

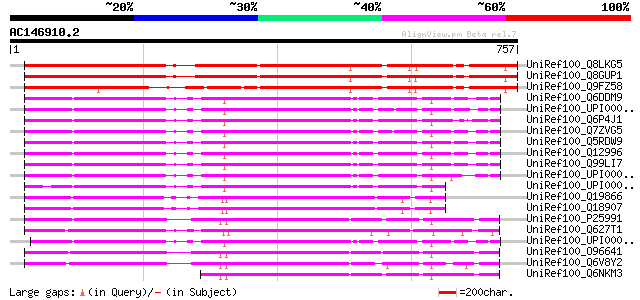
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146910.2 + phase: 1 /pseudo/partial
(757 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LKG5 Cleavage stimulation factor 77 [Arabidopsis tha... 935 0.0
UniRef100_Q8GUP1 Hypothetical protein At1g17760 [Arabidopsis tha... 935 0.0
UniRef100_Q9FZ58 F11A6.10 protein [Arabidopsis thaliana] 840 0.0
UniRef100_Q6DDM9 Cstf3-prov protein [Xenopus laevis] 365 4e-99
UniRef100_UPI0000361856 UPI0000361856 UniRef100 entry 364 5e-99
UniRef100_Q6P4J1 Hypothetical protein MGC76035 [Xenopus tropicalis] 360 7e-98
UniRef100_Q7ZVG5 Cleavage stimulation factor, 3' pre-RNA, subuni... 356 2e-96
UniRef100_Q5RDW9 Hypothetical protein DKFZp468C1613 [Pongo pygma... 356 2e-96
UniRef100_Q12996 Cleavage stimulation factor 77kDa subunit [Homo... 356 2e-96
UniRef100_Q99LI7 Cleavage stimulation factor, 3' pre-RNA, subuni... 355 3e-96
UniRef100_UPI00003ACE52 UPI00003ACE52 UniRef100 entry 354 6e-96
UniRef100_UPI00003ACE50 UPI00003ACE50 UniRef100 entry 338 5e-91
UniRef100_Q19866 Hypothetical protein F28C6.6 [Caenorhabditis el... 335 4e-90
UniRef100_Q18907 Cleavage stimulation factor [Caenorhabditis ele... 334 5e-90
UniRef100_P25991 Suppressor of forked protein [Drosophila melano... 333 9e-90
UniRef100_Q627T1 Hypothetical protein CBG00546 [Caenorhabditis b... 332 3e-89
UniRef100_UPI00001CF24F UPI00001CF24F UniRef100 entry 332 3e-89
UniRef100_O96641 Suppressor of forked protein [Drosophila virilis] 330 1e-88
UniRef100_Q6V8Y2 Suppressor of forked [Drosophila subobscura] 329 2e-88
UniRef100_Q6NKM3 SD14665p [Drosophila melanogaster] 211 6e-53
>UniRef100_Q8LKG5 Cleavage stimulation factor 77 [Arabidopsis thaliana]
Length = 734
Score = 935 bits (2417), Expect = 0.0
Identities = 489/765 (63%), Positives = 578/765 (74%), Gaps = 83/765 (10%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
IA+ATPIYEQLL LYPT+A+FWKQYVEA MAVNNDDA KQIFSRCLL CLQVPLW+CYIR
Sbjct: 22 IAQATPIYEQLLSLYPTSARFWKQYVEAQMAVNNDDATKQIFSRCLLTCLQVPLWQCYIR 81
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
FIRKV DKKGAEGQEET KAFEFML+Y+G+DIASGP+W EYIAFLKSLPA + E+ HR
Sbjct: 82 FIRKVYDKKGAEGQEETTKAFEFMLNYIGTDIASGPIWTEYIAFLKSLPALNLNEDLHRK 141
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T +RKVY RAI+TPTHH+EQLWKDY++FE++V+++LAKGL++EYQPK+NSARAVYRERKK
Sbjct: 142 TALRKVYHRAILTPTHHVEQLWKDYENFENTVNRQLAKGLVNEYQPKFNSARAVYRERKK 201
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+ +EIDWNMLAVPPTG+ K K +LS F N
Sbjct: 202 YIEEIDWNMLAVPPTGTSKEETQWVAWKKFLS---------FEKGNP------------- 239
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSIDAA 322
+RID ASS KR+I+ YEQCLM LYHYPDVWYDYA WH K+GS DAA
Sbjct: 240 --------------QRIDTASSTKRIIYAYEQCLMCLYHYPDVWYDYAEWHVKSGSTDAA 285
Query: 323 IKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRFL 382
IKVFQR+LKA+PDSEML+YA+AE+EESRGAIQ+AKK+YEN+LG S N+ LAHIQ++RFL
Sbjct: 286 IKVFQRALKAIPDSEMLKYAFAEMEESRGAIQSAKKLYENILGASTNS--LAHIQYLRFL 343
Query: 383 RRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPV 442
RR EGVE ARKYFLDARKSPSCTYHVY+A+A++AFC+DK+PK+AHN+FE GLK +M EPV
Sbjct: 344 RRAEGVEAARKYFLDARKSPSCTYHVYIAFATMAFCIDKEPKVAHNIFEEGLKLYMSEPV 403
Query: 443 YILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQR 502
YIL+YADFL RLNDD+NIRALFERALS+LP+EDS EVWKRF++FEQTYGDLAS+LKVEQR
Sbjct: 404 YILKYADFLTRLNDDRNIRALFERALSTLPVEDSAEVWKRFIQFEQTYGDLASILKVEQR 463
Query: 503 RKEAF---GEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQEWLVKNTKKVEKSI 559
KEA GEE ++ ESSLQDVVSRYS+MDLWPC+SNDLD+L+RQE LVKN K
Sbjct: 464 MKEALSGKGEEGSSPPESSLQDVVSRYSYMDLWPCTSNDLDHLARQELLVKNLNKKA--- 520
Query: 560 MLNGTTFIDKGPVASIS-TTSSKVVYPDTSKMLIYDP--------KHNPGTGAAGT---- 606
G T + P A S +SSKVVYPDTS+M++ DP NP +A +
Sbjct: 521 ---GKTNLPHVPAAIGSVASSSKVVYPDTSQMVVQDPTKKSEFASSANPVAASASSTFPS 577
Query: 607 -----------NAFDEILKATPPALVAFLANLPSVDGPTPNVDIVLSICLQSDLPTGQSV 655
+ FDEI K TPPALVAFLANLP VDGPTPNVD+VLSICLQSD PTGQ+V
Sbjct: 578 TVTATATHGSASTFDEIPKTTPPALVAFLANLPIVDGPTPNVDVVLSICLQSDFPTGQTV 637
Query: 656 KVGIPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQ 715
K ++ G P+ ++ SG ++ G+S P ++ KRK D QEEDDT +VQ
Sbjct: 638 KQSFAAK---GNPPSQNDPSGPTR------GVSQRLPRDRRATKRKDSDRQEEDDTATVQ 688
Query: 716 SQPLPQDAFRIRQFQKARAGSTSQ---TGSVSYGSALSGDLSGST 757
SQPLP D FR+RQ +KAR +TS TGS SYGSA SG+LSGST
Sbjct: 689 SQPLPTDVFRLRQMRKARGIATSSQTPTGSTSYGSAFSGELSGST 733
>UniRef100_Q8GUP1 Hypothetical protein At1g17760 [Arabidopsis thaliana]
Length = 734
Score = 935 bits (2416), Expect = 0.0
Identities = 489/765 (63%), Positives = 577/765 (74%), Gaps = 83/765 (10%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
IA+ATPIYEQLL LYPT+A+FWKQYVEA MAVNNDDA KQIFSRCLL CLQVPLW+CYIR
Sbjct: 22 IAQATPIYEQLLSLYPTSARFWKQYVEAQMAVNNDDATKQIFSRCLLTCLQVPLWQCYIR 81
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
FIRKV DKKGAEGQEET KAFEFML+Y+G+DIASGP+W EYIAFLKSLPA + E+ HR
Sbjct: 82 FIRKVYDKKGAEGQEETTKAFEFMLNYIGTDIASGPIWTEYIAFLKSLPALNLNEDLHRK 141
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T +RKVY RAI+TPTHH+EQLWKDY++FE++V+++LAKGL++EYQPK+NSARAVYRERKK
Sbjct: 142 TALRKVYHRAILTPTHHVEQLWKDYENFENTVNRQLAKGLVNEYQPKFNSARAVYRERKK 201
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+ +EIDWNMLAVPPTG+ K K +LS F N
Sbjct: 202 YIEEIDWNMLAVPPTGTSKEETQWVAWKKFLS---------FEKGNP------------- 239
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSIDAA 322
+RID ASS KR+I+ YEQCLM LYHYPDVWYDYA WH K+GS DAA
Sbjct: 240 --------------QRIDTASSTKRIIYAYEQCLMCLYHYPDVWYDYAEWHVKSGSTDAA 285
Query: 323 IKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRFL 382
IKVFQR+LKA+PDSEML+YA+AE+EESRGAIQ+AKK+YEN+LG S N+ LAHIQ++RFL
Sbjct: 286 IKVFQRALKAIPDSEMLKYAFAEMEESRGAIQSAKKLYENILGASTNS--LAHIQYLRFL 343
Query: 383 RRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPV 442
RR EGVE ARKYFLDARKSPSCTYHVY+A+A++AFC+DK+PK+AHN+FE GLK +M EPV
Sbjct: 344 RRAEGVEAARKYFLDARKSPSCTYHVYIAFATMAFCIDKEPKVAHNIFEEGLKLYMSEPV 403
Query: 443 YILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQR 502
YIL+YADFL RLNDD+NIRALFERALS+LP+EDS EVWKRF++FEQTYGDLAS+LKVEQR
Sbjct: 404 YILKYADFLTRLNDDRNIRALFERALSTLPVEDSAEVWKRFIQFEQTYGDLASILKVEQR 463
Query: 503 RKEAF---GEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQEWLVKNTKKVEKSI 559
KEA GEE ++ ESSLQDVVSRYS+MDLWPC+SNDLD+L+RQE LVKN K
Sbjct: 464 MKEALSGKGEEGSSPPESSLQDVVSRYSYMDLWPCTSNDLDHLARQELLVKNLNKKA--- 520
Query: 560 MLNGTTFIDKGPVASIS-TTSSKVVYPDTSKMLIYDP--------KHNPGTGAA------ 604
G T + P A S +SSKVVYPDTS+M++ DP NP +A
Sbjct: 521 ---GKTNLPHVPAAIGSVASSSKVVYPDTSQMVVQDPTKKSEFASSANPVAASASNTFPS 577
