

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146863.3 + phase: 0
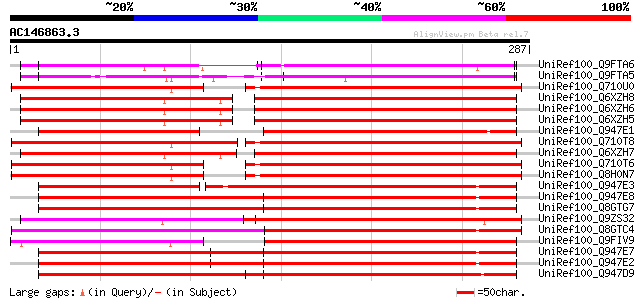
(287 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FTA6 T7N9.23 [Arabidopsis thaliana] 171 2e-41
UniRef100_Q9FTA5 T7N9.24 [Arabidopsis thaliana] 163 6e-39
UniRef100_Q710U0 TIR/NBS/LRR protein [Populus deltoides] 160 5e-38
UniRef100_Q6XZH8 Nematode resistance protein [Solanum tuberosum] 160 5e-38
UniRef100_Q6XZH6 Nematode resistance-like protein [Solanum tuber... 160 5e-38
UniRef100_Q6XZH5 Nematode resistance-like protein [Solanum tuber... 159 6e-38
UniRef100_Q947E1 Resistance gene analog NBS7 [Helianthus annuus] 157 4e-37
UniRef100_Q710T8 TIR/NBS/LRR protein [Populus deltoides] 155 9e-37
UniRef100_Q6XZH7 Nematode resistance-like protein [Solanum tuber... 155 1e-36
UniRef100_Q710T6 Part I of G08 TIR/NBS/LRR protein [Populus delt... 155 2e-36
UniRef100_Q8H0N7 TIR-NBS disease resistance protein [Populus tri... 154 3e-36
UniRef100_Q947E3 Resistance gene analog NBS5 [Helianthus annuus] 153 6e-36
UniRef100_Q947E8 Resistance gene analog PU3 [Helianthus annuus] 151 2e-35
UniRef100_Q8GTG7 NBS-LRR resistance protein RAS5-1 [Helianthus a... 150 4e-35
UniRef100_Q9ZS32 NL25 [Solanum tuberosum] 150 5e-35
UniRef100_Q8GTC4 NLS-TIR-NBS disease resistance protein [Populus... 149 1e-34
UniRef100_Q9FIV9 TMV resistance protein N [Arabidopsis thaliana] 148 2e-34
UniRef100_Q947E7 Resistance gene analog NBS1 [Helianthus annuus] 148 2e-34
UniRef100_Q947E2 Resistance gene analog NBS6 [Helianthus annuus] 148 2e-34
UniRef100_Q947D9 Resistance gene analog NBS9 [Helianthus annuus] 147 2e-34
>UniRef100_Q9FTA6 T7N9.23 [Arabidopsis thaliana]
Length = 1560
Score = 171 bits (434), Expect = 2e-41
Identities = 99/268 (36%), Positives = 142/268 (52%), Gaps = 36/268 (13%)
Query: 17 AIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPSEVRKQSGGYG 76
A+EDS +VV S N+A S +CL ELA + G+ +LPIFY+V+P +RKQ+G Y
Sbjct: 65 AMEDSVALVVVLSPNYAKSHWCLEELAMLCDLKSSLGRLVLPIFYEVEPCMLRKQNGPYE 124
Query: 77 ESLAKLEE--IAPQVQRWREALQLVGNISGWDLCHKPQHAELENIIEHINILGCKFSSRT 134
+ + ++QRWR AL ++GNI G+
Sbjct: 125 MDFEEHSKRFSEEKIQRWRRALNIIGNIPGF----------------------------- 155
Query: 135 KDLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIE 194
+ + YDVF+SFRG DTR NF DHL+ AL+ + + FRD+ +++G+ I+ L +E
Sbjct: 156 --VYRLKYDVFLSFRGADTRDNFGDHLYKALKDK-VRVFRDNEGMERGDEISSSLKAGME 212
Query: 195 ASQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEAL 254
S ++V S+NY+ S WCL EL + + ILPIFY VDPS V+KQS +
Sbjct: 213 DSAASVIVISRNYSGSRWCLDELAMLCKMKSSLDRRILPIFYHVDPSHVRKQSDHIKKDF 272
Query: 255 SKH--GFKHGLNMVHRWRETLTQVGNIS 280
+H F V WRE LT VGN++
Sbjct: 273 EEHQVRFSEEKEKVQEWREALTLVGNLA 300
Score = 96.3 bits (238), Expect = 8e-19
Identities = 48/140 (34%), Positives = 74/140 (52%), Gaps = 1/140 (0%)
Query: 140 INYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLY 199
+ +DVF+SF+ D R FT+ L+ L + + + +D + + L A+E S
Sbjct: 14 LEWDVFLSFQR-DARHKFTERLYEVLVKEQVRVWNNDDVERGNHELGASLVEAMEDSVAL 72
Query: 200 IVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKHGF 259
+VV S NYA S WCL EL + G+ +LPIFY+V+P ++KQ+G Y +H
Sbjct: 73 VVVLSPNYAKSHWCLEELAMLCDLKSSLGRLVLPIFYEVEPCMLRKQNGPYEMDFEEHSK 132
Query: 260 KHGLNMVHRWRETLTQVGNI 279
+ + RWR L +GNI
Sbjct: 133 RFSEEKIQRWRRALNIIGNI 152
Score = 79.0 bits (193), Expect = 1e-13
Identities = 54/147 (36%), Positives = 72/147 (48%), Gaps = 16/147 (10%)
Query: 7 GDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPS 66
GD + +EDS ++V S+N++ S +CL ELA + + ILPIFY VDPS
Sbjct: 200 GDEISSSLKAGMEDSAASVIVISRNYSGSRWCLDELAMLCKMKSSLDRRILPIFYHVDPS 259
Query: 67 EVRKQSG----GYGESLAKLEEIAPQVQRWREALQLVGNISGW------------DLCHK 110
VRKQS + E + E +VQ WREAL LVGN++G+ +L K
