oryza sativa] 209 6e-53
UniRef100_O23965 Dc3 promoter-binding factor-2 [Helianthus annuus] 189 9e-47
UniRef100_Q9M7Q5 Abscisic acid responsive elements-binding facto... 164 2e-39
UniRef100_Q9M7Q2 Abscisic acid responsive elements-binding facto... 159 1e-37
UniRef100_Q9LT88 Abscisic acid responsive elements-binding facto... 156 6e-37
UniRef100_Q9FX98 F14J22.7 protein [Arabidopsis thaliana] 143 7e-33
UniRef100_Q8RZ35 Putative abscisic acid insensitive 5 [Oryza sat... 143 7e-33
UniRef100_Q8LK78 ABA response element binding factor [Triticum a... 139 1e-31
UniRef100_Q8LK79 ABA response element binding factor [Triticum a... 138 2e-31
UniRef100_Q9C5Q4 BZIP protein DPBF2 [Arabidopsis thaliana] 135 1e-30
UniRef100_Q8RYD6 Basic leucine zipper transcription factor [Arab... 134 3e-30
UniRef100_Q9LXP1 BZIP transcription factor-like protein [Arabido... 134 4e-30
UniRef100_Q9M4H1 Putative ripening-related bZIP protein [Vitis v... 114 3e-24
>UniRef100_Q9LES3 Promoter-binding factor-like protein [Arabidopsis thaliana]
Length = 297
Score = 302 bits (773), Expect = 9e-81
Identities = 184/325 (56%), Positives = 214/325 (65%), Gaps = 32/325 (9%)
Query: 1 MGSQGGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGE 60
M SQ G V ++K+ L+R SLY+LTLDEVQNHLG+ GK LGSMNLDELLKSV SVEA +
Sbjct: 1 MDSQRGIVEQAKSQSLNRQSSLYSLTLDEVQNHLGSSGKALGSMNLDELLKSVCSVEANQ 60
Query: 61 VSDFGGSDVAATAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKKRGVDRDR 120
S +A G Q G + Q SLTL DLSKKTVDEVWKD+Q K G
Sbjct: 61 PSS-----MAVNGGAAAQE----GLSRQGSLTLPRDLSKKTVDEVWKDIQQNKNGGSAHE 111
Query: 121 KSREKQQTLGEMTLEDFLVKAGVVGESFHGKES----GLLRVDSNEDSRQKVSHGLHWMQ 176
+ R+KQ TLGEMTLED L+KAGVV E+ G G S Q ++ W+Q
Sbjct: 112 R-RDKQPTLGEMTLEDLLLKAGVVTETIPGSNHDGPVGGGSAGSGAGLGQNITQVGPWIQ 170
Query: 177 YPVHSVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAAISPSSLMGTLSDTQTLGR 236
Y H +P A P+ V+ QA+ +S SSLMG LSDTQT GR
Sbjct: 171 Y--------------HQLPSMPQPQAFM-PYPVSDMQAM---VSQSSLMGGLSDTQTPGR 212
Query: 237 KRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNE 296
KRVASG VVEKTVERRQKRMIKNRESAARSRAR+QAYT ELE+KVSRLEEENERLR+Q E
Sbjct: 213 KRVASGEVVEKTVERRQKRMIKNRESAARSRARKQAYTHELEIKVSRLEEENERLRKQKE 272
Query: 297 MEKEVPTAPPPEPKNQLRRTNSASF 321
+EK +P+ PPP+PK QLRRT+SA F
Sbjct: 273 VEKILPSVPPPDPKRQLRRTSSAPF 297
>UniRef100_Q9C5Q3 BZIP protein DPBF3 [Arabidopsis thaliana]
Length = 297
Score = 300 bits (767), Expect = 5e-80
Identities = 183/325 (56%), Positives = 213/325 (65%), Gaps = 32/325 (9%)
Query: 1 MGSQGGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGE 60
M SQ G V ++K+ L+R SLY+LTLDEVQNHLG+ GK LGSMNLDELLKSV SVEA +
Sbjct: 1 MDSQRGIVEQAKSQSLNRQSSLYSLTLDEVQNHLGSSGKALGSMNLDELLKSVCSVEANQ 60
Query: 61 VSDFGGSDVAATAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKKRGVDRDR 120
S +A G Q G + Q SLTL DLSKKTVDEVWKD+Q K G
Sbjct: 61 PSS-----MAVNGGAAAQE----GLSRQGSLTLPRDLSKKTVDEVWKDIQQNKNGGSAHE 111
Query: 121 KSREKQQTLGEMTLEDFLVKAGVVGESFHGKES----GLLRVDSNEDSRQKVSHGLHWMQ 176
+ R+KQ TLGEMTLED L+KAGVV E+ G G S Q ++ W+Q
Sbjct: 112 R-RDKQPTLGEMTLEDLLLKAGVVTETIPGSNHDGPVGGGSAGSGAGLGQNITQVGPWIQ 170
Query: 177 YPVHSVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAAISPSSLMGTLSDTQTLGR 236
Y H +P A P+ V+ QA+ +S SSLMG LSDTQ GR
Sbjct: 171 Y--------------HQLPSMPQPQAFM-PYPVSDMQAM---VSQSSLMGGLSDTQIPGR 212
Query: 237 KRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNE 296
KRVASG VVEKTVERRQKRMIKNRESAARSRAR+QAYT ELE+KVSRLEEENERLR+Q E