Query: 605 ---------GTNAFDEILKATPPALVAFLANLPSVDGPTPNVDIVLSICLQSDLPTGQSV 655
+ FDEI K TPPALVAFLANLP VDGPTPNVD+VLSICLQSD PTGQ+V
Sbjct: 578 TVTATATHGSASTFDEIPKTTPPALVAFLANLPIVDGPTPNVDVVLSICLQSDFPTGQTV 637
Query: 656 KVGIPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQ 715
K ++ G P+ ++ SG ++ G+S P ++ KRK D QEEDDT +VQ
Sbjct: 638 KQSFAAK---GNPPSQNDPSGPTR------GVSQRLPRDRRATKRKDSDRQEEDDTATVQ 688
Query: 716 SQPLPQDAFRIRQFQKARAGSTSQ---TGSVSYGSALSGDLSGST 757
SQPLP D FR+RQ +KAR +TS TGS SYGSA SG+LSGST
Sbjct: 689 SQPLPTDVFRLRQMRKARGIATSSQTPTGSTSYGSAFSGELSGST 733
>UniRef100_Q9FZ58 F11A6.10 protein [Arabidopsis thaliana]
Length = 793
Score = 840 bits (2171), Expect = 0.0
Identities = 457/769 (59%), Positives = 548/769 (70%), Gaps = 102/769 (13%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
IA+ATPIYEQLL LYPT+A+FWKQYVEA MAVNNDDA KQIFSRCLL CLQVPLW+CYIR
Sbjct: 92 IAQATPIYEQLLSLYPTSARFWKQYVEAQMAVNNDDATKQIFSRCLLTCLQVPLWQCYIR 151
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLP----AAHPQEE 138
FIRKV DKKGAEGQEET KAFEFML+Y+G+DIASGP+W EYIAFLKSLP A + E+
Sbjct: 152 FIRKVYDKKGAEGQEETTKAFEFMLNYIGTDIASGPIWTEYIAFLKSLPIPMQALNLNED 211
Query: 139 THRMTVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYR 198
HR T +RKVY RAI+TPTHH+EQLWKDY++FE++V+++LAKGL++EYQPK+NSARAVYR
Sbjct: 212 LHRKTALRKVYHRAILTPTHHVEQLWKDYENFENTVNRQLAKGLVNEYQPKFNSARAVYR 271
Query: 199 ERKKFFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLF 258
ERKK+ +EIDW ++L+V PP
Sbjct: 272 ERKKYIEEIDW------------------------NMLAV------------PP------ 289
Query: 259 LLTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGS 318
TG + +D + +D + CLM LYHYPDVWYDYA WH K+GS
Sbjct: 290 --TGTSKLDFNISQQLIGPLVDFFDFFGLRLM----CLMCLYHYPDVWYDYAEWHVKSGS 343
Query: 319 IDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALAHIQF 378
DAAIKVFQR+LKA+P ++R A S +Q+AKK+YEN+LG S N+ LAHIQ+
Sbjct: 344 TDAAIKVFQRALKAIPALLVVRILVAY---SCMILQSAKKLYENILGASTNS--LAHIQY 398
Query: 379 IRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFM 438
+RFLRR EGVE ARKYFLDARKSPSCTYHVY+A+A++AFC+DK+PK+AHN+FE GLK +M
Sbjct: 399 LRFLRRAEGVEAARKYFLDARKSPSCTYHVYIAFATMAFCIDKEPKVAHNIFEEGLKLYM 458
Query: 439 HEPVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLK 498
EPVYIL+YADFL RLNDD+NIRALFERALS+LP+EDS EVWKRF++FEQTYGDLAS+LK
Sbjct: 459 SEPVYILKYADFLTRLNDDRNIRALFERALSTLPVEDSAEVWKRFIQFEQTYGDLASILK 518
Query: 499 VEQRRKEAF---GEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQEWLVKNTKKV 555
VEQR KEA GEE ++ ESSLQDVVSRYS+MDLWPC+SNDLD+L+RQE LVKN K
Sbjct: 519 VEQRMKEALSGKGEEGSSPPESSLQDVVSRYSYMDLWPCTSNDLDHLARQELLVKNLNKK 578
Query: 556 EKSIMLNGTTFIDKGPVASIS-TTSSKVVYPDTSKMLIYDP--------KHNPGTGAA-- 604
G T + P A S +SSKVVYPDTS+M++ DP NP +A
Sbjct: 579 A------GKTNLPHVPAAIGSVASSSKVVYPDTSQMVVQDPTKKSEFASSANPVAASASN 632
Query: 605 -------------GTNAFDEILKATPPALVAFLANLPSVDGPTPNVDIVLSICLQSDLPT 651
+ FDEI K TPPALVAFLANLP VDGPTPNVD+VLSICLQSD PT
Sbjct: 633 TFPSTVTATATHGSASTFDEIPKTTPPALVAFLANLPIVDGPTPNVDVVLSICLQSDFPT 692
Query: 652 GQSVKVGIPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDT 711
GQ+VK ++ G P+ ++ SG ++ G+S P ++ KRK D QEEDDT
Sbjct: 693 GQTVKQSFAAK---GNPPSQNDPSGPTR------GVSQRLPRDRRATKRKDSDRQEEDDT 743
Query: 712 KSVQSQPLPQDAFRIRQFQKARAGSTSQ---TGSVSYGSALSGDLSGST 757
+VQSQPLP D FR+RQ +KAR +TS TGS SYGSA SG+LSGST
Sbjct: 744 ATVQSQPLPTDVFRLRQMRKARGIATSSQTPTGSTSYGSAFSGELSGST 792
>UniRef100_Q6DDM9 Cstf3-prov protein [Xenopus laevis]
Length = 718
Score = 365 bits (936), Expect = 4e-99
Identities = 237/735 (32%), Positives = 369/735 (49%), Gaps = 88/735 (11%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I +A YE+L+ +P++ +FWK Y+EA + N D ++++F RCL+ L + LW+CY+
Sbjct: 48 IDKARKTYERLVAQFPSSGRFWKLYIEAEVKAKNYDKVEKLFQRCLMKVLHIDLWKCYVS 107
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++R+ K + +E+ +A++F L +G +I S +W++YI FLK + A E R+
Sbjct: 108 YVRETKGKLPSY-KEKMAQAYDFALDKIGMEIMSYQIWVDYINFLKGVEAVGSYAENQRI 166
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VR+VYQR + P +IEQLW+DY+ +E ++ LAK +I + Y +AR V +E +
Sbjct: 167 TAVRRVYQRGCVNPMINIEQLWRDYNKYEEGINVHLAKKMIEDRSRDYMNARRVAKEYET 226
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+D N +VPP + + ++ + + K ++ W +
Sbjct: 227 VMKGLDRNAPSVPPQNTPQEAQQVEMWKKYIQ----------WEKS-------------- 262
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSI--- 319
N + ++T I KRV+F YEQCL+ L H+PD+WY+ A + ++ +
Sbjct: 263 -NPLRTEDQTLI----------TKRVMFAYEQCLLVLGHHPDIWYEAAQYLEQSSKLLAE 311
Query: 320 -----------DAAIKVFQRSLKALPDSEMLRY-AYAELEESRGAIQAAKKIYENLLGDS 367
D A +++R++ L ML Y AYA+ EESR + IY LL
Sbjct: 312 KGDMNNAKLFSDEAANIYERAISTLLKKNMLLYFAYADYEESRMKYEKTHSIYNRLLSIE 371
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ L +IQ+++F RR EG++ R F AR+ P +HVYV A + + KD +A
Sbjct: 372 DIDPTLVYIQYMKFARRAEGIKSGRMIFKKAREDPRTRHHVYVTAALMEYYCSKDTSVAF 431
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS--SLPLEDSVEVWKRFVK 485
+FE GLK + P Y+L Y D+L LN+D N R LFER L+ SLP E S E+W RF+
Sbjct: 432 KIFELGLKKYGDIPEYVLAYIDYLSHLNEDNNTRVLFERVLTSGSLPPEKSGEIWARFLA 491
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDLAS+LKVE+RR AF EE E++L +V RY FMDL+PCS+++L L
Sbjct: 492 FESNIGDLASILKVEKRRYTAFKEE-YEGKETAL--LVDRYKFMDLYPCSTSELKALG-- 546
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVY--PDTSKMLIYDPKH--NPGT 601
K+ + + + ++ + K Y PDTS+M+ + P+H PG
Sbjct: 547 ---YKDVSRAKLTALIPDPVVAPSIAPSQKDDADRKPEYPKPDTSQMIPFQPRHLAPPGL 603
Query: 602 GAAGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLPTGQSVKVG 658
F PPA V + LP GP VD +L I + LP
Sbjct: 604 HPVPGGVF-----PVPPAAVVLMKLLPPPICFQGPFVQVDEILEILRRCKLPDTVEEAAR 658
Query: 659 IPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQP 718
I + A SE +G + + + + + KR DS +E++ SV P
Sbjct: 659 I---ITGSQAEMNSEGNGPVEVNAILN----------KSVKRPNEDSDDEEEKGSV--VP 703
Query: 719 LPQDAFRIRQFQKAR 733
D +R RQ ++ R
Sbjct: 704 PVHDIYRTRQQKRIR 718
>UniRef100_UPI0000361856 UPI0000361856 UniRef100 entry
Length = 711
Score = 364 bits (935), Expect = 5e-99
Identities = 240/735 (32%), Positives = 367/735 (49%), Gaps = 90/735 (12%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I +A YE+L+ +P++ +FWK ++EA + N D ++++F RCL+ L + LW+CY+
Sbjct: 43 IDKARKTYERLVTQFPSSGRFWKLFIEAEIKAKNYDKVEKLFQRCLMKVLHIDLWKCYLS 102
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++R+ K + +E+ +A++F L +G +I S +W++YI FLK + A E R+
Sbjct: 103 YVRETKGKLPSY-KEKMAQAYDFALDKIGMEIMSYQIWVDYINFLKGVEAVGSYAENQRI 161
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VR+VYQR + P +IEQLW+DY +E ++ LAK +I + Y +AR V +E +