Sbjct: 260 HVRKQSDHIKKDFEEHQVRFSEEKEKVQEWREALTLVGNLAGYVCDKDSKDDDMIELVVK 319
Query: 111 PQHAELENIIEHINILGCKFSSRTKDL 137
AEL N E + S KDL
Sbjct: 320 RVLAELSNTPEKVGEFIVGLESPLKDL 346
>UniRef100_Q9FTA5 T7N9.24 [Arabidopsis thaliana]
Length = 1590
Score = 163 bits (412), Expect = 6e-39
Identities = 99/267 (37%), Positives = 152/267 (56%), Gaps = 25/267 (9%)
Query: 17 AIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCIL-PIFYDVDPSEVRKQSGGY 75
AIEDS F+VV S N+A+S L ELA + C + KC++ PIFY V+P EV++Q+G +
Sbjct: 103 AIEDSVAFVVVLSPNYANSHLRLEELAKL--CDL---KCLMVPIFYKVEPREVKEQNGPF 157
Query: 76 GESLAKLEEI--APQVQRWREALQLVGNISGWDLCHKPQHAELENIIEHINILGCKFSSR 133
+ + + ++QRW+ A+ VGNISG+ +C E+E + +R
Sbjct: 158 EKDFEEHSKRFGEEKIQRWKGAMTTVGNISGF-ICGYEIQLEMETGV---------VPNR 207
Query: 134 TKDLVEINYDVFVSFRGPDTRFNFTDHLFAAL-QRRGINAFRDDTKLKKGEFIAPGLFRA 192
K Y VF+SFRG DTR NF + L+ AL +++ + FRD+ ++KG+ I P LF A
Sbjct: 208 LK------YSVFLSFRGFDTRTNFCERLYIALNEKQNVRVFRDNEGMEKGDKIDPSLFEA 261
Query: 193 IEASQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGE 252
IE S +++ S NYA+S+WCL EL + + ++PIFY V+P +V+KQSG + +
Sbjct: 262 IEDSAASVIILSTNYANSSWCLDELALLCDLRSSLKRPMIPIFYGVNPEDVRKQSGEFRK 321
Query: 253 ALSKHGFKHGLNMVHRWRETLTQVGNI 279
+ + RW+ + VGNI
Sbjct: 322 DFEEKAKSFDEETIQRWKRAMNLVGNI 348
Score = 88.6 bits (218), Expect = 2e-16
Identities = 53/144 (36%), Positives = 81/144 (55%), Gaps = 8/144 (5%)
Query: 140 INYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEF---IAPGLFRAIEAS 196
+ +D F+SF+ DT NFTD L+ AL + + + DD + + + P L AIE S
Sbjct: 49 VKWDAFLSFQR-DTSHNFTDRLYEALVKEELRVWNDDLERVDHDHDHELRPSLVEAIEDS 107
Query: 197 QLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSK 256
++VV S NYA+S L EL + C K ++PIFY V+P EV++Q+G + + +
Sbjct: 108 VAFVVVLSPNYANSHLRLEELAKL--CDLKC--LMVPIFYKVEPREVKEQNGPFEKDFEE 163
Query: 257 HGFKHGLNMVHRWRETLTQVGNIS 280
H + G + RW+ +T VGNIS
Sbjct: 164 HSKRFGEEKIQRWKGAMTTVGNIS 187
Score = 78.6 bits (192), Expect = 2e-13
Identities = 48/152 (31%), Positives = 79/152 (51%), Gaps = 7/152 (4%)
Query: 7 GDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPS 66
GD + F AIEDS +++ S N+A+S +CL ELA + + ++PIFY V+P
Sbjct: 251 GDKIDPSLFEAIEDSAASVIILSTNYANSSWCLDELALLCDLRSSLKRPMIPIFYGVNPE 310
Query: 67 EVRKQSGGYGESLAKLEEIAPQ--VQRWREALQLVGNISGWDLCHKP-----QHAELENI 119
+VRKQSG + + + + + +QRW+ A+ LVGNI G+ K + E +
Sbjct: 311 DVRKQSGEFRKDFEEKAKSFDEETIQRWKRAMNLVGNIPGYVCTAKTVGDDNEGINREKV 370
Query: 120 IEHINILGCKFSSRTKDLVEINYDVFVSFRGP 151
+ I+++ K + ++ EI D V P
Sbjct: 371 DDMIDLVVKKVVAAVRNRPEIVADYTVGLESP 402
>UniRef100_Q710U0 TIR/NBS/LRR protein [Populus deltoides]
Length = 216
Score = 160 bits (404), Expect = 5e-38
Identities = 83/154 (53%), Positives = 107/154 (68%), Gaps = 3/154 (1%)
Query: 131 SSRTKDLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLF 190
SSR++ E YDVF+SFRG DTR FTDHL+ AL + GI+ FRDD +L +GE I+ L
Sbjct: 42 SSRSRP--EGTYDVFLSFRGEDTRHTFTDHLYTALIQAGIHTFRDDDELPRGEEISDHLI 99
Query: 191 RAIEASQLYIVVFSKNYASSTWCLRELEYILHCS-KKYGKHILPIFYDVDPSEVQKQSGG 249
RAI+ S++ IVVFSK YASS WCL EL IL C KK G+ +LPIFYD+DPS+V+KQ+G
Sbjct: 100 RAIQESKISIVVFSKGYASSRWCLNELVEILKCKRKKTGQIVLPIFYDIDPSDVRKQNGS 159
Query: 250 YGEALSKHGFKHGLNMVHRWRETLTQVGNISSSN 283
+ EA KH + +V WR+ L + GN+S N
Sbjct: 160 FAEAFVKHEERFEEKLVKEWRKALEEAGNLSGWN 193
Score = 95.9 bits (237), Expect = 1e-18
Identities = 50/109 (45%), Positives = 72/109 (65%), Gaps = 3/109 (2%)
Query: 2 DDYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCS-VLYGKCILPIF 60
D+ G+ + AI++S++ IVVFSK +A S +CL EL IL C G+ +LPIF
Sbjct: 86 DELPRGEEISDHLIRAIQESKISIVVFSKGYASSRWCLNELVEILKCKRKKTGQIVLPIF 145
Query: 61 YDVDPSEVRKQSGGYGESLAKLEEIAPQ--VQRWREALQLVGNISGWDL 107
YD+DPS+VRKQ+G + E+ K EE + V+ WR+AL+ GN+SGW+L
Sbjct: 146 YDIDPSDVRKQNGSFAEAFVKHEERFEEKLVKEWRKALEEAGNLSGWNL 194