Sbjct: 213 KRVASGEVVEKTVERRQKRMIKNRESAARSRARKQAYTHELEIKVSRLEEENERLRKQKE 272
Query: 297 MEKEVPTAPPPEPKNQLRRTNSASF 321
+EK +P+ PPP+PK QLRRT+SA F
Sbjct: 273 VEKILPSVPPPDPKRQLRRTSSAPF 297
>UniRef100_Q6AVM5 Putative ABA-responsive element-binding protein 3 [Oryza sativa]
Length = 335
Score = 247 bits (631), Expect = 3e-64
Identities = 159/328 (48%), Positives = 195/328 (58%), Gaps = 20/328 (6%)
Query: 2 GSQGGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEV 61
G G+ L+R GSLY LTL+EVQ+ LG +PL SMNLDELLKSV+ A
Sbjct: 20 GRDAGSAQRGPMQGLARQGSLYGLTLNEVQSQLG---EPLLSMNLDELLKSVFPDGADLD 76
Query: 62 SDFGGSDVAATAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQG-KKRGVDRD- 119
GG +A Q G Q S+T+ +LSKKTVDEVWK +Q KRG +
Sbjct: 77 GGGGGGGIAG------QSQPALGLQRQGSITMPPELSKKTVDEVWKGIQDVPKRGAEEGG 130
Query: 120 RKSREKQQTLGEMTLEDFLVKAGVVGE--SFHGKESGLLRVDSNEDSRQKVSHGLHWMQY 177
R RE+Q TLGEMTLEDFLVKAGVV + G + + ++ G W+Q
Sbjct: 131 RWRRERQPTLGEMTLEDFLVKAGVVTDPNDLPGNMDVVGGAAAAAAGTSDLNAGAQWLQQ 190
Query: 178 PVHSVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAAISP----SSLMGTLSDTQT 233
+ QH + A H QP VA LD S S ++G LSD QT
Sbjct: 191 YHQQALEPQHP---SIGAPYMATHLAPQPLAVATGAVLDPIYSDGQITSPMLGALSDPQT 247
Query: 234 LGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRR 293
GRKR A+G + +K VERRQKRMIKNRESAARSRAR+QAYT ELE KV RLEEENERL++
Sbjct: 248 PGRKRCATGEIADKLVERRQKRMIKNRESAARSRARKQAYTNELENKVLRLEEENERLKK 307
Query: 294 QNEMEKEVPTAPPPEPKNQLRRTNSASF 321
Q E+++ + +APPPEPK QLRRT+SA+F
Sbjct: 308 QKELDEILNSAPPPEPKYQLRRTSSAAF 335
>UniRef100_Q8GXQ0 Putative bZIP transcription factor AtbZIP12 / DPBF4 [Arabidopsis
thaliana]
Length = 262
Score = 241 bits (616), Expect = 1e-62
Identities = 161/321 (50%), Positives = 192/321 (59%), Gaps = 61/321 (19%)
Query: 1 MGSQGGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGE 60
MGS G + E + L+R SLY+L L EVQ HLG+ GKPLGSMNLDELLK+V A E
Sbjct: 1 MGSIRGNIEEPISQSLTRQNSLYSLKLHEVQTHLGSSGKPLGSMNLDELLKTVLP-PAEE 59
Query: 61 VSDFGGSDVAATAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKKRGVDRDR 120
G Q SLTL DLSKKTVDEVW+D+Q K G
Sbjct: 60 -----------------------GLVRQGSLTLPRDLSKKTVDEVWRDIQQDKNGNGTST 96
Query: 121 KSREKQQTLGEMTLEDFLVKAGVVGESFHGKESGLLRVDSNEDSRQKVSHGLHWMQYPVH 180
+ KQ TLGE+TLED L++AGVV E+ +E+ ++ + SN W++Y H
Sbjct: 97 TTTHKQPTLGEITLEDLLLRAGVVSETVVPQEN-VVNIASNGQ----------WVEYH-H 144
Query: 181 SVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAAISPSSLMGTLSDT-QTLGRKRV 239
QQQQ GF + V +MG LSDT Q GRKRV
Sbjct: 145 QPQQQQ---------GFMTYPVCEMQDMV--------------MMGGLSDTPQAPGRKRV 181
Query: 240 ASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEMEK 299
A G +VEKTVERRQKRMIKNRESAARSRAR+QAYT ELE+KVSRLEEENE+LRR E+EK
Sbjct: 182 A-GEIVEKTVERRQKRMIKNRESAARSRARKQAYTHELEIKVSRLEEENEKLRRLKEVEK 240
Query: 300 EVPTAPPPEPKNQLRRTNSAS 320
+P+ PPP+PK +LRRTNSAS
Sbjct: 241 ILPSEPPPDPKWKLRRTNSAS 261
>UniRef100_Q9C5Q2 BZIP protein DPBF4 [Arabidopsis thaliana]
Length = 262
Score = 241 bits (614), Expect = 2e-62
Identities = 161/321 (50%), Positives = 192/321 (59%), Gaps = 61/321 (19%)
Query: 1 MGSQGGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGE 60
MGS G + E + L+R SLY+L L EVQ HLG+ GKPLGSMNLDELLK+V A E
Sbjct: 1 MGSIRGNIEEPISQSLTRQNSLYSLKLHEVQTHLGSSGKPLGSMNLDELLKTVLP-PAEE 59
Query: 61 VSDFGGSDVAATAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKKRGVDRDR 120
G Q SLTL DLSKKTVDEVW+D+Q K G