Sbjct: 162 TAVRRVYQRGCVNPMINIEQLWRDYSKYEEGINVHLAKKMIEDRSRDYMNARRVAKEYET 221
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+D N +VPP S + ++ + + K ++ W +
Sbjct: 222 VMKGLDRNAPSVPPQNSPQEAQQVEMWKKYIQ----------WEKS-------------- 257
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSI--- 319
N + ++T I KRV+F YEQCL+ L H+PDVWY+ A + ++ +
Sbjct: 258 -NPLRTEDQTLI----------TKRVMFAYEQCLLVLGHHPDVWYEAAQYLEQSSKLLAE 306
Query: 320 -----------DAAIKVFQRSLKALPDSEMLRY-AYAELEESRGAIQAAKKIYENLLGDS 367
D A +++R++ L ML Y ++A+ EESR + IY LL
Sbjct: 307 KGDMNNSKLFSDEAANIYERAIGTLLKKNMLLYFSFADYEESRMKYEKVHSIYNKLLVIE 366
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ L +IQ+++F RR EG++ R F AR+ P +HV+V+ A + + KD +A
Sbjct: 367 DIDPTLVYIQYMKFARRAEGIKSGRSIFKKAREDPRTRHHVFVSAALMEYYCSKDKSVAF 426
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS--LPLEDSVEVWKRFVK 485
+FE GLK + P YIL Y D+L LN+D N R LFER L+S L E S EVW RF+
Sbjct: 427 KIFELGLKKYGDIPEYILAYIDYLSHLNEDNNTRVLFERVLTSGNLSPEKSGEVWARFLA 486
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDLAS+LKVE+RR AF +E E++L +V RY FMDL+PCSS++L L
Sbjct: 487 FESNIGDLASILKVERRRFSAFKDE-YEGKETAL--LVDRYKFMDLYPCSSSELKALG-- 541
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVY--PDTSKMLIYDPKH--NPGT 601
K+ + + + ML P K Y PDT++M+ + P+H PG
Sbjct: 542 ---YKDVSRAKLAAMLPEAVVAPSAPTLK-DEADRKPEYPKPDTNQMIPFQPRHLAPPGL 597
Query: 602 GAAGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLPTGQSVKVG 658
F PP V + LP GP VD ++ + LP V
Sbjct: 598 HPVPGGVF-----PVPPPAVVLMKLLPPPTCFTGPFVQVDELMETFRRCMLPETVDAAVE 652
Query: 659 IPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQP 718
+ + P A E +G ++H V L KR DS EEDD +V P
Sbjct: 653 L---ITGRPLDAAGEGNGPMENHAVAKSL-----------KRPNADSDEEDDKGAV--AP 696
Query: 719 LPQDAFRIRQFQKAR 733
D +R RQ ++ R
Sbjct: 697 PIHDIYRSRQQKRIR 711
>UniRef100_Q6P4J1 Hypothetical protein MGC76035 [Xenopus tropicalis]
Length = 718
Score = 360 bits (925), Expect = 7e-98
Identities = 237/737 (32%), Positives = 372/737 (50%), Gaps = 92/737 (12%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I +A YE+L+ +P++ +FWK Y+EA + N D ++++F RCL+ L + LW+CY+
Sbjct: 48 IDKARKTYERLVAQFPSSGRFWKLYIEAEVKAKNYDKVEKLFQRCLMKVLHIDLWKCYVS 107
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++R+ K + +E+ +A++F L +G +I S +W++YI FLK + A E R+
Sbjct: 108 YVRETKGKLPSY-KEKMAQAYDFALDKIGMEIMSYQIWVDYINFLKGVEAVGSYAENQRI 166
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VR+VYQR + P +IEQLW+DY+ +E ++ LAK +I + Y +AR V +E +
Sbjct: 167 TAVRRVYQRGCVNPMINIEQLWRDYNKYEEGINIHLAKKMIEDRSRDYMNARRVAKEYET 226
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+D N +VPP + + ++ + + K ++ W +
Sbjct: 227 VMKGLDRNAPSVPPQNTPQEAQQVEMWKKYIQ----------WEKS-------------- 262
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSI--- 319
N + ++T I KRV+F YEQCL+ L H+PD+WY+ + ++ +
Sbjct: 263 -NPLRTEDQTLI----------TKRVMFAYEQCLLVLGHHPDIWYEAGQYLEQSSKLLAE 311
Query: 320 -----------DAAIKVFQRSLKALPDSEMLRY-AYAELEESRGAIQAAKKIYENLLGDS 367
D A +++R++ L ML Y AYA+ EESR + IY LL
Sbjct: 312 KGDMNNAKLFSDEAANIYERAISTLLKKNMLLYFAYADYEESRMKYEKTHSIYNRLLAIE 371
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ L +IQ+++F RR EG++ R F AR+ P +HVYV A + + KD +A
Sbjct: 372 DIDPTLVYIQYMKFARRAEGIKSGRMIFKKAREDPRTRHHVYVTAALMEYYCSKDTSVAF 431
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS--SLPLEDSVEVWKRFVK 485
+FE GLK + P Y+L Y D+L LN+D N R LFER L+ SLP E S E+W RF+
Sbjct: 432 KIFELGLKKYGDIPEYVLAYIDYLSHLNEDNNTRVLFERVLTSGSLPPEKSGEIWARFLA 491
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDLAS+LKVE+RR AF EE E++L +V RY FMDL+PCS+++L L
Sbjct: 492 FESNIGDLASILKVEKRRYTAFKEE-YEGKETAL--LVDRYKFMDLYPCSTSELKALG-- 546
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVY--PDTSKMLIYDPKH--NPGT 601
K+ + + + ++ + K Y PDTS+M+ + P+H PG
Sbjct: 547 ---YKDVSRAKLAALIPDPVIAPSIAPSLKDDVDRKPEYPKPDTSQMIPFQPRHLAPPGL 603
Query: 602 GAAGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLP--TGQSVK 656
F PPA V + LP GP VD ++ I + LP ++V+
Sbjct: 604 HPVPGGVF-----PVPPAAVILMKLLPPPICFQGPFVQVDEIMEILRRCKLPDTVEEAVR 658
Query: 657 VGIPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQS 716
+ SQ A + G+ PV+ + + KR DS +E++ SV
Sbjct: 659 IITGSQ-------AEMNMEGNG---PVE-----VNTLLNKSVKRPNEDSDDEEEKGSV-- 701
Query: 717 QPLPQDAFRIRQFQKAR 733
P D +R RQ ++ R
Sbjct: 702 VPPVHDIYRTRQQKRIR 718
>UniRef100_Q7ZVG5 Cleavage stimulation factor, 3' pre-RNA, subunit 3 [Brachydanio
rerio]
Length = 716
Score = 356 bits (913), Expect = 2e-96
Identities = 236/735 (32%), Positives = 367/735 (49%), Gaps = 89/735 (12%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I +A YE+L+ +P++ +FWK Y+EA + N D ++++F RCL+ L + LW+CY+
Sbjct: 47 IDKARKTYERLVAQFPSSGRFWKLYIEAEIKAKNYDKVEKLFQRCLMKVLHIDLWKCYLS 106
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++R+ K + +E+ +A++F L +G +I S +W++YI FLK + A E R+
Sbjct: 107 YVRETKGKLPSY-KEKMPQAYDFALDKIGMEIMSYQIWVDYINFLKGVEAVGSYAENQRI 165
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VR+VYQR + P +IEQLW+DY +E ++ LAK +I + Y +AR V +E +
Sbjct: 166 TAVRRVYQRGCVNPMINIEQLWRDYSKYEEGINVHLAKKMIEDRSRDYMNARRVAKEYET 225
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+D N +VPP S + ++ + + K ++ W +
Sbjct: 226 VMKGLDRNAPSVPPQNSPQEAQQVEMWKKYIQ----------WEKS-------------- 261
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSI--- 319
N + ++T I KRV+F YEQCL+ L H+PD+WY+ A + ++ +
Sbjct: 262 -NPLRTEDQTLI----------TKRVMFAYEQCLLVLGHHPDIWYEAAQYLEQSSKLLAE 310
Query: 320 -----------DAAIKVFQRSLKALPDSEMLRY-AYAELEESRGAIQAAKKIYENLLGDS 367
D A +++R++ L ML Y ++A+ EESR + IY LL
Sbjct: 311 KGDMNNAKLFSDEAANIYERAIGTLLKKNMLLYFSFADYEESRMKHEKVHSIYNRLLAIE 370
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ L +IQ+++F RR EG++ R F AR+ P +HVYV A + + KD +A
Sbjct: 371 DIDPTLVYIQYMKFARRAEGIKSGRSIFKKAREDPRTRHHVYVTAALMEYYCSKDKSVAF 430
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS--SLPLEDSVEVWKRFVK 485
+FE GLK + P YIL Y D+L LN+D N R LFER L+ SL E S E+W RF+
Sbjct: 431 KIFELGLKKYGDIPEYILAYIDYLSHLNEDNNTRVLFERVLTSGSLSPEKSGEIWARFLA 490
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDLAS+LKVE+RR AF +E E++L +V RY FMDL+PCS ++L L
Sbjct: 491 FESNIGDLASILKVERRRFMAFKDE-YEGKETAL--LVDRYKFMDLYPCSPSELKALG-- 545
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVY--PDTSKMLIYDPKH--NPGT 601
K+ + + + ++ P A K Y PDT +M+ + P+H PG
Sbjct: 546 ---YKDVSRAKYASLIPEAVVAPSTP-ALKDEADRKPEYPKPDTCQMIPFQPRHLAPPGL 601
Query: 602 GAAGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLPTGQSVKVG 658
F PPA V + LP GP VD ++ + LP V
Sbjct: 602 HPVPGGVF-----PVPPAAVVLMKLLPPPSCFTGPFVQVDEMMEALRRCVLPDTVDAAVE 656