>UniRef100_Q6XZH8 Nematode resistance protein [Solanum tuberosum]
Length = 1136
Score = 160 bits (404), Expect = 5e-38
Identities = 73/145 (50%), Positives = 103/145 (70%)
Query: 136 DLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEA 195
D++ +YDVF+SFRG D R F DHL+ AL+++ IN F+DD KL+KG+FI+P L +IE
Sbjct: 12 DIIRWSYDVFLSFRGEDVRKTFVDHLYLALEQKCINTFKDDEKLEKGKFISPELVSSIEE 71
Query: 196 SQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALS 255
S++ +++FSKNYA+STWCL EL I+ C G+ ++P+FYDVDPS V+KQ +GEA S
Sbjct: 72 SRIALIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFS 131
Query: 256 KHGFKHGLNMVHRWRETLTQVGNIS 280
KH + + V +WR L + NIS
Sbjct: 132 KHEARFQEDKVQKWRAALEEAANIS 156
Score = 97.8 bits (242), Expect = 3e-19
Identities = 51/122 (41%), Positives = 77/122 (62%), Gaps = 5/122 (4%)
Query: 7 GDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPS 66
G + + +IE+S++ +++FSKN+A+S +CL EL I+ C + G+ ++P+FYDVDPS
Sbjct: 58 GKFISPELVSSIEESRIALIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPS 117
Query: 67 EVRKQSGGYGESLAKLEE--IAPQVQRWREALQLVGNISGWDLCHKPQHAE---LENIIE 121
VRKQ +GE+ +K E +VQ+WR AL+ NISGWDL + E +E I E
Sbjct: 118 TVRKQKSIFGEAFSKHEARFQEDKVQKWRAALEEAANISGWDLPNTANGHEARVMEKIAE 177
Query: 122 HI 123
I
Sbjct: 178 DI 179
>UniRef100_Q6XZH6 Nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 160 bits (404), Expect = 5e-38
Identities = 73/145 (50%), Positives = 103/145 (70%)
Query: 136 DLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEA 195
+++ +YDVF+SFRG D R F DHL+ ALQ++ IN F+DD KL+KG+FI+P L +IE
Sbjct: 12 EIIRWSYDVFLSFRGEDVRKTFVDHLYLALQQKCINTFKDDEKLEKGKFISPELMSSIEE 71
Query: 196 SQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALS 255
S++ +++FSKNYA+STWCL EL I+ C G+ ++P+FYDVDPS V+KQ +GEA S
Sbjct: 72 SRIALIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFS 131
Query: 256 KHGFKHGLNMVHRWRETLTQVGNIS 280
KH + + V +WR L + NIS
Sbjct: 132 KHEARFQEDKVQKWRAALEEAANIS 156
Score = 98.2 bits (243), Expect = 2e-19
Identities = 51/122 (41%), Positives = 77/122 (62%), Gaps = 5/122 (4%)
Query: 7 GDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPS 66
G + + +IE+S++ +++FSKN+A+S +CL EL I+ C + G+ ++P+FYDVDPS
Sbjct: 58 GKFISPELMSSIEESRIALIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPS 117
Query: 67 EVRKQSGGYGESLAKLEE--IAPQVQRWREALQLVGNISGWDLCHKPQHAE---LENIIE 121
VRKQ +GE+ +K E +VQ+WR AL+ NISGWDL + E +E I E
Sbjct: 118 TVRKQKSIFGEAFSKHEARFQEDKVQKWRAALEEAANISGWDLPNTSNGHEARVMEKIAE 177
Query: 122 HI 123
I
Sbjct: 178 DI 179
>UniRef100_Q6XZH5 Nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 159 bits (403), Expect = 6e-38
Identities = 73/145 (50%), Positives = 103/145 (70%)
Query: 136 DLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEA 195
+++ +YDVF+SFRG D R F DHL+ ALQ++ IN F+DD KL+KG+FI+P L +IE
Sbjct: 12 EIIRWSYDVFLSFRGEDVRKTFVDHLYLALQQKCINTFKDDEKLEKGKFISPELVSSIEE 71
Query: 196 SQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALS 255
S++ +++FSKNYA+STWCL EL I+ C G+ ++P+FYDVDPS V+KQ +GEA S
Sbjct: 72 SRIALIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFS 131
Query: 256 KHGFKHGLNMVHRWRETLTQVGNIS 280
KH + + V +WR L + NIS
Sbjct: 132 KHEARFQEDKVQKWRAALEEAANIS 156
Score = 97.8 bits (242), Expect = 3e-19
Identities = 51/122 (41%), Positives = 77/122 (62%), Gaps = 5/122 (4%)
Query: 7 GDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPS 66
G + + +IE+S++ +++FSKN+A+S +CL EL I+ C + G+ ++P+FYDVDPS
Sbjct: 58 GKFISPELVSSIEESRIALIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPS 117
Query: 67 EVRKQSGGYGESLAKLEE--IAPQVQRWREALQLVGNISGWDLCHKPQHAE---LENIIE 121
VRKQ +GE+ +K E +VQ+WR AL+ NISGWDL + E +E I E
Sbjct: 118 TVRKQKSIFGEAFSKHEARFQEDKVQKWRAALEEAANISGWDLPNTSNGHEARVMEKIAE 177
Query: 122 HI 123
I
Sbjct: 178 DI 179