Sbjct: 60 -----------------------GLVRQGSLTLPRDLSKKTVDEVWRDIQQDKNGNGTST 96
Query: 121 KSREKQQTLGEMTLEDFLVKAGVVGESFHGKESGLLRVDSNEDSRQKVSHGLHWMQYPVH 180
+ KQ TLGE+TLED L++AGVV E+ +E+ ++ + SN W++Y H
Sbjct: 97 TTTHKQPTLGEITLEDLLLRAGVVTETVVPQEN-VVNIASNGQ----------WVEYH-H 144
Query: 181 SVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAAISPSSLMGTLSDT-QTLGRKRV 239
QQQQ GF + V +MG LSDT Q GRKRV
Sbjct: 145 QPQQQQ---------GFMTYPVCEMQDMV--------------MMGGLSDTPQAPGRKRV 181
Query: 240 ASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEMEK 299
A G +VEKTVERRQKRMIKNRESAARSRAR+QAYT ELE+KVSRLEEENE+LRR E+EK
Sbjct: 182 A-GEIVEKTVERRQKRMIKNRESAARSRARKQAYTHELEIKVSRLEEENEKLRRLKEVEK 240
Query: 300 EVPTAPPPEPKNQLRRTNSAS 320
+P+ PPP+PK +LRRTNSAS
Sbjct: 241 ILPSEPPPDPKWKLRRTNSAS 261
>UniRef100_O81241 Dc3 promoter-binding factor-3 [Helianthus annuus]
Length = 246
Score = 226 bits (575), Expect = 8e-58
Identities = 136/252 (53%), Positives = 162/252 (63%), Gaps = 34/252 (13%)
Query: 88 QESLTLSGDLSKKTVDEVWKDMQ-GKKRGVDRD----------RKSREKQQTLGEMTLED 136
Q S+ L+ DL KKTVDEVW+ +Q GK +G + R +RE+Q TLGEMTLED
Sbjct: 11 QSSINLAQDLRKKTVDEVWQGIQQGKNKGSNNSSGSGNNDGDKRGNRERQPTLGEMTLED 70
Query: 137 FLVKAGVVGESFHGKESGLLRVDSNEDSRQKVSHGLHWMQYPVHSVQQQQHQYEKHTMPG 196
FL+KAGVV SG VD N+++ WMQY V + QQ H
Sbjct: 71 FLLKAGVV------TGSGKKNVDVNQENANHQQ--AQWMQYQVAPIPQQHVYMSGH---- 118
Query: 197 FAAVHAIQQPFQVAGNQALDA-------AISPSSLMGTLSDTQTLGRKRVASGIVVEKTV 249
H +QQ + N +D A+SPS LM LSDTQT GRKRVASG V+EKTV
Sbjct: 119 ----HPVQQSLSIGANPMMDMVYPETQMAMSPSHLMHNLSDTQTPGRKRVASGDVIEKTV 174
Query: 250 ERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEMEKEVPTAPPPEP 309
ERRQKRMIKNRESAARSRAR+QAYT ELE K+SRLEEENE L+RQ E+ +P+APPP+P
Sbjct: 175 ERRQKRMIKNRESAARSRARKQAYTHELENKISRLEEENELLKRQKEVGMVLPSAPPPKP 234
Query: 310 KNQLRRTNSASF 321
K QLRRT+SASF
Sbjct: 235 KYQLRRTSSASF 246
>UniRef100_Q8LGU9 Basic leucine zipper transcription factor [Arabidopsis thaliana]
Length = 226
Score = 209 bits (533), Expect = 6e-53
Identities = 143/284 (50%), Positives = 169/284 (59%), Gaps = 61/284 (21%)
Query: 38 GKPLGSMNLDELLKSVWSVEAGEVSDFGGSDVAATAGGNMQHNQLGGFNSQESLTLSGDL 97
GKPLGSMNLDELLK+V A E G Q SLTL DL
Sbjct: 2 GKPLGSMNLDELLKTVLP-PAEE-----------------------GLVRQGSLTLPRDL 37
Query: 98 SKKTVDEVWKDMQGKKRGVDRDRKSREKQQTLGEMTLEDFLVKAGVVGESFHGKESGLLR 157
SKKTVDEVW+D+Q K G + KQ TLGE+TLED L++AGVV E+ +E+ ++
Sbjct: 38 SKKTVDEVWRDIQQDKNGNGTSTTTTHKQPTLGEITLEDLLLRAGVVTETVVPQEN-VVN 96
Query: 158 VDSNEDSRQKVSHGLHWMQYPVHSVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDA 217
+ SN W++Y H QQQQ GF + V
Sbjct: 97 IASNGQ----------WVEYH-HQPQQQQ---------GFMTYPVCEMQDMV-------- 128
Query: 218 AISPSSLMGTLSDT-QTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQE 276
+MG LSDT Q GRKRVA G +VEKTVERRQKRMIKNRESAARSRAR+QAYT E
Sbjct: 129 ------MMGGLSDTPQAPGRKRVA-GEIVEKTVERRQKRMIKNRESAARSRARKQAYTHE 181
Query: 277 LELKVSRLEEENERLRRQNEMEKEVPTAPPPEPKNQLRRTNSAS 320
LE+KVSRLEEENE+LRR E+EK +P+ PPP+PK +LRRTNSAS
Sbjct: 182 LEIKVSRLEEENEKLRRLKEVEKILPSEPPPDPKWKLRRTNSAS 225
>UniRef100_Q5VQQ8 Putative bZIP protein DPBF3 [Oryza sativa]
Length = 366
Score = 209 bits (533), Expect = 6e-53
Identities = 137/301 (45%), Positives = 175/301 (57%), Gaps = 27/301 (8%)
Query: 16 LSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDFGGSDVAATAGG 75
L R GS+Y+LTLDEVQ+ LG +PL SMNLDELL+SV+ +D A
Sbjct: 21 LGRQGSMYSLTLDEVQSQLG---EPLHSMNLDELLRSVFP------DGLAIADGAGATTS 71