Query: 659 IPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQP 718
+ + +SE +G ++H V + + KR DS EE+D S+ P
Sbjct: 657 L---ITGKQLEMSSEGNGPVENHSVAN----------KSLKRPNADSDEEEDKGSI--AP 701
Query: 719 LPQDAFRIRQFQKAR 733
D +R RQ ++ R
Sbjct: 702 PIHDIYRARQQKRIR 716
>UniRef100_Q5RDW9 Hypothetical protein DKFZp468C1613 [Pongo pygmaeus]
Length = 717
Score = 356 bits (913), Expect = 2e-96
Identities = 234/735 (31%), Positives = 367/735 (49%), Gaps = 88/735 (11%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I +A YE+L+ +P++ +FWK Y+EA + N D ++++F RCL+ L + LW+CY+
Sbjct: 47 IDKARKTYERLVAQFPSSGRFWKLYIEAEIKAKNYDKVEKLFQRCLMKVLHIDLWKCYLS 106
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++R+ K + +E+ +A++F L +G +I S +W++YI FLK + A E R+
Sbjct: 107 YVRETKGKLPSY-KEKMAQAYDFALDKIGMEIMSYQIWVDYINFLKGVEAVGSYAENQRI 165
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VR+VYQR + P +IEQLW+DY+ +E ++ LAK +I + Y +AR V +E +
Sbjct: 166 TAVRRVYQRGCVNPMINIEQLWRDYNKYEEGINIHLAKKMIEDRSRDYMNARRVAKEYET 225
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+D N +VPP + + ++ + + K ++ W +
Sbjct: 226 VMKGLDRNAPSVPPQNTPQEAQQVDMWKKYIQ----------WEKS-------------- 261
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSI--- 319
N + ++T I KRV+F YEQCL+ L H+PD+WY+ A + ++ +
Sbjct: 262 -NPLRTEDQTLI----------TKRVMFAYEQCLLVLGHHPDIWYEAAQYLEQSSKLLAE 310
Query: 320 -----------DAAIKVFQRSLKALPDSEMLRY-AYAELEESRGAIQAAKKIYENLLGDS 367
D A +++R++ L ML Y AYA+ EESR + IY LL
Sbjct: 311 KGDMNNAKLFSDEAANIYERAISTLLKKNMLLYFAYADYEESRMKYEKVHSIYNRLLAIE 370
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ L +IQ+++F RR EG++ R F AR+ +HVYV A + + KD +A
Sbjct: 371 DIDPTLVYIQYMKFARRAEGIKSGRMIFKKAREDTRTRHHVYVTAALMEYYCSKDKSVAF 430
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS--SLPLEDSVEVWKRFVK 485
+FE GLK + P Y+L Y D+L LN+D N R LFER L+ SLP E S E+W RF+
Sbjct: 431 KIFELGLKKYGDIPEYVLAYIDYLSHLNEDNNTRVLFERVLTSGSLPPEKSGEIWARFLA 490
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDLAS+LKVE+RR AF EE E++L +V RY FMDL+PCS+++L L
Sbjct: 491 FESNIGDLASILKVEKRRFTAFKEE-YEGKETAL--LVDRYKFMDLYPCSASELKALG-- 545
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVY--PDTSKMLIYDPKH--NPGT 601
K+ + + + ++ K Y PDT +M+ + P+H PG
Sbjct: 546 ---YKDVSRAKLAAIIPDPVVAPSIVPVLKDEVDRKPEYPKPDTQQMIPFQPRHLAPPGL 602
Query: 602 GAAGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLPTGQSVKVG 658
F PPA V + LP GP VD ++ I + +P V
Sbjct: 603 HPVPGGVF-----PVPPAAVVLMKLLPPPICFQGPFVQVDELMEIFRRCKIPNTVEEAVR 657
Query: 659 IPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQP 718
I + G E +G +S+ V + + KR DS E+++ +V P
Sbjct: 658 I---ITGGAPELAVEGNGPVESNAVLT----------KAVKRPNEDSDEDEEKGAV--VP 702
Query: 719 LPQDAFRIRQFQKAR 733
D +R RQ ++ R
Sbjct: 703 PVHDIYRARQQKRIR 717
>UniRef100_Q12996 Cleavage stimulation factor 77kDa subunit [Homo sapiens]
Length = 717
Score = 356 bits (913), Expect = 2e-96
Identities = 234/735 (31%), Positives = 367/735 (49%), Gaps = 88/735 (11%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I +A YE+L+ +P++ +FWK Y+EA + N D ++++F RCL+ L + LW+CY+
Sbjct: 47 IDKARKTYERLVAQFPSSGRFWKLYIEAEIKAKNYDKVEKLFQRCLMKVLHIDLWKCYLS 106
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++R+ K + +E+ +A++F L +G +I S +W++YI FLK + A E R+
Sbjct: 107 YVRETKGKLPSY-KEKMAQAYDFALDKIGMEIMSYQIWVDYINFLKGVEAVGSYAENQRI 165
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VR+VYQR + P +IEQLW+DY+ +E ++ LAK +I + Y +AR V +E +
Sbjct: 166 TAVRRVYQRGCVNPMINIEQLWRDYNKYEEGINIHLAKKMIEDRSRDYMNARRVAKEYET 225
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+D N +VPP + + ++ + + K ++ W +
Sbjct: 226 VMKGLDRNAPSVPPQNTPQEAQQVDMWKKYIQ----------WEKS-------------- 261
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSI--- 319
N + ++T I KRV+F YEQCL+ L H+PD+WY+ A + ++ +
Sbjct: 262 -NPLRTEDQTLI----------TKRVMFAYEQCLLVLGHHPDIWYEAAQYLEQSSKLLAE 310
Query: 320 -----------DAAIKVFQRSLKALPDSEMLRY-AYAELEESRGAIQAAKKIYENLLGDS 367
D A +++R++ L ML Y AYA+ EESR + IY LL
Sbjct: 311 KGDMNNAKLFSDEAANIYERAISTLLKKNMLLYFAYADYEESRMKYEKVHSIYNRLLAIE 370
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ L +IQ+++F RR EG++ R F AR+ +HVYV A + + KD +A
Sbjct: 371 DIDPTLVYIQYMKFARRAEGIKSGRMIFKKAREDTRTRHHVYVTAALMEYYCSKDKSVAF 430
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS--SLPLEDSVEVWKRFVK 485
+FE GLK + P Y+L Y D+L LN+D N R LFER L+ SLP E S E+W RF+
Sbjct: 431 KIFELGLKKYGDIPEYVLAYIDYLSHLNEDNNTRVLFERVLTSGSLPPEKSGEIWARFLA 490
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDLAS+LKVE+RR AF EE E++L +V RY FMDL+PCS+++L L
Sbjct: 491 FESNIGDLASILKVEKRRFTAFKEE-YEGKETAL--LVDRYKFMDLYPCSASELKALG-- 545
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVY--PDTSKMLIYDPKH--NPGT 601
K+ + + + ++ K Y PDT +M+ + P+H PG
Sbjct: 546 ---YKDVSRAKLAAIIPDPVVAPSIVPVLKDEVDRKPEYPKPDTQQMIPFQPRHLAPPGL 602
Query: 602 GAAGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLPTGQSVKVG 658
F PPA V + LP GP VD ++ I + +P V
Sbjct: 603 HPVPGGVF-----PVPPAAVVLMKLLPPPICFQGPFVQVDELMEIFRRCKIPNTVEEAVR 657
Query: 659 IPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQP 718
I + G E +G +S+ V + + KR DS E+++ +V P
Sbjct: 658 I---ITGGAPELAVEGNGPVESNAVLT----------KAVKRPNEDSDEDEEKGAV--VP 702
Query: 719 LPQDAFRIRQFQKAR 733
D +R RQ ++ R
Sbjct: 703 PVHDIYRARQQKRIR 717
>UniRef100_Q99LI7 Cleavage stimulation factor, 3' pre-RNA, subunit 3 [Mus musculus]
Length = 717
Score = 355 bits (911), Expect = 3e-96
Identities = 234/735 (31%), Positives = 366/735 (48%), Gaps = 88/735 (11%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I +A YE+L+ +P++ +FWK Y+EA + N D ++++F RCL+ L + LW+CY+
Sbjct: 47 IDKARKTYERLVAQFPSSGRFWKLYIEAEIKAKNYDKVEKLFQRCLMKVLHIDLWKCYLS 106
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++R+ K + +E+ +A++F L +G +I S +W++YI FLK + A E R+
Sbjct: 107 YVRETKGKLPSY-KEKMAQAYDFALDKIGMEIMSYQIWVDYINFLKGVEAVGSYAENQRI 165
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VR+VYQR + P +IEQLW+DY+ +E ++ LAK +I + Y +AR V +E +
Sbjct: 166 TAVRRVYQRGCVNPMINIEQLWRDYNKYEEGINIHLAKKMIEDRSRDYMNARRVAKEYET 225
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+D N +VPP + + ++ + + K ++ W +
Sbjct: 226 VMKGLDRNAPSVPPQNTPQEAQQVDMWKKYIQ----------WEKS-------------- 261
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSI--- 319
N + ++T I KRV+F YEQCL+ L H+PD+WY+ A + ++ +
Sbjct: 262 -NPLRTEDQTLI----------TKRVMFAYEQCLLVLGHHPDIWYEAAQYLEQSSKLLAE 310
Query: 320 -----------DAAIKVFQRSLKALPDSEMLRY-AYAELEESRGAIQAAKKIYENLLGDS 367
D A +++R++ L ML Y AYA+ EESR + IY LL
Sbjct: 311 KGDMNNAKLFSDEAANIYERAISTLLKKNMLLYFAYADYEESRMKYEKVHSIYNRLLAIE 370