>UniRef100_Q947E1 Resistance gene analog NBS7 [Helianthus annuus]
Length = 259
Score = 157 bits (396), Expect = 4e-37
Identities = 73/140 (52%), Positives = 101/140 (72%), Gaps = 1/140 (0%)
Query: 141 NYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYI 200
NYDVF+SFRG DTR +F DHL+ AL++RGI ++DD L +GE I P L +AI+ S++ +
Sbjct: 21 NYDVFLSFRGDDTRKSFVDHLYTALEQRGIYTYKDDETLPRGESIGPALLKAIQESRVAV 80
Query: 201 VVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKHGFK 260
+VFSKNYA S+WCL EL +I+ C G+ ++PIFY VDPS+V+KQ G YGEA +KH +
Sbjct: 81 IVFSKNYADSSWCLDELAHIMECMDTRGQIVMPIFYHVDPSDVRKQKGKYGEAFTKHERE 140
Query: 261 HGLNMVHRWRETLTQVGNIS 280
+ L V WR+ L + G +S
Sbjct: 141 NKLK-VESWRKALEKAGKLS 159
Score = 101 bits (252), Expect = 2e-20
Identities = 48/90 (53%), Positives = 67/90 (74%), Gaps = 1/90 (1%)
Query: 17 AIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPSEVRKQSGGYG 76
AI++S+V ++VFSKN+ADS +CL ELA+I+ C G+ ++PIFY VDPS+VRKQ G YG
Sbjct: 72 AIQESRVAVIVFSKNYADSSWCLDELAHIMECMDTRGQIVMPIFYHVDPSDVRKQKGKYG 131
Query: 77 ESLAKLE-EIAPQVQRWREALQLVGNISGW 105
E+ K E E +V+ WR+AL+ G +SGW
Sbjct: 132 EAFTKHERENKLKVESWRKALEKAGKLSGW 161
>UniRef100_Q710T8 TIR/NBS/LRR protein [Populus deltoides]
Length = 1147
Score = 155 bits (393), Expect = 9e-37
Identities = 81/154 (52%), Positives = 105/154 (67%), Gaps = 3/154 (1%)
Query: 131 SSRTKDLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLF 190
SSR++ E YDVF+SFRG DTR FTDHL+ AL + GI+ FRDD +L +GE I+
Sbjct: 30 SSRSRP--EGAYDVFLSFRGEDTRKTFTDHLYTALVQAGIHTFRDDDELPRGEEISDHFL 87
Query: 191 RAIEASQLYIVVFSKNYASSTWCLRELEYILHCSK-KYGKHILPIFYDVDPSEVQKQSGG 249
RAI+ S++ I VFSK YASS WCL EL IL C K K G+ +LPIFYD+DPS+V+KQ+G
Sbjct: 88 RAIQESKISIAVFSKGYASSRWCLNELVEILKCKKRKTGQIVLPIFYDIDPSDVRKQNGS 147
Query: 250 YGEALSKHGFKHGLNMVHRWRETLTQVGNISSSN 283
+ EA KH + +V WR+ L + GN+S N
Sbjct: 148 FAEAFVKHEERFEEKLVKEWRKALEEAGNLSGWN 181
Score = 95.1 bits (235), Expect = 2e-18
Identities = 53/128 (41%), Positives = 78/128 (60%), Gaps = 3/128 (2%)
Query: 2 DDYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLY-GKCILPIF 60
D+ G+ + AI++S++ I VFSK +A S +CL EL IL C G+ +LPIF
Sbjct: 74 DELPRGEEISDHFLRAIQESKISIAVFSKGYASSRWCLNELVEILKCKKRKTGQIVLPIF 133
Query: 61 YDVDPSEVRKQSGGYGESLAKLEEIAPQ--VQRWREALQLVGNISGWDLCHKPQHAELEN 118
YD+DPS+VRKQ+G + E+ K EE + V+ WR+AL+ GN+SGW+L E +
Sbjct: 134 YDIDPSDVRKQNGSFAEAFVKHEERFEEKLVKEWRKALEEAGNLSGWNLNDMANGHEAKF 193
Query: 119 IIEHINIL 126
I E I ++
Sbjct: 194 IKEIIKVV 201
>UniRef100_Q6XZH7 Nematode resistance-like protein [Solanum tuberosum]
Length = 1121
Score = 155 bits (392), Expect = 1e-36
Identities = 70/145 (48%), Positives = 103/145 (70%)
Query: 136 DLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEA 195
+++ +YDVF+SFRG + R F DHL+ AL+++ IN F+DD KL+KG+FI+P L +IE
Sbjct: 12 EIIRWSYDVFLSFRGENVRKTFVDHLYLALEQKCINTFKDDEKLEKGKFISPELMSSIEE 71
Query: 196 SQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALS 255
S++ +++FSKNYA+STWCL EL I+ C G+ ++P+FYDVDPS V++Q +GEA S
Sbjct: 72 SRIALIIFSKNYANSTWCLDELTKIIECKNVKGQIVVPVFYDVDPSTVRRQKNIFGEAFS 131
Query: 256 KHGFKHGLNMVHRWRETLTQVGNIS 280
KH + + V +WR L + NIS
Sbjct: 132 KHEARFEEDKVKKWRAALEEAANIS 156
Score = 95.9 bits (237), Expect = 1e-18
Identities = 49/124 (39%), Positives = 78/124 (62%), Gaps = 5/124 (4%)
Query: 7 GDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPS 66
G + + +IE+S++ +++FSKN+A+S +CL EL I+ C + G+ ++P+FYDVDPS
Sbjct: 58 GKFISPELMSSIEESRIALIIFSKNYANSTWCLDELTKIIECKNVKGQIVVPVFYDVDPS 117
Query: 67 EVRKQSGGYGESLAKLEE--IAPQVQRWREALQLVGNISGWDLCHKPQHAE---LENIIE 121
VR+Q +GE+ +K E +V++WR AL+ NISGWDL + E +E I E
Sbjct: 118 TVRRQKNIFGEAFSKHEARFEEDKVKKWRAALEEAANISGWDLPNTSNGHEARVIEKITE 177
Query: 122 HINI 125
I +
Sbjct: 178 DIMV 181
>UniRef100_Q710T6 Part I of G08 TIR/NBS/LRR protein [Populus deltoides]