Query: 76 NMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKKR-------GVDRDRKSREKQQT 128
+ QH G Q S+T+ +LSKKTVDEVWK +Q + G R+ RE+Q T
Sbjct: 72 SQQHQPGSGLLRQGSITMPPELSKKTVDEVWKGIQAAPKRNAETGGGGGGGRRRRERQPT 131
Query: 129 LGEMTLEDFLVKAGVVGESFHGKESGLLRVDS-----NEDSRQKVSHGLHWM-QYPVHSV 182
LGE+TLEDFLVKAGVV + + S + VD ++ G HW+ QY
Sbjct: 132 LGEVTLEDFLVKAGVVTQGSLKELSDVGNVDPVGRGVTATGTVDLAPGSHWIEQYKQQIA 191
Query: 183 QQQQHQYEKHTMPG-FAAVHAIQQPFQVAGNQALDAAISP----SSLMGTLSDTQTLGRK 237
H + + + G + + QP V L+ + S S ++G +SD+QT GRK
Sbjct: 192 STDAHHHGQQGVQGAYFPNRLVPQPLNVGPGAILEPSYSDGQTSSGMIGGMSDSQTPGRK 251
Query: 238 RVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEM 297
R SG V +K +ERRQKRMIKNRESAARSRAR+QAYT ELE KVSRLEEEN RL+RQ E
Sbjct: 252 RGMSGDVADKLMERRQKRMIKNRESAARSRARKQAYTNELENKVSRLEEENVRLKRQKES 311
Query: 298 E 298
+
Sbjct: 312 D 312
>UniRef100_O23965 Dc3 promoter-binding factor-2 [Helianthus annuus]
Length = 174
Score = 189 bits (480), Expect = 9e-47
Identities = 112/193 (58%), Positives = 128/193 (66%), Gaps = 22/193 (11%)
Query: 132 MTLEDFLVKAGVVGESFHGK--ESGLLRVDSNEDSRQKVSHGLHWMQYPVHSVQQQQHQY 189
MTLEDFLVKAGVV ES GK E G L + W+ Y H+VQQQ
Sbjct: 1 MTLEDFLVKAGVVAESSPGKVNEEGNLEPQETQ-----------WIGYQSHAVQQQNMIM 49
Query: 190 EKHTMPGFAAVHAIQQPFQVAGNQALDAA-ISPSSLMGTLSDTQTLGRKRVASGIVVEKT 248
H + +Q V GN +D +SP+SLMG+LSD GRKR ASG V+EKT
Sbjct: 50 AGH--------YQVQPSVTVPGNSLMDVGYMSPTSLMGSLSDRHMSGRKRFASGDVMEKT 101
Query: 249 VERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEMEKEVPTAPPPE 308
VERRQKRMIKNRESAARSRAR+QAYT ELE KVSRLEEENERLRR+ E+EK +P PPP+
Sbjct: 102 VERRQKRMIKNRESAARSRARKQAYTHELENKVSRLEEENERLRREKEVEKVIPWVPPPK 161
Query: 309 PKNQLRRTNSASF 321
PK QLRRT+SA F
Sbjct: 162 PKYQLRRTSSAPF 174
>UniRef100_Q9M7Q5 Abscisic acid responsive elements-binding factor [Arabidopsis
thaliana]
Length = 392
Score = 164 bits (416), Expect = 2e-39
Identities = 143/385 (37%), Positives = 184/385 (47%), Gaps = 82/385 (21%)
Query: 5 GGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDF 64
G T +++ PL+R SLY+LT DE+Q+ LG GK GSMN+DELLK++W+ E +
Sbjct: 12 GDTSRGNESKPLARQSSLYSLTFDELQSTLGEPGKDFGSMNMDELLKNIWTAEDTQAFMT 71
Query: 65 GGSDVAA------TAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKK-RGVD 117
S VAA GGN G Q SLTL LS+KTVDEVWK + K+ +
Sbjct: 72 TTSSVAAPGPSGFVPGGN-------GLQRQGSLTLPRTLSQKTVDEVWKYLNSKEGSNGN 124
Query: 118 RDRKSREKQQTLGEMTLEDFLVKAGVVGE------------------------------- 146
+ E+QQTLGEMTLEDFL++AGVV E
Sbjct: 125 TGTDALERQQTLGEMTLEDFLLRAGVVKEDNTQQNENSSSGFYANNGAAGLEFGFGQPNQ 184
Query: 147 ---SFHGKESGLLRVDSNEDSRQKVSHGLHWMQYPVHSVQQQQHQ------YEKHTMPGF 197
SF+G S ++ ++ KV + Q P QQ HQ + K F
Sbjct: 185 NSISFNGNNSSMI-MNQAPGLGLKVGGTMQQQQQPHQQQLQQPHQRLPPTIFPKQANVTF 243
Query: 198 AA-VHAIQQPF-----------QVAGNQALDAAISP---SSLMGTLSD--TQTLGRKRVA 240
AA V+ + + + G A SP S+ T S GR R
Sbjct: 244 AAPVNMVNRGLFETSADGPANSNMGGAGGTVTATSPGTSSAENNTWSSPVPYVFGRGR-R 302
Query: 241 SGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQ------ 294
S +EK VERRQKRMIKNRESAARSRAR+QAYT ELE ++ L+ N+ L+++
Sbjct: 303 SNTGLEKVVERRQKRMIKNRESAARSRARKQAYTLELEAEIESLKLVNQDLQKKQAEIMK 362
Query: 295 --NEMEKEVPTAPPPEPKNQ-LRRT 316
N KE PP K Q LRRT
Sbjct: 363 THNSELKEFSKQPPLLAKRQCLRRT 387
>UniRef100_Q9M7Q2 Abscisic acid responsive elements-binding factor [Arabidopsis
thaliana]
Length = 431
Score = 159 bits (402), Expect = 1e-37
Identities = 138/416 (33%), Positives = 189/416 (45%), Gaps = 110/416 (26%)