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ L +IQ+++F RR EG++ R F AR+ +HVYV A + + KD +A
Sbjct: 371 DIDPTLVYIQYMKFARRAEGIKSGRMIFKKAREDARTRHHVYVTAALMEYYCSKDKSVAF 430
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS--SLPLEDSVEVWKRFVK 485
+FE GLK + P Y+L Y D+L LN+D N R LFER L+ SLP E S E+W RF+
Sbjct: 431 KIFELGLKKYGDIPEYVLAYIDYLSHLNEDNNTRVLFERVLTSGSLPPEKSGEIWARFLA 490
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDLAS+LKVE+RR AF EE E++L +V RY FMDL+PCS+++L L
Sbjct: 491 FESNIGDLASILKVEKRRFTAFREE-YEGKETAL--LVDRYKFMDLYPCSASELKALG-- 545
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVY--PDTSKMLIYDPKH--NPGT 601
K+ + + + ++ K Y PDT +M+ + P+H PG
Sbjct: 546 ---YKDVSRAKLAAIIPDPVVAPSIVPVLKDEVDRKPEYPKPDTQQMIPFQPRHLAPPGL 602
Query: 602 GAAGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLPTGQSVKVG 658
F PPA V + LP GP VD ++ I + +P V
Sbjct: 603 HPVPGGVF-----PVPPAAVVLMKLLPPPICFQGPFVQVDELMEIFRRCKIPNTVEEAVR 657
Query: 659 IPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQP 718
I + G E +G +S V + + KR DS E+++ +V P
Sbjct: 658 I---ITGGAPELAVEGNGPVESSAVLT----------KAVKRPNEDSDEDEEKGAV--VP 702
Query: 719 LPQDAFRIRQFQKAR 733
D +R RQ ++ R
Sbjct: 703 PVHDIYRARQQKRIR 717
>UniRef100_UPI00003ACE52 UPI00003ACE52 UniRef100 entry
Length = 708
Score = 354 bits (908), Expect = 6e-96
Identities = 233/740 (31%), Positives = 366/740 (48%), Gaps = 98/740 (13%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I +A YE+L+ +P++ +FWK Y+EA + N D ++++F RCL+ L + LW+CY+
Sbjct: 38 IDKARKTYERLVAQFPSSGRFWKLYIEAEIKAKNYDKVEKLFQRCLMKVLHIDLWKCYLS 97
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++R+ K + +E+ +A++F L +G +I S +W++YI FLK + A E R+
Sbjct: 98 YVRETKGKLPSY-KEKMAQAYDFALDKIGMEIMSYQIWVDYINFLKGVEAVGSYAENQRI 156
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VR+VYQR + P +IEQLW+DY+ +E ++ LAK +I + Y +AR V +E +
Sbjct: 157 TAVRRVYQRGCVNPMINIEQLWRDYNKYEEGINIHLAKKMIEDRSRDYMNARRVAKEYET 216
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+D N +VPP + + ++ + + K ++ W +
Sbjct: 217 VMKGLDRNAPSVPPQNTPQEAQQVDMWKKYIQ----------WEKS-------------- 252
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSI--- 319
N + ++T I KRV+F YEQCL+ L H+PD+WY+ A + ++ +
Sbjct: 253 -NPLRTEDQTLI----------TKRVMFAYEQCLLVLGHHPDIWYEAAQYLEQSSKLLAE 301
Query: 320 -----------DAAIKVFQRSLKALPDSEMLRY-AYAELEESRGAIQAAKKIYENLLGDS 367
D A +++R++ L ML Y AYA+ EESR + IY LL
Sbjct: 302 KGDMNNAKLFSDEAANIYERAISTLLKKNMLLYFAYADYEESRMKYEKVHSIYNRLLAIE 361
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ L +IQ+++F RR EG++ R F AR+ +HVYV A + + KD +A
Sbjct: 362 DIDPTLVYIQYMKFARRAEGIKSGRMIFKKAREDTRTRHHVYVTAALMEYYCSKDKSVAF 421
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS--SLPLEDSVEVWKRFVK 485
+FE GLK + P Y+L Y D+L LN+D N R LFER L+ SLP E S E+W RF+
Sbjct: 422 KIFELGLKKYGDIPEYVLAYIDYLSHLNEDNNTRVLFERVLTSGSLPPEKSGEIWARFLA 481
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDLAS+LKVE+RR AF EE E++L +V RY FMDL+PCS+++L L
Sbjct: 482 FESNIGDLASILKVEKRRFTAFKEE-YEGKETAL--LVDRYKFMDLYPCSASELKALG-- 536
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVY--PDTSKMLIYDPKH--NPGT 601
K+ + + + ++ K Y PDT +M+ + P+H PG
Sbjct: 537 ---YKDVSRAKLAAIIPDPVVAPSIVPVLKDEVDRKPEYPKPDTQQMIPFQPRHLAPPGL 593
Query: 602 GAAGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLP--TGQSVK 656
F PPA V + LP GP VD ++ I + LP ++V+
Sbjct: 594 HPVPGGVF-----PVPPAAVVLMKLLPPPICFQGPFVQVDELMEIFRRCKLPDTVDEAVR 648
Query: 657 V---GIPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKS 713
+ G+P G P + + KR DS ++++ S
Sbjct: 649 IITGGLPEIAVEGNGPVENNAM------------------LNKAVKRSNEDSDDDEEKGS 690
Query: 714 VQSQPLPQDAFRIRQFQKAR 733
V P D +R RQ ++ R
Sbjct: 691 V--VPPVHDIYRARQQKRIR 708
>UniRef100_UPI00003ACE50 UPI00003ACE50 UniRef100 entry
Length = 653
Score = 338 bits (866), Expect = 5e-91
Identities = 214/652 (32%), Positives = 330/652 (49%), Gaps = 84/652 (12%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I +A YE+L+ +P++ +FWK Y+EA ++F RCL+ L + LW+CY+
Sbjct: 36 IDKARKTYERLVAQFPSSGRFWKLYIEA-----------ELFQRCLMKVLHIDLWKCYLS 84
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++R+ K + +E+ +A++F L +G +I S +W++YI FLK + A E R+
Sbjct: 85 YVRETKGKLPSY-KEKMAQAYDFALDKIGMEIMSYQIWVDYINFLKGVEAVGSYAENQRI 143
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VR+VYQR + P +IEQLW+DY+ +E ++ LAK +I + Y +AR V +E +
Sbjct: 144 TAVRRVYQRGCVNPMINIEQLWRDYNKYEEGINIHLAKKMIEDRSRDYMNARRVAKEYET 203
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+D N +VPP + + ++ + + K ++ W +
Sbjct: 204 VMKGLDRNAPSVPPQNTPQEAQQVDMWKKYIQ----------WEKS-------------- 239
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSI--- 319
N + ++T I KRV+F YEQCL+ L H+PD+WY+ A + ++ +
Sbjct: 240 -NPLRTEDQTLI----------TKRVMFAYEQCLLVLGHHPDIWYEAAQYLEQSSKLLAE 288
Query: 320 -----------DAAIKVFQRSLKALPDSEMLRY-AYAELEESRGAIQAAKKIYENLLGDS 367
D A +++R++ L ML Y AYA+ EESR + IY LL
Sbjct: 289 KGDMNNAKLFSDEAANIYERAISTLLKKNMLLYFAYADYEESRMKYEKVHSIYNRLLAIE 348
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ L +IQ+++F RR EG++ R F AR+ +HVYV A + + KD +A
Sbjct: 349 DIDPTLVYIQYMKFARRAEGIKSGRMIFKKAREDTRTRHHVYVTAALMEYYCSKDKSVAF 408
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS--SLPLEDSVEVWKRFVK 485
+FE GLK + P Y+L Y D+L LN+D N R LFER L+ SLP E S E+W RF+
Sbjct: 409 KIFELGLKKYGDIPEYVLAYIDYLSHLNEDNNTRVLFERVLTSGSLPPEKSGEIWARFLA 468
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDLAS+LKVE+RR AF EE E++L +V RY FMDL+PCS+++L L
Sbjct: 469 FESNIGDLASILKVEKRRFTAFKEE-YEGKETAL--LVDRYKFMDLYPCSASELKALG-- 523
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVY--PDTSKMLIYDPKH--NPGT 601
K+ + + + ++ K Y PDT +M+ + P+H PG
Sbjct: 524 ---YKDVSRAKLAAIIPDPVVAPSIVPVLKDEVDRKPEYPKPDTQQMIPFQPRHLAPPGL 580
Query: 602 GAAGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLP 650
F PPA V + LP GP VD ++ I + LP
Sbjct: 581 HPVPGGVF-----PVPPAAVVLMKLLPPPICFQGPFVQVDELMEIFRRCKLP 627
>UniRef100_Q19866 Hypothetical protein F28C6.6 [Caenorhabditis elegans]
Length = 735
Score = 335 bits (858), Expect = 4e-90
Identities = 211/661 (31%), Positives = 342/661 (50%), Gaps = 77/661 (11%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I + YE L++ +P + ++WK Y+E + N + ++++FSRCL++ L + LW+CYI
Sbjct: 35 IDQERDFYESLVKQFPNSGRYWKAYIEHELRSKNFENVEKLFSRCLVSVLNIDLWKCYIH 94
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++ + ++ + +EE KA++F L VG D+ + ++ EYIAFLK +PA E R+
Sbjct: 95 YVFETKGQRD-QYREEMAKAYDFALEKVGMDVQAYSIFTEYIAFLKKVPAVGQYAENQRI 153
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VRK+YQ+A+ TP H++E +W DY ++E +++ LA+ LI+E +Y +AR V ++ ++