Length = 226
Score = 155 bits (391), Expect = 2e-36
Identities = 82/154 (53%), Positives = 105/154 (67%), Gaps = 3/154 (1%)
Query: 131 SSRTKDLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLF 190
SSR++ E +YDVF+SFRG DTR FTDHL+ AL + GI FRDD +L +GE I+ L
Sbjct: 6 SSRSRP--EGDYDVFLSFRGEDTRKTFTDHLYTALVQAGIYTFRDDDELPRGEEISYHLL 63
Query: 191 RAIEASQLYIVVFSKNYASSTWCLRELEYILHCSK-KYGKHILPIFYDVDPSEVQKQSGG 249
RAI+ S++ IVVFSK YASS WCL EL IL C K G+ +LPIFYD+DPS V+KQ+G
Sbjct: 64 RAIQESKISIVVFSKGYASSRWCLNELVEILKCKNGKTGQIVLPIFYDIDPSYVRKQNGS 123
Query: 250 YGEALSKHGFKHGLNMVHRWRETLTQVGNISSSN 283
+ EA KH + +V WR+ L + GN+S N
Sbjct: 124 FAEAFVKHEERFEETLVKEWRKALAEAGNLSGWN 157
Score = 93.2 bits (230), Expect = 7e-18
Identities = 50/109 (45%), Positives = 71/109 (64%), Gaps = 3/109 (2%)
Query: 2 DDYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHC-SVLYGKCILPIF 60
D+ G+ + AI++S++ IVVFSK +A S +CL EL IL C + G+ +LPIF
Sbjct: 50 DELPRGEEISYHLLRAIQESKISIVVFSKGYASSRWCLNELVEILKCKNGKTGQIVLPIF 109
Query: 61 YDVDPSEVRKQSGGYGESLAKLEEIAPQ--VQRWREALQLVGNISGWDL 107
YD+DPS VRKQ+G + E+ K EE + V+ WR+AL GN+SGW+L
Sbjct: 110 YDIDPSYVRKQNGSFAEAFVKHEERFEETLVKEWRKALAEAGNLSGWNL 158
>UniRef100_Q8H0N7 TIR-NBS disease resistance protein [Populus trichocarpa]
Length = 567
Score = 154 bits (389), Expect = 3e-36
Identities = 81/154 (52%), Positives = 106/154 (68%), Gaps = 3/154 (1%)
Query: 131 SSRTKDLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLF 190
SSR++ E YDVF+SFRG DTR FTDHL+ AL + GI+ FRDD +L +GE I+ L
Sbjct: 6 SSRSRP--EGAYDVFLSFRGEDTRKTFTDHLYTALVQAGIHTFRDDDELPRGEEISDHLL 63
Query: 191 RAIEASQLYIVVFSKNYASSTWCLRELEYILHC-SKKYGKHILPIFYDVDPSEVQKQSGG 249
RA++ S++ IVVFSK YASS WCL EL IL C ++K G+ +LPIFYD+DPS V+KQ+G
Sbjct: 64 RAVQESKISIVVFSKGYASSRWCLNELVEILKCKNRKTGQIVLPIFYDIDPSYVRKQNGS 123
Query: 250 YGEALSKHGFKHGLNMVHRWRETLTQVGNISSSN 283
+ EA KH +V WR+ L + GN+S N
Sbjct: 124 FAEAFVKHEECFEEKLVKEWRKALEEAGNLSGWN 157
Score = 94.7 bits (234), Expect = 2e-18
Identities = 49/109 (44%), Positives = 72/109 (65%), Gaps = 3/109 (2%)
Query: 2 DDYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHC-SVLYGKCILPIF 60
D+ G+ + A+++S++ IVVFSK +A S +CL EL IL C + G+ +LPIF
Sbjct: 50 DELPRGEEISDHLLRAVQESKISIVVFSKGYASSRWCLNELVEILKCKNRKTGQIVLPIF 109
Query: 61 YDVDPSEVRKQSGGYGESLAKLEEIAPQ--VQRWREALQLVGNISGWDL 107
YD+DPS VRKQ+G + E+ K EE + V+ WR+AL+ GN+SGW+L
Sbjct: 110 YDIDPSYVRKQNGSFAEAFVKHEECFEEKLVKEWRKALEEAGNLSGWNL 158
>UniRef100_Q947E3 Resistance gene analog NBS5 [Helianthus annuus]
Length = 285
Score = 153 bits (386), Expect = 6e-36
Identities = 75/172 (43%), Positives = 109/172 (62%), Gaps = 3/172 (1%)
Query: 109 HKPQHAELENIIEHINILGCKFSSRTKDLVEINYDVFVSFRGPDTRFNFTDHLFAALQRR 168
H P +E E E + SS + + ++VF+SFRG DTR NF DHL+ L ++
Sbjct: 16 HTPMSSEEEE--EDVQSRSMASSSSSSSSHSLKHEVFLSFRGEDTRKNFVDHLYKDLVQQ 73
Query: 169 GINAFRDDTKLKKGEFIAPGLFRAIEASQLYIVVFSKNYASSTWCLRELEYILHCSKKYG 228
GI ++DD L++GE I P L +AI+ S++ ++VFS+NYA S+WCL EL+ I+ C G
Sbjct: 74 GIQTYKDDETLRRGESIRPALLKAIQESRIAVIVFSENYADSSWCLDELQQIIECMDTNG 133
Query: 229 KHILPIFYDVDPSEVQKQSGGYGEALSKHGFKHGLNMVHRWRETLTQVGNIS 280
+ ++PIFY VDPS+V+KQ+G YG+A KH K V WR+ L + GN+S
Sbjct: 134 QIVIPIFYHVDPSDVRKQNGKYGKAFRKHK-KENKQKVESWRKALEKAGNLS 184
Score = 97.1 bits (240), Expect = 5e-19
Identities = 44/90 (48%), Positives = 67/90 (73%), Gaps = 1/90 (1%)
Query: 17 AIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPSEVRKQSGGYG 76
AI++S++ ++VFS+N+ADS +CL EL I+ C G+ ++PIFY VDPS+VRKQ+G YG
Sbjct: 97 AIQESRIAVIVFSENYADSSWCLDELQQIIECMDTNGQIVIPIFYHVDPSDVRKQNGKYG 156
Query: 77 ESLAK-LEEIAPQVQRWREALQLVGNISGW 105
++ K +E +V+ WR+AL+ GN+SGW
Sbjct: 157 KAFRKHKKENKQKVESWRKALEKAGNLSGW 186
>UniRef100_Q947E8 Resistance gene analog PU3 [Helianthus annuus]