Query: 2 GSQGGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEV 61
GS +PL+R S+Y+LT DE+QN LG GK GSMN+DELLKS+W+ E +
Sbjct: 20 GSSNQMKPTGSVMPLARQSSVYSLTFDELQNTLGGPGKDFGSMNMDELLKSIWTAEEAQA 79
Query: 62 SDFGGSDVAATA-----------GGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWK--- 107
S AATA GGN+Q Q SLTL +S+KTVDEVWK
Sbjct: 80 MAM-TSAPAATAVAQPGAGIPPPGGNLQR--------QGSLTLPRTISQKTVDEVWKCLI 130
Query: 108 ----DMQGKKRGVDRDRKSREKQQTLGEMTLEDFLVKAGVVGE----------------S 147
+M+G G +QQTLGEMTLE+FL +AGVV E
Sbjct: 131 TKDGNMEGSSGGGGESNVPPGRQQTLGEMTLEEFLFRAGVVREDNCVQQMGQVNGNNNNG 190
Query: 148 FHGKESGL--------------LRVDSNEDSR---------QKVSHGLHWMQYPVHSVQQ 184
F+G + + + DS K+ + Q +QQ
Sbjct: 191 FYGNSTAAGGLGFGFGQPNQNSITFNGTNDSMILNQPPGLGLKMGGTMQQQQQQQQLLQQ 250
Query: 185 QQHQYEKHTMP-----------------GFAA-VHAIQQPFQVAGNQALD---------- 216
QQ Q ++ P F+A V+ + F A N +++
Sbjct: 251 QQQQMQQLNQPHPQQRLPQTIFPKQANVAFSAPVNITNKGFAGAANNSINNNNGLASYGG 310
Query: 217 -----AAISP---SSLMGTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRA 268
AA SP S+ +LS + + S +EK +ERRQ+RMIKNRESAARSRA
Sbjct: 311 TGVTVAATSPGTSSAENNSLSPVPYVLNRGRRSNTGLEKVIERRQRRMIKNRESAARSRA 370
Query: 269 RRQAYTQELELKVSRLEEENERLRRQN----EME----KEVPTAPPPEPKNQLRRT 316
R+QAYT ELE ++ +L++ N+ L+++ EM+ KE P + LRRT
Sbjct: 371 RKQAYTLELEAEIEKLKKTNQELQKKQAEMVEMQKNELKETSKRPWGSKRQCLRRT 426
>UniRef100_Q9LT88 Abscisic acid responsive elements-binding factor [Arabidopsis
thaliana]
Length = 442
Score = 156 bits (395), Expect = 6e-37
Identities = 129/386 (33%), Positives = 178/386 (45%), Gaps = 102/386 (26%)
Query: 2 GSQGGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEV 61
GS +PL+R S+Y+LT DE+QN LG GK GSMN+DELLKS+W+ E +
Sbjct: 20 GSSNQMKPTGSVMPLARQSSVYSLTFDELQNTLGGPGKDFGSMNMDELLKSIWTAEEAQA 79
Query: 62 SDFGGSDVAATA-----------GGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWK--- 107
S AATA GGN+Q Q SLTL +S+KTVDEVWK
Sbjct: 80 MAM-TSAPAATAVAQPGAGIPPPGGNLQR--------QGSLTLPRTISQKTVDEVWKCLI 130
Query: 108 ----DMQGKKRGVDRDRKSREKQQTLGEMTLEDFLVKAGVVGE----------------S 147
+M+G G +QQTLGEMTLE+FL +AGVV E
Sbjct: 131 TKDGNMEGSSGGGGESNVPPGRQQTLGEMTLEEFLFRAGVVREDNCVQQMGQVNGNNNNG 190
Query: 148 FHGKESGL--------------LRVDSNEDSR---------QKVSHGLHWMQYPVHSVQQ 184
F+G + + + DS K+ + Q +QQ
Sbjct: 191 FYGNSTAAGGLGFGFGQPNQNSITFNGTNDSMILNQPPGLGLKMGGTMQQQQQQQQLLQQ 250
Query: 185 QQHQYEKHTMP-----------------GFAA-VHAIQQPFQVAGNQALD---------- 216
QQ Q ++ P F+A V+ + F A N +++
Sbjct: 251 QQQQMQQLNQPHPQQRLPQTIFPKQANVAFSAPVNITNKGFAGAANNSINNNNGLASYGG 310
Query: 217 -----AAISP---SSLMGTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRA 268
AA SP S+ +LS + + S +EK +ERRQ+RMIKNRESAARSRA
Sbjct: 311 TGVTVAATSPGTSSAENNSLSPVPYVLNRGRRSNTGLEKVIERRQRRMIKNRESAARSRA 370
Query: 269 RRQAYTQELELKVSRLEEENERLRRQ 294
R+QAYT ELE ++ +L++ N+ L+++
Sbjct: 371 RKQAYTLELEAEIEKLKKTNQELQKK 396
>UniRef100_Q9FX98 F14J22.7 protein [Arabidopsis thaliana]
Length = 343
Score = 143 bits (360), Expect = 7e-33
Identities = 123/331 (37%), Positives = 157/331 (47%), Gaps = 73/331 (22%)
Query: 5 GGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDF 64
G T +++ PL+R SLY+LT DE+Q+ LG GK GSMN+DELLK++W+ E +
Sbjct: 12 GDTSRGNESKPLARQSSLYSLTFDELQSTLGEPGKDFGSMNMDELLKNIWTAEDTQAFMT 71
Query: 65 GGSDVAA------TAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKK-RGVD 117
S VAA GGN G Q SLTL LS+KTVDEVWK + K+ +