Sbjct: 154 TAVRKIYQKALATPMHNLELIWNDYCTYEKAINITLAEKLIAERGKEYQNARRVEKDLQQ 213
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
++ ++VPP G+ K + L K N + W
Sbjct: 214 MTRGLNRQAVSVPPKGTATEFKQVELWK----------NLIAWEKT-------------- 249
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKA------ 316
N + E +RV++TYEQ L+ L +YPD+WY+ A + +A
Sbjct: 250 -NPLQTEE----------YGQHARRVVYTYEQSLLCLGYYPDIWYEAAMFLQEASHTLDE 298
Query: 317 -GSIDAA-------IKVFQRSLKAL-PDSEMLRYAYAELEESRGAIQAAKKIYENLLGDS 367
G + A I +++R++ L +S++L +AYA+ +E +A K IY+ LLG
Sbjct: 299 KGDVKMAQVLKLETISLYERAITGLMKESKLLYFAYADFQEEHKQFEAVKNIYDRLLGIE 358
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
L ++Q +RF+RR+EG AR F AR+ Y V+VA A + + KD ++A
Sbjct: 359 HINPTLTYVQLMRFIRRSEGPNNARLVFKRAREDKRTGYQVFVAAALLEYNCMKDKEVAI 418
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS--LPLEDSVEVWKRFVK 485
VF+ GLK + +EP + L YADFL LN+D N R +FER L+S LP + S+ +W RF+
Sbjct: 419 RVFKLGLKKYENEPEFGLAYADFLSNLNEDNNTRVVFERILTSSKLPADKSIRIWDRFLD 478
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDLAS+LKVE+RRK A+ E + + V+ RY FMDL PCS L +
Sbjct: 479 FESCVGDLASILKVEKRRKTAYEEAQKDQTMNHSMLVIDRYKFMDLMPCSGEQLKLIGYN 538
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVY----------PDTSKMLIYDP 595
+K T+ + + GP A+ + + PD S+M+ + P
Sbjct: 539 --ALKGTESIAGPSFVGSKNVPTHGPQAASAIMGGAGGHADVARYGFPRPDISQMIPFKP 596
Query: 596 KHNPGTGAAGTNAFDEILKAT--PPALVAFLANL----PSVDGPTPNVDIVLSICLQSDL 649
+ N T +F + PP VA L +L GP NV+++ ++ L
Sbjct: 597 RVN------CTASFHPVPGGVFPPPQSVAHLMSLLPPPTCFIGPFINVELLCNMINNMQL 650
Query: 650 P 650
P
Sbjct: 651 P 651
>UniRef100_Q18907 Cleavage stimulation factor [Caenorhabditis elegans]
Length = 735
Score = 334 bits (857), Expect = 5e-90
Identities = 211/661 (31%), Positives = 342/661 (50%), Gaps = 77/661 (11%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I + YE L++ +P + ++WK Y+E + N + ++++FSRCL++ L + LW+CYI
Sbjct: 35 IDQERDFYESLVKQFPNSGRYWKAYIEHELRSKNFENVEKLFSRCLVSVLNIDLWKCYIH 94
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++ + ++ + +EE KA++F L VG D+ + ++ EYIAFLK +PA E R+
Sbjct: 95 YVFETKGQRD-QYREEMAKAYDFALEKVGMDVQAYSIFTEYIAFLKKVPAVGQYAENQRI 153
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VRK+YQ+A+ TP H++E +W DY ++E +++ LA+ LI+E +Y +AR V ++ ++
Sbjct: 154 TAVRKIYQKALATPMHNLELIWNDYCTYEKAINITLAEKLIAERGKEYQNARRVEKDLQQ 213
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
++ ++VPP G+ K + L K N + W
Sbjct: 214 MTRGLNRQAVSVPPKGTATEFKQVELWK----------NLIAWEKT-------------- 249
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKA------ 316
N + E +RV++TYEQ L+ L +YPD+WY+ A + +A
Sbjct: 250 -NPLQTEE----------YGQHARRVVYTYEQSLLCLGYYPDIWYEAAMFLQEASHTLDE 298
Query: 317 -GSIDAA-------IKVFQRSLKAL-PDSEMLRYAYAELEESRGAIQAAKKIYENLLGDS 367
G + A I +++R++ L +S++L +AYA+ +E +A K IY+ LLG
Sbjct: 299 KGDVKMAQVLKLETISLYERAITGLMKESKLLYFAYADFQEEHKQFEAVKNIYDRLLGIE 358
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
L ++Q +RF+RR+EG AR F AR+ Y V+VA A + + KD ++A
Sbjct: 359 HINPTLTYVQLMRFVRRSEGPNNARLVFKRAREDKRTGYQVFVAAALLEYNCMKDKEVAI 418
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS--LPLEDSVEVWKRFVK 485
VF+ GLK + +EP + L YADFL LN+D N R +FER L+S LP + S+ +W RF+
Sbjct: 419 RVFKLGLKKYENEPEFGLAYADFLSNLNEDNNTRVVFERILTSSKLPADKSIRIWDRFLD 478
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDLAS+LKVE+RRK A+ E + + V+ RY FMDL PCS L +
Sbjct: 479 FESCVGDLASILKVEKRRKTAYEEAQKDQTMNHSMLVIDRYKFMDLMPCSGEQLKLIGYN 538
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVY----------PDTSKMLIYDP 595
+K T+ + + GP A+ + + PD S+M+ + P
Sbjct: 539 --ALKGTESIAGPSFVGSKNVPTHGPQAASAIMGGAGGHADVARYGFPRPDISQMIPFKP 596
Query: 596 KHNPGTGAAGTNAFDEILKAT--PPALVAFLANL----PSVDGPTPNVDIVLSICLQSDL 649
+ N T +F + PP VA L +L GP NV+++ ++ L
Sbjct: 597 RVN------CTASFHPVPGGVFPPPQSVAHLMSLLPPPTCFIGPFINVELLCNMINNMQL 650
Query: 650 P 650
P
Sbjct: 651 P 651
>UniRef100_P25991 Suppressor of forked protein [Drosophila melanogaster]
Length = 733
Score = 333 bits (855), Expect = 9e-90
Identities = 221/733 (30%), Positives = 363/733 (49%), Gaps = 76/733 (10%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I E +YE L+ ++PT A++WK Y+E M + ++++F RCL+ L + LW+ Y+
Sbjct: 48 IHEVRSLYESLVNVFPTTARYWKLYIEMEMRSRYYERVEKLFQRCLVKILNIDLWKLYLT 107
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++++ +E+ +A++F L +G D+ S +W +YI FL+ + A E ++
Sbjct: 108 YVKETKSGLSTH-KEKMAQAYDFALEKIGMDLHSFSIWQDYIYFLRGVEAVGNYAENQKI 166
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VR+VYQ+A++TP IEQLWKDY +FE +++ +++ + E Y +AR V +E +
Sbjct: 167 TAVRRVYQKAVVTPIVGIEQLWKDYIAFEQNINPIISEKMSLERSKDYMNARRVAKELEY 226
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
++ N+ AVPPT + + K + L K +++
Sbjct: 227 HTKGLNRNLPAVPPTLTKEEVKQVELWKRFITY--------------------------- 259
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWH-------AK 315
E S + D A +RV+F EQCL+ L H+P VW+ + + +
Sbjct: 260 --------EKSNPLRTEDTALVTRRVMFATEQCLLVLTHHPAVWHQASQFLDTSARVLTE 311
Query: 316 AGSIDAA-------IKVFQRSLKA-LPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDS 367
G + AA + +RS+ L + +L +AYA+ EE R + +Y LL
Sbjct: 312 KGDVQAAKIFADECANILERSINGVLNRNALLYFAYADFEEGRLKYEKVHTMYNKLLQLP 371
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ L ++Q+++F RR EG++ AR F AR+ YH++VA A + + KD ++A
Sbjct: 372 DIDPTLVYVQYMKFARRAEGIKSARSIFKKAREDVRSRYHIFVAAALMEYYCSKDKEIAF 431
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS--LPLEDSVEVWKRFVK 485
+FE GLK F P Y++ Y D+L LN+D N R LFER LSS L SVEVW RF++
Sbjct: 432 RIFELGLKRFGGSPEYVMCYIDYLSHLNEDNNTRVLFERVLSSGGLSPHKSVEVWNRFLE 491
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDL+S++KVE+RR F E +V RY F+DL+PC+S +L ++
Sbjct: 492 FESNIGDLSSIVKVERRRSAVF-ENLKEYEGKETAQLVDRYKFLDLYPCTSTELKSIGYA 550
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVYPDTSKMLIYDPK--HNPGTGA 603
E + KV G + G V + S + + PD S+M+ + P+ +PG
Sbjct: 551 ENVGIILNKVG-----GGAQSQNTGEVETDSEATPPLPRPDFSQMIPFKPRPCAHPGAHP 605
Query: 604 AGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLPTGQSVKVGIP 660
F + PPAL A A LP S GP +V+++ I ++ +LP G
Sbjct: 606 LAGGVFPQ-----PPALAALCATLPPPNSFRGPFVSVELLFDIFMRLNLPDSAPQPNGDN 660
Query: 661 SQLPA--GPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQP 718
P A + + +S VQ ++ + P R R+ L ++ D + + P
Sbjct: 661 ELSPKIFDLAKSVHWIVDTSTYTGVQHSVTAVPPRR-----RRLLPGGDDSDDELQTAVP 715
Query: 719 LPQDAFRIRQFQK 731
D +R+RQ ++
Sbjct: 716 PSHDIYRLRQLKR 728
>UniRef100_Q627T1 Hypothetical protein CBG00546 [Caenorhabditis briggsae]
Length = 736
Score = 332 bits (851), Expect = 3e-89