Length = 770
Score = 151 bits (382), Expect = 2e-35
Identities = 71/140 (50%), Positives = 99/140 (70%), Gaps = 1/140 (0%)
Query: 141 NYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYI 200
N+DVF+SFRG DTR +F DHL+AAL ++GI ++DD L +GE I P L +AI+ S++ +
Sbjct: 82 NHDVFLSFRGEDTRNSFVDHLYAALVQQGIQTYKDDQTLPRGERIGPALLKAIQESRIAV 141
Query: 201 VVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKHGFK 260
VVFS+NYA S+WCL EL +I+ C G+ ++PIFY VDPS+V+KQ G YG+A KH +
Sbjct: 142 VVFSQNYADSSWCLDELAHIMECMDTRGQIVIPIFYFVDPSDVRKQKGKYGKAFRKHK-R 200
Query: 261 HGLNMVHRWRETLTQVGNIS 280
V WR+ L + GN+S
Sbjct: 201 ENKQKVESWRKALEKAGNLS 220
Score = 99.8 bits (247), Expect = 7e-20
Identities = 54/127 (42%), Positives = 84/127 (65%), Gaps = 3/127 (2%)
Query: 17 AIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPSEVRKQSGGYG 76
AI++S++ +VVFS+N+ADS +CL ELA+I+ C G+ ++PIFY VDPS+VRKQ G YG
Sbjct: 133 AIQESRIAVVVFSQNYADSSWCLDELAHIMECMDTRGQIVIPIFYFVDPSDVRKQKGKYG 192
Query: 77 ESLAK-LEEIAPQVQRWREALQLVGNISGWDLCHKPQHAE-LENIIEHINI-LGCKFSSR 133
++ K E +V+ WR+AL+ GN+SGW + A+ ++ I+ I+ L ++
Sbjct: 193 KAFRKHKRENKQKVESWRKALEKAGNLSGWVINENSHEAKCIKEIVATISSRLPTLSTNV 252
Query: 134 TKDLVEI 140
KDL+ I
Sbjct: 253 NKDLIGI 259
>UniRef100_Q8GTG7 NBS-LRR resistance protein RAS5-1 [Helianthus annuus]
Length = 448
Score = 150 bits (379), Expect = 4e-35
Identities = 71/140 (50%), Positives = 99/140 (70%), Gaps = 1/140 (0%)
Query: 141 NYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYI 200
N+DVF+SFRG DTR +F DHL+AAL ++GI A++DD L +GE I P L +AI+ S++ +
Sbjct: 83 NHDVFLSFRGEDTRNSFVDHLYAALAQQGIQAYKDDETLPRGERIGPALLKAIQESRIAV 142
Query: 201 VVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKHGFK 260
VVFS+NYA S+WCL EL +I+ C G+ ++PIFY VDPS+V+KQ G YG+A K +
Sbjct: 143 VVFSQNYADSSWCLDELAHIMECMDTRGQIVIPIFYFVDPSDVRKQKGKYGKAFRKRK-R 201
Query: 261 HGLNMVHRWRETLTQVGNIS 280
V WR+ L + GN+S
Sbjct: 202 ENRQKVESWRKALEKAGNLS 221
Score = 100 bits (248), Expect = 6e-20
Identities = 54/127 (42%), Positives = 85/127 (66%), Gaps = 3/127 (2%)
Query: 17 AIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPSEVRKQSGGYG 76
AI++S++ +VVFS+N+ADS +CL ELA+I+ C G+ ++PIFY VDPS+VRKQ G YG
Sbjct: 134 AIQESRIAVVVFSQNYADSSWCLDELAHIMECMDTRGQIVIPIFYFVDPSDVRKQKGKYG 193
Query: 77 ESLAKLE-EIAPQVQRWREALQLVGNISGWDLCHKPQHAE-LENIIEHINI-LGCKFSSR 133
++ K + E +V+ WR+AL+ GN+SGW + A+ ++ I+ I+ L ++
Sbjct: 194 KAFRKRKRENRQKVESWRKALEKAGNLSGWVINENSHEAKCIKEIVATISSRLPTLSTNV 253
Query: 134 TKDLVEI 140
KDL+ I
Sbjct: 254 NKDLIGI 260
>UniRef100_Q9ZS32 NL25 [Solanum tuberosum]
Length = 533
Score = 150 bits (378), Expect = 5e-35
Identities = 72/159 (45%), Positives = 105/159 (65%), Gaps = 5/159 (3%)
Query: 130 FSSRTKDLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGL 189
++S +++ Y VF+SFRG DTR FT HLF L+ RGI F+DD +L+KG+ I L
Sbjct: 9 YASDSQNCTHWKYHVFLSFRGDDTRKTFTSHLFEGLKHRGIFTFQDDKRLEKGDSIPEEL 68
Query: 190 FRAIEASQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGG 249
+AIE SQ+ +V+FSKNYA+S WCL EL I+ C + + ++P+FYDVDPS+V+ Q+G
Sbjct: 69 LKAIEESQVALVIFSKNYATSRWCLNELVKIMECKEVKKQIVMPVFYDVDPSDVRHQTGS 128
Query: 250 YGEALSKHGFKH-----GLNMVHRWRETLTQVGNISSSN 283
+ EA SKH ++ G+ MV WR L+ ++S +N
Sbjct: 129 FAEAFSKHKSRYKDDVDGMQMVQGWRTALSAAADLSGTN 167
Score = 90.9 bits (224), Expect = 3e-17
Identities = 47/137 (34%), Positives = 81/137 (58%), Gaps = 7/137 (5%)
Query: 7 GDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPS 66
GD + + AIE+SQV +V+FSKN+A S +CL EL I+ C + + ++P+FYDVDPS
Sbjct: 61 GDSIPEELLKAIEESQVALVIFSKNYATSRWCLNELVKIMECKEVKKQIVMPVFYDVDPS 120
Query: 67 EVRKQSGGYGESLAKLE-------EIAPQVQRWREALQLVGNISGWDLCHKPQHAELENI 119
+VR Q+G + E+ +K + + VQ WR AL ++SG ++ + + + +
Sbjct: 121 DVRHQTGSFAEAFSKHKSRYKDDVDGMQMVQGWRTALSAAADLSGTNVPGRIESECIREL 180