Sbjct: 72 TTSSVAAPGPSGFVPGGN-------GLQRQGSLTLPRTLSQKTVDEVWKYLNSKEGSNGN 124
Query: 118 RDRKSREKQQTLGEMTLEDFLVKAGVVGE------------------------------- 146
+ E+QQTLGEMTLEDFL++AGVV E
Sbjct: 125 TGTDALERQQTLGEMTLEDFLLRAGVVKEDNTQQNENSSSGFYANNGAAGLEFGFGQPNQ 184
Query: 147 ---SFHGKESGLLRVDSNEDSRQKVSHGLHWMQYPVHSVQQQQHQ------YEKHTMPGF 197
SF+G S ++ ++ KV + Q P QQ HQ + K F
Sbjct: 185 NSISFNGNNSSMI-MNQAPGLGLKVGGTMQQQQQPHQQQLQQPHQRLPPTIFPKQANVTF 243
Query: 198 AA-VHAIQQPF-----------QVAGNQALDAAISP---SSLMGTLSD--TQTLGRKRVA 240
AA V+ + + + G A SP S+ T S GR R
Sbjct: 244 AAPVNMVNRGLFETSADGPANSNMGGAGGTVTATSPGTSSAENNTWSSPVPYVFGRGR-R 302
Query: 241 SGIVVEKTVERRQKRMIKNRESAARSRARRQ 271
S +EK VERRQKRMIKNRESAARSRAR+Q
Sbjct: 303 SNTGLEKVVERRQKRMIKNRESAARSRARKQ 333
>UniRef100_Q8RZ35 Putative abscisic acid insensitive 5 [Oryza sativa]
Length = 388
Score = 143 bits (360), Expect = 7e-33
Identities = 119/333 (35%), Positives = 164/333 (48%), Gaps = 43/333 (12%)
Query: 5 GGTV--SESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVS 62
GGT E + PL+R S+ +LTL+E+QN L G+ GSMN+DE + ++W+ E + +
Sbjct: 26 GGTKVGGEEEIAPLARQSSILSLTLEELQNSLCEPGRNFGSMNMDEFVANIWNAEEFQAT 85
Query: 63 DFG---GSDVAATAGGNMQHNQLGGFN--SQESLTLSGDLSKKTVDEVWKDMQGKKRGVD 117
G + A GG Q S +L L +KTV+EVW ++
Sbjct: 86 TGGCKGAMEEAKVVDSGSGSGDAGGSGLCRQGSFSLPLPLCQKTVEEVWTEINQAPAHTS 145
Query: 118 RDRKS------------REKQQTLGEMTLEDFLVKAGVVGESFHGKES-GLLRVDSNEDS 164
+ ++Q TLGEMTLEDFLVKAGVV SF G+ + G V+ +
Sbjct: 146 APASALQPHAGSGGVAANDRQVTLGEMTLEDFLVKAGVVRGSFTGQAAMGSGMVNGPVNP 205
Query: 165 RQKVSHGLHWMQYPVHSVQQQQHQY-EKHTMPGFAAVHAIQQPFQVAGNQALDAAISPSS 223
Q+ G M +PV V + PG AI P A Q +SP S
Sbjct: 206 MQQGQGGP--MMFPVGPVNAMYPVMGDGMGYPGGYNGMAIVPPPPPA--QGAMVVVSPGS 261
Query: 224 LMGTLSDTQTLGRKRVASGIVVE---------------KTVERRQKRMIKNRESAARSRA 268
G + T + +G+++E KTVERRQ+RMIKNRESAARSRA
Sbjct: 262 SDGMSAMTHADMMNCIGNGMMIENGTRKRPHREDGCAEKTVERRQRRMIKNRESAARSRA 321
Query: 269 RRQAYTQELELKVSRLEEENERLRRQNEMEKEV 301
R+QAYT ELE +++ L++EN RL+ E EK V
Sbjct: 322 RKQAYTVELEAELNYLKQENARLK---EAEKTV 351
>UniRef100_Q8LK78 ABA response element binding factor [Triticum aestivum]
Length = 391
Score = 139 bits (349), Expect = 1e-31
Identities = 105/329 (31%), Positives = 162/329 (48%), Gaps = 44/329 (13%)
Query: 10 ESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDFGG--- 66
E + + +R S++ LTLDE+Q + G+ GSMN+DE + ++W+ + + + GG
Sbjct: 26 EEQAVAPARQSSIFALTLDELQYSVCEAGRNFGSMNMDEFMSNIWNADEFQAATGGGLVG 85
Query: 67 SDVAAT--AGGNMQHNQLGGFN--SQESLTLSGDLSKKTVDEVWKDMQGKKRGVDRDRKS 122
+VA AGG GG N QES +L L +KTVDEVW ++ + R V ++
Sbjct: 86 MEVAPVVGAGGGGGGLDAGGSNLARQESFSLPPPLCRKTVDEVWAEINREPRPVHAQPQA 145
Query: 123 ---------------REKQQTLGEMTLEDFLVKAGVVGESFHGKES----GLLRVDSNED 163
++Q TLGE+TLE FLVKAGVV S G ++ G++ N
Sbjct: 146 ARPSQQPPVQPSVPANDRQGTLGELTLEQFLVKAGVVRGSGAGGQAPVPVGMVHGQMNPA 205
Query: 164 SRQKVSHGLHWMQYPVHSVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAAISPSS 223
+ + + + P + + G+A + + P G +SP S
Sbjct: 206 QQGQQPGPMMYPIAPANGMFPAMGDGMGFIPNGYAGMVVVPPPPPPQGGVV--GIVSPGS 263
Query: 224 LMGTLSDTQTLGRKRVASGIVVE----------------KTVERRQKRMIKNRESAARSR 267
G + TQ + G ++E +++ERR +RMIKNRESAARSR
Sbjct: 264 SDGRSAMTQADMMNCMGEGAMMENGGTRKRGAPEDQSCERSIERRHRRMIKNRESAARSR 323
Query: 268 ARRQAYTQELELKVSRLEEENERLRRQNE 296
AR+QAYT ELE +++ L+EEN RL+ + +
Sbjct: 324 ARKQAYTVELEAELNHLKEENARLKAEEK 352
>UniRef100_Q8LK79 ABA response element binding factor [Triticum aestivum]
Length = 351