Identities = 233/749 (31%), Positives = 371/749 (49%), Gaps = 89/749 (11%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I + YE L+ +P + ++W+ Y+E + N ++ +F RCL N L + LW+CYI
Sbjct: 35 IDQEREFYESLVTQFPNSGRYWRAYIEHELRSKNFVEVEALFQRCLANVLNIDLWKCYIV 94
Query: 83 FIRKVNDKKGA--EGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETH 140
+ V++ KG + +E K F+F L +G D+ + ++ +YIAFLK +PA E
Sbjct: 95 Y---VSETKGTIEKYRETMAKTFDFALEKIGMDVLAFSIYQDYIAFLKKVPAIGQYAENQ 151
Query: 141 RMTVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRER 200
R+T VR+VYQ+A+ TP H+++ +W DY +FE +++ LA+ LI+E Y ++R V ++
Sbjct: 152 RITAVRRVYQKALATPMHNLDTIWADYCAFEKNINMTLAEKLIAERGKDYQNSRRVEKDL 211
Query: 201 KKFFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLL 260
+ ++ ++VPP G+ SK + L K +++ W
Sbjct: 212 QAMTRGLNRQAVSVPPKGTASESKQVELWKKYIA----------WEKT------------ 249
Query: 261 TGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGS-- 318
N + E + +RV++TYEQ L+ L +YPD+WY+ A + +A
Sbjct: 250 ---NPLGTEEYGQLA----------RRVVYTYEQALLSLGYYPDIWYEAAMFLQEASQTL 296
Query: 319 -------IDAAIKV-----FQRSLKAL-PDSEMLRYAYAELEESRGAIQAAKKIYENLLG 365
+ A+KV ++R++ L +S++L +AYA+ +E + A K+IY LL
Sbjct: 297 DEKGDVKLAQALKVECIGLYERAITGLMKESKLLYFAYADFQEEQKKFDAVKEIYNRLLT 356
Query: 366 DSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKM 425
+ L ++Q +RF+RRTEG AR F AR+ Y V+VA A + + KD +
Sbjct: 357 IEQINPTLTYVQLMRFIRRTEGPNNARLVFKRAREDKRTGYQVFVAAAHMEYNCMKDADV 416
Query: 426 AHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS--LPLEDSVEVWKRF 483
A VF+ GLK + +EP + L YADFL +N+D N R +FER L+S LP + + +W RF
Sbjct: 417 AIRVFKLGLKKYENEPEFGLAYADFLSNMNEDNNTRVVFERILTSSKLPADKQIRIWDRF 476
Query: 484 VKFEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDL---- 539
+ FE GDLAS+LKVE+RRK A+ E + V+ RY FMDL PCS + L
Sbjct: 477 LDFESCVGDLASILKVEKRRKIAYEEAQKDLGMNHSMLVIDRYKFMDLMPCSGDQLRLIG 536
Query: 540 -DNLSRQEWLVKNTKKVEKSIMLNG---TTFIDKGPVASISTTSSKVVYPDTSKMLIYDP 595
D L + +T K + G + I G PD S+M+ + P
Sbjct: 537 YDALKGNDTPSTSTSSNPKPVPTRGPQAASAIMGGAGGHADVARYGFPRPDISQMIPFKP 596
Query: 596 KHNPGTGAAGTNAFDEILKATPPALVAFLANLPSVD---GPTPNVDIVLSICLQSDLPTG 652
+ N A + + PPA ++ LP GP NV+ + S+ Q+ LP+
Sbjct: 597 RVN---CTASFHPVPGGVYPPPPAAAHLMSLLPPPTCFIGPFVNVEALCSVLSQTQLPSL 653
Query: 653 QSVKVGIPSQLPAGPAPATSEL-----SGSSKSHPVQ-SGLSHMQPGRKQYGKRKQLDSQ 706
Q K P A +L + S S V+ S L+ + KRK+ DS
Sbjct: 654 QYDKSENGMIGPMNEADVKKDLYQLLATTSDPSAVVRSSALADL--------KRKRGDSD 705
Query: 707 EEDDTKSVQSQPL----PQDAFRIRQFQK 731
EED+ +QS + +DA++ R +K
Sbjct: 706 EEDEYSHLQSSTIGSIGSRDAYKRRMNKK 734
>UniRef100_UPI00001CF24F UPI00001CF24F UniRef100 entry
Length = 685
Score = 332 bits (850), Expect = 3e-89
Identities = 228/728 (31%), Positives = 355/728 (48%), Gaps = 89/728 (12%)
Query: 31 EQLLQLYPTAAKFWKQYV-EAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIRFIRKVND 89
E+ L+ P W + EA + N D ++++F RCL+ L + LW+CY+ ++R+
Sbjct: 22 EKKLEENPYDLDAWSILIREAQIKAKNYDKVEKLFQRCLMKVLHIDLWKCYLSYVRETKG 81
Query: 90 KKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRMTVVRKVY 149
K + +E+ +A++F L +G +I S +W++YI FLK + A E R+T VR+VY
Sbjct: 82 KLPSY-KEKMAQAYDFALDKIGMEIMSYQIWVDYINFLKGVEAVGSYAENQRITAVRRVY 140
Query: 150 QRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKKFFDEIDW 209
QR + P +IEQLW+DY+ +E ++ LAK +I + Y +AR V +E + +D
Sbjct: 141 QRGCVNPMINIEQLWRDYNKYEEGINIHLAKKMIEDRSRDYMNARRVAKEYETVMKGLDR 200
Query: 210 NMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTGRNAVDVM 269
N +VPP + + ++ + + K ++ W + N +
Sbjct: 201 NAPSVPPQNTPQEAQQVDMWKKYIQ----------WEKS---------------NPLRTE 235
Query: 270 EETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSI---------- 319
++T I KRV+F YEQCL+ L H+PD+WY+ A + ++ +
Sbjct: 236 DQTLI----------TKRVMFAYEQCLLVLGHHPDIWYEAAQYLEQSSKLLAEKGDMNNA 285
Query: 320 ----DAAIKVFQRSLKALPDSEMLRY-AYAELEESRGAIQAAKKIYENLLGDSENATALA 374
D A +++R++ L ML Y AYA+ EESR + IY LL + L
Sbjct: 286 KLFSDEAANIYERAISTLLKKNMLLYFAYADYEESRMKYEKVHSIYNRLLAIEDIDPTLV 345
Query: 375 HIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGL 434
+IQ+++F RR EG++ R F AR+ +HVYV A + + KD +A +FE GL
Sbjct: 346 YIQYMKFARRAEGIKSGRMIFKKAREDARTRHHVYVTAALMEYYCSKDKSVAFKIFELGL 405
Query: 435 KHFMHEPVYILEYADFLIRLNDDQNIRALFERALS--SLPLEDSVEVWKRFVKFEQTYGD 492
K + P Y+L Y D+L LN+D N R LFER L+ SLP E S E+W RF+ FE GD
Sbjct: 406 KKYGDIPEYVLAYIDYLSHLNEDNNTRVLFERVLTSGSLPPEKSGEIWARFLAFESNIGD 465
Query: 493 LASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQEWLVKNT 552
LAS+LKVE+RR AF EE E++L +V RY FMDL+PCS+++L L K+
Sbjct: 466 LASILKVEKRRFTAFREE-YEGKETAL--LVDRYKFMDLYPCSASELKALG-----YKDV 517
Query: 553 KKVEKSIMLNGTTFIDKGPVASISTTSSKVVY--PDTSKMLIYDPKH--NPGTGAAGTNA 608
+ + + ++ K Y PDT +M+ + P+H PG
Sbjct: 518 SRAKLAAIIPDPVVAPSIVPVLKDEVDRKPEYPKPDTQQMIPFQPRHLAPPGLHPVPGGV 577
Query: 609 FDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLPTGQSVKVGIPSQLPA 665
F PPA V + LP GP VD ++ I + +P V I +
Sbjct: 578 F-----PVPPAAVVLMKLLPPPICFQGPFVQVDELMEIFRRCKIPNTVEEAVRI---ITG 629
Query: 666 GPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQPLPQDAFR 725
G E +G +S V + + KR DS E+++ +V P D +R
Sbjct: 630 GAPELAVEGNGPVESSAVLT----------KAVKRPNEDSDEDEEKGAV--VPPVHDIYR 677
Query: 726 IRQFQKAR 733
RQ ++ R
Sbjct: 678 ARQQKRIR 685
>UniRef100_O96641 Suppressor of forked protein [Drosophila virilis]
Length = 737
Score = 330 bits (845), Expect = 1e-88
Identities = 221/734 (30%), Positives = 364/734 (49%), Gaps = 74/734 (10%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I E +YE L+ ++PT A++WK Y+E M + ++++F RCL+ L + LW+ Y+
Sbjct: 48 IHEVRSLYESLVNVFPTTARYWKLYIEMEMRSRYYERVEKLFQRCLVKILNIDLWKLYLT 107
Query: 83 FIRKVNDKKG-AEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHR 141
++++ K G + +E+ +A++F L +G D+ S +W +YI FL+ + A E +
Sbjct: 108 YVKET--KAGLSTHKEKMAQAYDFALEKIGMDLHSFSIWQDYIYFLRGVEAVGNYAENQK 165
Query: 142 MTVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERK 201
+T VR+VYQ+A++TP IEQLWKDY +FE +++ +++ + E Y +AR V +E +
Sbjct: 166 ITAVRRVYQKAVVTPIVGIEQLWKDYIAFEQNINPIISEKMSLERSKDYMNARRVAKELE 225
Query: 202 KFFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLT 261
++ N+ AVPPT + + +K + L K +++
Sbjct: 226 YHTKGLNRNLPAVPPTLTKEETKQVELWKRFITY-------------------------- 259
Query: 262 GRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWH-------A 314
E S + D A +RV+F EQCL+ L H+P VW+ + +
Sbjct: 260 ---------EKSNPLRTEDTALVTRRVMFATEQCLLVLTHHPAVWHQASQFLDTSARVLT 310
Query: 315 KAGSIDAA-------IKVFQRSLKA-LPDSEMLRYAYAELEESRGAIQAAKKIYENLLGD 366
+ G + AA + +RS+ L + +L +AYA+ EE R + +Y LL
Sbjct: 311 EKGDVQAAKIFADECANILERSINGVLNRNALLYFAYADFEEGRLKYEKVHSMYNKLLTL 370