Query: 120 IEHINILGCKFSSRTKD 136
++ ++ CK SS + +
Sbjct: 181 VDAVSSKLCKTSSSSSE 197
>UniRef100_Q8GTC4 NLS-TIR-NBS disease resistance protein [Populus tremula]
Length = 516
Score = 149 bits (375), Expect = 1e-34
Identities = 78/143 (54%), Positives = 96/143 (66%), Gaps = 2/143 (1%)
Query: 142 YDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYIV 201
YDVF SFRG DTR FTDHL+ AL + GI+ FRDD +L +GE I+ L +AI S++ IV
Sbjct: 1 YDVFFSFRGKDTRKTFTDHLYTALVQAGIHTFRDDDELPRGEEISDHLLKAIRESKICIV 60
Query: 202 VFSKNYASSTWCLRELEYILHCS-KKYGKHILPIFYDVDPSEVQKQSGGYGEALSKHGFK 260
VFSK YASS WCL EL IL C +K G+ LPIFYD+DPS V+KQ+G + EA KH +
Sbjct: 61 VFSKGYASSRWCLDELVEILKCKYRKTGQIALPIFYDIDPSYVRKQTGSFAEAFVKHE-E 119
Query: 261 HGLNMVHRWRETLTQVGNISSSN 283
V WRE L + GN+S N
Sbjct: 120 RSKEKVKEWREALEEAGNLSGWN 142
Score = 95.9 bits (237), Expect = 1e-18
Identities = 59/142 (41%), Positives = 82/142 (57%), Gaps = 2/142 (1%)
Query: 2 DDYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLY-GKCILPIF 60
D+ G+ + AI +S++ IVVFSK +A S +CL EL IL C G+ LPIF
Sbjct: 36 DELPRGEEISDHLLKAIRESKICIVVFSKGYASSRWCLDELVEILKCKYRKTGQIALPIF 95
Query: 61 YDVDPSEVRKQSGGYGESLAKLEEIAPQ-VQRWREALQLVGNISGWDLCHKPQHAELENI 119
YD+DPS VRKQ+G + E+ K EE + + V+ WREAL+ GN+SGW+L E I
Sbjct: 96 YDIDPSYVRKQTGSFAEAFVKHEERSKEKVKEWREALEEAGNLSGWNLKDHEAKFIQEII 155
Query: 120 IEHINILGCKFSSRTKDLVEIN 141
+ + L K+ K LV I+
Sbjct: 156 KDVLTKLDPKYLHVPKHLVGID 177
>UniRef100_Q9FIV9 TMV resistance protein N [Arabidopsis thaliana]
Length = 1130
Score = 148 bits (373), Expect = 2e-34
Identities = 73/140 (52%), Positives = 96/140 (68%), Gaps = 1/140 (0%)
Query: 142 YDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYIV 201
YDVFVSFRG D R NF HL+ +L+R GI+ F DD +L++GE+I+P L AIE S++ IV
Sbjct: 14 YDVFVSFRGADVRKNFLSHLYDSLRRCGISTFMDDVELQRGEYISPELLNAIETSKILIV 73
Query: 202 VFSKNYASSTWCLRELEYILHCSKKYGKH-ILPIFYDVDPSEVQKQSGGYGEALSKHGFK 260
V +K+YASS WCL EL +I+ K H + PIF VDPS+++ Q G Y ++ SKH
Sbjct: 74 VLTKDYASSAWCLDELVHIMKSHKNNPSHMVFPIFLYVDPSDIRWQQGSYAKSFSKHKNS 133
Query: 261 HGLNMVHRWRETLTQVGNIS 280
H LN + WRE LT+V NIS
Sbjct: 134 HPLNKLKDWREALTKVANIS 153
Score = 78.6 bits (192), Expect = 2e-13
Identities = 45/112 (40%), Positives = 67/112 (59%), Gaps = 5/112 (4%)
Query: 1 MDDYH--PGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLY-GKCIL 57
MDD G+ + + +AIE S++ IVV +K++A S +CL EL +I+ +
Sbjct: 46 MDDVELQRGEYISPELLNAIETSKILIVVLTKDYASSAWCLDELVHIMKSHKNNPSHMVF 105
Query: 58 PIFYDVDPSEVRKQSGGYGESLAKLEEIAP--QVQRWREALQLVGNISGWDL 107
PIF VDPS++R Q G Y +S +K + P +++ WREAL V NISGWD+
Sbjct: 106 PIFLYVDPSDIRWQQGSYAKSFSKHKNSHPLNKLKDWREALTKVANISGWDI 157
>UniRef100_Q947E7 Resistance gene analog NBS1 [Helianthus annuus]
Length = 339
Score = 148 bits (373), Expect = 2e-34
Identities = 72/169 (42%), Positives = 105/169 (61%), Gaps = 1/169 (0%)
Query: 112 QHAELENIIEHINILGCKFSSRTKDLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGIN 171
QH + + E + SS + + ++VF+SFRG D R NF DHL+ L ++GI
Sbjct: 51 QHTPMSSEEEDVQSRSMASSSSSSSSHSLKHEVFLSFRGEDVRKNFVDHLYKDLVQQGIQ 110
Query: 172 AFRDDTKLKKGEFIAPGLFRAIEASQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHI 231
++DD L +GE I P L +AI+ S++ +VVFS+NYA S+WCL EL +I+ C G+ +
Sbjct: 111 TYKDDETLPRGERIGPALLKAIQESRIALVVFSENYADSSWCLDELAHIMECMDTRGQIV 170
Query: 232 LPIFYDVDPSEVQKQSGGYGEALSKHGFKHGLNMVHRWRETLTQVGNIS 280
+PIFY VDPS+V+KQ G YG+A KH + V WR+ L + GN+S
Sbjct: 171 IPIFYFVDPSDVRKQKGKYGKAFRKHK-RENKQKVESWRKALEKAGNLS 218
Score = 99.4 bits (246), Expect = 1e-19
Identities = 54/127 (42%), Positives = 84/127 (65%), Gaps = 3/127 (2%)
Query: 17 AIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPSEVRKQSGGYG 76
AI++S++ +VVFS+N+ADS +CL ELA+I+ C G+ ++PIFY VDPS+VRKQ G YG
Sbjct: 131 AIQESRIALVVFSENYADSSWCLDELAHIMECMDTRGQIVIPIFYFVDPSDVRKQKGKYG 190