Score = 138 bits (347), Expect = 2e-31
Identities = 101/314 (32%), Positives = 152/314 (48%), Gaps = 45/314 (14%)
Query: 25 LTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDFGG-------SDVAATAGGNM 77
LTLDE+Q + G+ GSMN+DE + ++W+ E + + GG V A AGG
Sbjct: 2 LTLDELQYSVCEAGRNFGSMNMDEFMSNIWNAEEFQAATGGGLVGMEVAPVVGAGAGGGG 61
Query: 78 QHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKKRGVDRDRK---------------S 122
QES +L L +KTV+EVW ++ + R V + +
Sbjct: 62 ADAGGSNLARQESFSLPPPLCRKTVEEVWAEINREPRQVHAQPQGARASQQPPVQPPVAA 121
Query: 123 REKQQTLGEMTLEDFLVKAGVVGESFHGKES----GLLRVDSNEDSRQKVSHGLHWMQYP 178
++Q TLGEMTLE FLVKAGVV S G ++ G++ N + + + + P
Sbjct: 122 NDRQGTLGEMTLEQFLVKAGVVRGSGAGGQAPVPVGMVHAQMNPVQQGQQPGPMMYPMAP 181
Query: 179 VHSVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAAISPSSLMGTLSDTQTLGRKR 238
+ + Q G+A + + P G + +SP S G + TQ
Sbjct: 182 ANGMFQVMGDGMGFVPNGYAGMAVVPPPPPPQGGMGI---VSPGSSDGRSAMTQADMMNC 238
Query: 239 VASGIVVE----------------KTVERRQKRMIKNRESAARSRARRQAYTQELELKVS 282
+ G ++E +++ERR +RMIKNRESAARSRAR+QAYT ELE +++
Sbjct: 239 MGDGAMMENGGARKRGAPEDQSCERSIERRHRRMIKNRESAARSRARKQAYTVELEAELN 298
Query: 283 RLEEENERLRRQNE 296
L+EEN RL+ + +
Sbjct: 299 HLKEENARLKAEEK 312
>UniRef100_Q9C5Q4 BZIP protein DPBF2 [Arabidopsis thaliana]
Length = 331
Score = 135 bits (341), Expect = 1e-30
Identities = 116/312 (37%), Positives = 160/312 (51%), Gaps = 64/312 (20%)
Query: 11 SKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDFGGSDVA 70
S+ P+ R S+ +LTLDE+Q GK G+MN+DE L ++W+ ++ GG+
Sbjct: 30 SEEEPVGRQNSILSLTLDEIQM---KSGKSFGAMNMDEFLANLWTTVEENDNEGGGA--- 83
Query: 71 ATAGGNMQHN---QLGGFNSQESLTLSGDLSKKTVDEVWKDMQG--------KKRGVDRD 119
HN + Q SL+L L KKTVDEVW ++Q G + D
Sbjct: 84 --------HNDGEKPAVLPRQGSLSLPVPLCKKTVDEVWLEIQNGVQQHPPSSNSGQNSD 135
Query: 120 RKSREKQQTLGEMTLEDFLVKAGVVGESFHGKESGLLRVDSNEDS-RQKVSHGLHWMQYP 178
R +QQTLGE+TLEDFLVKAGVV E +R+ S++ + GLH
Sbjct: 136 ENIR-RQQTLGEITLEDFLVKAGVVQEPL----KTTMRMSSSDFGYNPEFGVGLHC---- 186
Query: 179 VHSVQQQQHQYEKHTMPGFAAVHAIQQPF-QVAGNQALDAAISPSSLMGTLSDTQTLG-- 235
Q Q+ Y + +V++ +PF V G S S + G Q L
Sbjct: 187 -----QNQNNYGDNR-----SVYSENRPFYSVLGE-------SSSCMTGNGRSNQYLTGL 229
Query: 236 -----RKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENER 290
+KR+ G E +ERRQ+RMIKNRESAARSRARRQAYT ELEL+++ L EEN +
Sbjct: 230 DAFRIKKRIIDG-PPEILMERRQRRMIKNRESAARSRARRQAYTVELELELNNLTEENTK 288
Query: 291 LR---RQNEMEK 299
L+ +NE ++
Sbjct: 289 LKEIVEENEKKR 300
>UniRef100_Q8RYD6 Basic leucine zipper transcription factor [Arabidopsis thaliana]
Length = 331
Score = 134 bits (337), Expect = 3e-30
Identities = 113/311 (36%), Positives = 158/311 (50%), Gaps = 62/311 (19%)
Query: 11 SKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDFGGSDVA 70
S+ P+ R S+ +LTLDE+Q GK G+MN+DE L ++W+ ++ GG+
Sbjct: 30 SEEEPVGRQNSILSLTLDEIQM---KSGKSFGAMNMDEFLANLWTTVEENDNEGGGA--- 83
Query: 71 ATAGGNMQHN---QLGGFNSQESLTLSGDLSKKTVDEVWKDMQG-------KKRGVDRDR 120
HN + Q SL+L L KKTVDEVW ++Q
Sbjct: 84 --------HNDGEKPAVLPRQGSLSLPVPLCKKTVDEVWLEIQNGVQQHPPSSNSGQNSA 135
Query: 121 KSREKQQTLGEMTLEDFLVKAGVVGESFHGKESGLLRVDSNEDS-RQKVSHGLHWMQYPV 179
++ +QQTLGE+TLEDFLVKAGVV E +R+ S++ + GLH
Sbjct: 136 ENIRRQQTLGEITLEDFLVKAGVVQEPL----KTTMRMSSSDFGYNPEFGVGLHC----- 186
Query: 180 HSVQQQQHQYEKHTMPGFAAVHAIQQPF-QVAGNQALDAAISPSSLMGTLSDTQTLG--- 235
Q Q+ Y + +V++ +PF V G S S + G Q L
Sbjct: 187 ----QNQNNYGDNR-----SVYSENRPFYSVLGE-------SSSCMTGNGRSNQYLTGLD 230