Query: 367 SENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMA 426
+ L ++Q+++F RR EG++ AR F AR+ YH++VA A + + KD ++A
Sbjct: 371 PDIDPTLVYVQYMKFARRAEGIKSARGIFKKAREDVRSRYHIFVAAALMEYYCSKDKEIA 430
Query: 427 HNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS--LPLEDSVEVWKRFV 484
+FE GLK F P Y++ Y D+L LN+D N R LFER LSS L SVEVW RF+
Sbjct: 431 FRIFELGLKRFGGSPEYVMCYIDYLSHLNEDNNTRVLFERVLSSGGLSPHKSVEVWNRFL 490
Query: 485 KFEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSR 544
+FE GDL+S++KVE+RR F E +V RY F+DL+PC+S +L ++
Sbjct: 491 EFESNIGDLSSIVKVERRRSAVF-ENLKEYEGKETAQLVDRYKFLDLYPCTSTELKSIGY 549
Query: 545 QEWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVYPDTSKMLIYDPKH--NPGTG 602
E + KV + + + + + PD S+M+ + P+ +PG
Sbjct: 550 AENVGIILNKV-GGASGGANSHNNNNENETDGEATQPLPRPDFSQMIPFKPRSCAHPGAH 608
Query: 603 AAGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLPTGQSVKVGI 659
F + PPAL A A LP S GP +V+++ I ++ +LP G
Sbjct: 609 PLAGGVFPQ-----PPALAALCAALPPPNSFRGPFVSVELLFDIFMRLNLPESAPQPNGD 663
Query: 660 PSQLPA--GPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQ 717
P A + + +S VQ ++ M P R R+ L ++ D + S
Sbjct: 664 NDLTPKIFDLAKSVHWIVDTSTYTGVQHSVTSMAPRR-----RRLLPGGDDSDDELQTSA 718
Query: 718 PLPQDAFRIRQFQK 731
P D +R+RQ ++
Sbjct: 719 PPTNDIYRLRQLKR 732
>UniRef100_Q6V8Y2 Suppressor of forked [Drosophila subobscura]
Length = 732
Score = 329 bits (843), Expect = 2e-88
Identities = 223/745 (29%), Positives = 369/745 (48%), Gaps = 101/745 (13%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I E +YE L+ ++PT A++WK Y+E M + ++++F RCL+ L + LW+ Y+
Sbjct: 48 IHEVRSLYESLVSVFPTTARYWKLYIEMEMRSRYYERVEKLFQRCLVKILNIDLWKLYLT 107
Query: 83 FIRKVNDKKG-AEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHR 141
+++ + K G + +E+ +A++F L +G D+ S +W +YI FL+ + A E +
Sbjct: 108 YVK--DTKSGLSTHKEKMAQAYDFALEKIGMDLHSFSIWQDYIYFLRGVEAVGNYAENQK 165
Query: 142 MTVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERK 201
+T VR+VYQ+A++TP IE LWKDY SFE +++ +++ + E Y +AR V +E +
Sbjct: 166 ITAVRRVYQKAVVTPIVGIEHLWKDYISFEQNINPIISEKMSLERSKDYMNARRVAKELE 225
Query: 202 KFFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLT 261
++ N+ AVPPT + + K + L K +++
Sbjct: 226 YQTKGLNRNLPAVPPTLTKEEVKQVELWKRFITY-------------------------- 259
Query: 262 GRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWH-------A 314
E S + D A +RV+F EQCL+ L H+P VW+ + + +
Sbjct: 260 ---------EKSNPLRTEDTALVTRRVMFATEQCLLVLTHHPAVWHQASQFLDTSARVLS 310
Query: 315 KAGSIDAA-------IKVFQRSLKA-LPDSEMLRYAYAELEESRGAIQAAKKIYENLLGD 366
+ G + AA + +RS+ L + +L +AYA+ EE R + +Y L+
Sbjct: 311 EKGDVQAAKIFADECANILERSINGVLNRNALLYFAYADFEEGRLKYEKVHSMYNKLIMH 370
Query: 367 SENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMA 426
+ L ++Q+++F RR EG++ AR F AR+ C YH++VA A + + KD ++A
Sbjct: 371 PDIDPTLVYVQYMQFARRAEGIKSARGIFKKAREDVRCRYHIFVAAALMEYYCSKDKEIA 430
Query: 427 HNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS--LPLEDSVEVWKRFV 484
+FE GLK F P Y++ Y D+L LN+D N R LFER LSS L SVEVW RF+
Sbjct: 431 FRIFELGLKRFGGSPDYVMCYIDYLSHLNEDNNTRVLFERVLSSGGLSPHKSVEVWNRFL 490
Query: 485 KFEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSR 544
+FE GDL+S++KVE+RR F E +V RY F+DL+PC+ +L ++
Sbjct: 491 EFEANIGDLSSIVKVERRRSAVF-ENLKEYEGKETAQLVDRYKFLDLFPCTPTELKSIGY 549
Query: 545 QEWLVKNTKKVEKSIMLN---GTTFIDKGPVASISTTSSKVVYPDTSKMLIYDPK--HNP 599
E I+LN G + G + S ++ + PD +M+ Y P+ +P
Sbjct: 550 AE---------IGGIILNKVGGVQNQNNGDAEADSEATAPLPRPDFRQMIPYKPRPCAHP 600
Query: 600 GTGAAGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLPTGQSVK 656
G+ F + PPAL + A LP S GP +V+++ I ++ +LP
Sbjct: 601 GSHPLAGGVFPQ-----PPALASLCAGLPPPNSFRGPFVSVELLFDIFMRLNLPDS---- 651
Query: 657 VGIPSQLPAGPAPATSELSGSSKS----------HPVQSGLSHMQPGRKQYGKRKQLDSQ 706
+ LP G + ++ +KS VQ ++ + P R R+ L
Sbjct: 652 ----APLPNGDNELSLKIFDLAKSVHWIVDTSTYTGVQHSVTAIPPRR-----RRLLPGG 702
Query: 707 EEDDTKSVQSQPLPQDAFRIRQFQK 731
++ D S P D +R+RQ ++
Sbjct: 703 DDSDDDLHPSAPPAHDIYRLRQLKR 727
>UniRef100_Q6NKM3 SD14665p [Drosophila melanogaster]
Length = 464
Score = 211 bits (537), Expect = 6e-53
Identities = 148/470 (31%), Positives = 232/470 (48%), Gaps = 40/470 (8%)
Query: 286 KRVIFTYEQCLMYLYHYPDVWYDYATWH-------AKAGSIDAA-------IKVFQRSLK 331
+RV+F EQCL+ L H+P VW+ + + + G + AA + +RS+
Sbjct: 6 RRVMFATEQCLLVLTHHPAVWHQASQFLDTSARVLTEKGDVQAAKIFADECANILERSIN 65
Query: 332 A-LPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEP 390
L + +L +AYA+ EE R + +Y LL + L ++Q+++F RR EG++
Sbjct: 66 GVLNRNALLYFAYADFEEGRLKYEKVHTMYNKLLQLPDIDPTLVYVQYMKFARRAEGIKS 125
Query: 391 ARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADF 450
AR F AR+ YH++VA A + + KD ++A +FE GLK F P Y++ Y D+
Sbjct: 126 ARSIFKKAREDVRSRYHIFVAAALMEYYCSKDKEIAFRIFELGLKRFGGSPEYVMCYIDY 185
Query: 451 LIRLNDDQNIRALFERALSS--LPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFG 508
L LN+D N R LFER LSS L SVEVW RF++FE GDL+S++KVE+RR F
Sbjct: 186 LSHLNEDNNTRVLFERVLSSGGLSPHKSVEVWNRFLEFESNIGDLSSIVKVERRRSAVF- 244
Query: 509 EEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQEWLVKNTKKVEKSIMLNGTTFID 568
E +V RY F+DL+PC+S +L ++ E + KV G +
Sbjct: 245 ENLKEYEGKETAQLVDRYKFLDLYPCTSTELKSIGYAENVGIILNKVG-----GGAQSQN 299
Query: 569 KGPVASISTTSSKVVYPDTSKMLIYDPK--HNPGTGAAGTNAFDEILKATPPALVAFLAN 626
G V + S + + PD S+M+ + P+ +PG F + PPAL A A
Sbjct: 300 TGEVETDSEATPPLPRPDFSQMIPFKPRPCAHPGAHPLAGGVFPQ-----PPALAALCAT 354
Query: 627 LP---SVDGPTPNVDIVLSICLQSDLPTGQSVKVGIPSQLPA--GPAPATSELSGSSKSH 681
LP S GP +V+++ I ++ +LP G P A + + +S
Sbjct: 355 LPPPNSFRGPFVSVELLFDIFMRLNLPDSAPQPNGDNELSPKIFDLAKSVHWIVDTSTYT 414
Query: 682 PVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQPLPQDAFRIRQFQK 731
VQ ++ + P R R+ L ++ D + + P D +R+RQ ++
Sbjct: 415 GVQHSVTAVPPRR-----RRLLPGGDDSDDELQTAVPPSHDIYRLRQLKR 459
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,222,080,035
Number of Sequences: 2790947
Number of extensions: 50760733
Number of successful extensions: 139376
Number of sequences better than 10.0: 295
Number of HSP's better than 10.0 without gapping: 125
Number of HSP's successfully gapped in prelim test: 171
Number of HSP's that attempted gapping in prelim test: 137422
Number of HSP's gapped (non-prelim): 1484
length of query: 757
length of database: 848,049,833
effective HSP length: 135
effective length of query: 622
effective length of database: 471,271,988
effective search space: 293131176536
effective search space used: 293131176536
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 79 (35.0 bits)
Medicago: description of AC146910.2