Query: 77 ESLAK-LEEIAPQVQRWREALQLVGNISGWDLCHKPQHAE-LENIIEHINI-LGCKFSSR 133
++ K E +V+ WR+AL+ GN+SGW + A+ ++ I+ I+ L ++
Sbjct: 191 KAFRKHKRENKQKVESWRKALEKAGNLSGWVINENSHEAKCIKEIVATISSRLPTLSTNV 250
Query: 134 TKDLVEI 140
KDL+ I
Sbjct: 251 NKDLIGI 257
>UniRef100_Q947E2 Resistance gene analog NBS6 [Helianthus annuus]
Length = 303
Score = 148 bits (373), Expect = 2e-34
Identities = 72/169 (42%), Positives = 105/169 (61%), Gaps = 1/169 (0%)
Query: 112 QHAELENIIEHINILGCKFSSRTKDLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGIN 171
QH + + E + SS + + ++VF+SFRG D R NF DHL+ L ++GI
Sbjct: 15 QHTPMSSEEEDVQSRSMASSSSSSSSHSLKHEVFLSFRGEDVRKNFVDHLYKDLVQQGIQ 74
Query: 172 AFRDDTKLKKGEFIAPGLFRAIEASQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHI 231
++DD L +GE I P L +AI+ S++ +VVFS+NYA S+WCL EL +I+ C G+ +
Sbjct: 75 TYKDDETLPRGERIGPALLKAIQESRIALVVFSENYADSSWCLDELAHIMECMDTRGQIV 134
Query: 232 LPIFYDVDPSEVQKQSGGYGEALSKHGFKHGLNMVHRWRETLTQVGNIS 280
+PIFY VDPS+V+KQ G YG+A KH + V WR+ L + GN+S
Sbjct: 135 IPIFYFVDPSDVRKQKGKYGKAFRKHK-RENKQKVESWRKALEKAGNLS 182
Score = 99.4 bits (246), Expect = 1e-19
Identities = 54/127 (42%), Positives = 84/127 (65%), Gaps = 3/127 (2%)
Query: 17 AIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPSEVRKQSGGYG 76
AI++S++ +VVFS+N+ADS +CL ELA+I+ C G+ ++PIFY VDPS+VRKQ G YG
Sbjct: 95 AIQESRIALVVFSENYADSSWCLDELAHIMECMDTRGQIVIPIFYFVDPSDVRKQKGKYG 154
Query: 77 ESLAK-LEEIAPQVQRWREALQLVGNISGWDLCHKPQHAE-LENIIEHINI-LGCKFSSR 133
++ K E +V+ WR+AL+ GN+SGW + A+ ++ I+ I+ L ++
Sbjct: 155 KAFRKHKRENKQKVESWRKALEKAGNLSGWVINENSHEAKRIKEIVATISSRLPTLSTNV 214
Query: 134 TKDLVEI 140
KDL+ I
Sbjct: 215 NKDLIGI 221
>UniRef100_Q947D9 Resistance gene analog NBS9 [Helianthus annuus]
Length = 304
Score = 147 bits (372), Expect = 2e-34
Identities = 71/150 (47%), Positives = 100/150 (66%), Gaps = 1/150 (0%)
Query: 131 SSRTKDLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLF 190
SS + + ++VF+SFRG DTR NF DHL+ L ++GI ++DD L +GE I P L
Sbjct: 35 SSSSSSSHSLKHEVFLSFRGEDTRKNFVDHLYKDLVQQGIQTYKDDETLPRGERIGPALL 94
Query: 191 RAIEASQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGY 250
+AI+ S +VVFS+NYA S+WCL EL +I+ C G+ ++PIFY VDPS+V+KQ+G Y
Sbjct: 95 KAIQESHFAVVVFSENYADSSWCLDELAHIMECVDTRGQIVIPIFYHVDPSDVRKQNGKY 154
Query: 251 GEALSKHGFKHGLNMVHRWRETLTQVGNIS 280
G+A +KH K+ V WR L + GN+S
Sbjct: 155 GKAFTKHERKN-KQKVESWRNALEKAGNLS 183
Score = 99.8 bits (247), Expect = 7e-20
Identities = 55/127 (43%), Positives = 82/127 (64%), Gaps = 3/127 (2%)
Query: 17 AIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPSEVRKQSGGYG 76
AI++S +VVFS+N+ADS +CL ELA+I+ C G+ ++PIFY VDPS+VRKQ+G YG
Sbjct: 96 AIQESHFAVVVFSENYADSSWCLDELAHIMECVDTRGQIVIPIFYHVDPSDVRKQNGKYG 155
Query: 77 ESLAKLEEIAPQ-VQRWREALQLVGNISGWDLCHKPQHAE-LENIIEHI-NILGCKFSSR 133
++ K E Q V+ WR AL+ GN+SGW + A+ + +I+ I + L ++
Sbjct: 156 KAFTKHERKNKQKVESWRNALEKAGNLSGWVIDENSHEAQCISDIVGTISDRLSSLNTND 215
Query: 134 TKDLVEI 140
KDL+ I
Sbjct: 216 NKDLIGI 222
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.140 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 502,833,663
Number of Sequences: 2790947
Number of extensions: 20861026
Number of successful extensions: 44199
Number of sequences better than 10.0: 388
Number of HSP's better than 10.0 without gapping: 321
Number of HSP's successfully gapped in prelim test: 67
Number of HSP's that attempted gapping in prelim test: 42901
Number of HSP's gapped (non-prelim): 768
length of query: 287
length of database: 848,049,833
effective HSP length: 126
effective length of query: 161
effective length of database: 496,390,511
effective search space: 79918872271
effective search space used: 79918872271
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146863.3