Query: 236 ----RKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERL 291
+KR+ G E +ERRQ+RMIKNRESAARSRARRQAYT ELEL+++ L EEN +L
Sbjct: 231 AFRIKKRIIDG-PPEILMERRQRRMIKNRESAARSRARRQAYTVELELELNNLTEENTKL 289
Query: 292 R---RQNEMEK 299
+ +NE ++
Sbjct: 290 KEIVEENEKKR 300
>UniRef100_Q9LXP1 BZIP transcription factor-like protein [Arabidopsis thaliana]
Length = 296
Score = 134 bits (336), Expect = 4e-30
Identities = 111/301 (36%), Positives = 153/301 (49%), Gaps = 59/301 (19%)
Query: 11 SKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDFGGSDVA 70
S+ P+ R S+ +LTLDE+Q GK G+MN+DE L ++W+ ++ GG+
Sbjct: 30 SEEEPVGRQNSILSLTLDEIQM---KSGKSFGAMNMDEFLANLWTTVEENDNEGGGA--- 83
Query: 71 ATAGGNMQHN---QLGGFNSQESLTLSGDLSKKTVDEVWKDMQG-------KKRGVDRDR 120
HN + Q SL+L L KKTVDEVW ++Q
Sbjct: 84 --------HNDGEKPAVLPRQGSLSLPVPLCKKTVDEVWLEIQNGVQQHPPSSNSGQNSA 135
Query: 121 KSREKQQTLGEMTLEDFLVKAGVVGESFHGKESGLLRVDSNEDS-RQKVSHGLHWMQYPV 179
++ +QQTLGE+TLEDFLVKAGVV E +R+ S++ + GLH
Sbjct: 136 ENIRRQQTLGEITLEDFLVKAGVVQEPL----KTTMRMSSSDFGYNPEFGVGLHC----- 186
Query: 180 HSVQQQQHQYEKHTMPGFAAVHAIQQPF-QVAGNQALDAAISPSSLMGTLSDTQTLG--- 235
Q Q+ Y + +V++ +PF V G S S + G Q L
Sbjct: 187 ----QNQNNYGDNR-----SVYSENRPFYSVLGE-------SSSCMTGNGRSNQYLTGLD 230
Query: 236 ----RKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERL 291
+KR+ G E +ERRQ+RMIKNRESAARSRARRQAYT ELEL+++ L EEN +L
Sbjct: 231 AFRIKKRIIDG-PPEILMERRQRRMIKNRESAARSRARRQAYTVELELELNNLTEENTKL 289
Query: 292 R 292
+
Sbjct: 290 K 290
>UniRef100_Q9M4H1 Putative ripening-related bZIP protein [Vitis vinifera]
Length = 447
Score = 114 bits (286), Expect = 3e-24
Identities = 73/156 (46%), Positives = 94/156 (59%), Gaps = 14/156 (8%)
Query: 2 GSQGGTVSESK--TLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAG 59
GSQ G S +PL R GS+Y+LT DE Q+ +G +GK GSMN+DELLK++WS E
Sbjct: 12 GSQQGDGSGRPPGNMPLVRQGSIYSLTFDEFQSTMGGIGKDFGSMNMDELLKNIWSAEEA 71
Query: 60 EVSDFGGSDVAATAGG-NMQHNQLGG--FNSQESLTLSGDLSKKTVDEVWKDMQGKKRGV 116
+ + AATA ++Q + G Q SLTL LS+KTVDEVWKDM + G
Sbjct: 72 QTM---AAVAAATAPPISVQEGVVAGGYLQRQGSLTLPRTLSQKTVDEVWKDMSKEYGGG 128
Query: 117 DRDRKSR------EKQQTLGEMTLEDFLVKAGVVGE 146
+D ++Q TLGEMTLE+FLV+AGVV E
Sbjct: 129 AKDGSGAGGSNLPQRQPTLGEMTLEEFLVRAGVVRE 164
Score = 77.8 bits (190), Expect = 4e-13
Identities = 48/89 (53%), Positives = 60/89 (66%), Gaps = 9/89 (10%)
Query: 236 RKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQN 295
R R SG VEK +ERRQ+RMIKNRESAARSRAR+QAYT ELE +V++L+E+NE L ++
Sbjct: 355 RGRKCSG-AVEKVIERRQRRMIKNRESAARSRARKQAYTMELEAEVAKLKEKNEELEKKQ 413
Query: 296 ----EMEK----EVPTAPPPEPKNQLRRT 316
EM+K E+ K LRRT
Sbjct: 414 AEMMEMQKNQVMEMMNLQREVKKRCLRRT 442
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.310 0.127 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 515,176,172
Number of Sequences: 2790947
Number of extensions: 20983430
Number of successful extensions: 97109
Number of sequences better than 10.0: 1203
Number of HSP's better than 10.0 without gapping: 825
Number of HSP's successfully gapped in prelim test: 398
Number of HSP's that attempted gapping in prelim test: 94789
Number of HSP's gapped (non-prelim): 2358
length of query: 321
length of database: 848,049,833
effective HSP length: 127
effective length of query: 194
effective length of database: 493,599,564
effective search space: 95758315416
effective search space used: 95758315416
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146910.4


