

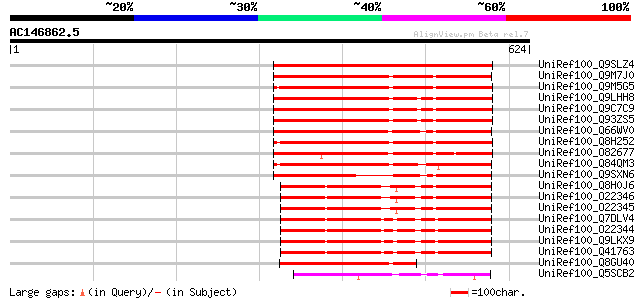
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.5 + phase: 0 /pseudo
(624 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SLZ4 Retinoblastoma-related protein [Pisum sativum] 507 e-142
UniRef100_Q9M7J0 Retinoblastoma-related protein 1 [Populus tremu... 413 e-114
UniRef100_Q9M5G5 Retinoblastoma-like protein [Euphorbia esula] 411 e-113
UniRef100_Q9LHH8 Retinoblastoma-related protein [Arabidopsis tha... 402 e-110
UniRef100_Q9C7C9 Retinoblastoma-related protein, putative; 44014... 402 e-110
UniRef100_Q93ZS5 Putative retinoblastoma-related protein [Arabid... 402 e-110
UniRef100_Q66WV0 Retinoblastoma protein [Nicotiana benthamiana] 398 e-109
UniRef100_Q8H252 Retinoblastoma-like protein [Cocos nucifera] 396 e-109
UniRef100_O82677 Retinoblastoma-related protein [Chenopodium rub... 392 e-107
UniRef100_Q84QM3 Putative Retinoblastoma protein [Oryza sativa] 353 6e-96
UniRef100_Q9SXN6 NtRb1 protein [Nicotiana tabacum] 318 4e-85
UniRef100_Q8H0J6 Retinoblastoma-related protein [Zea mays] 295 2e-78
UniRef100_O22346 Retinoblastoma-related protein 2b [Zea mays] 291 3e-77
UniRef100_O22345 Retinoblastoma-related protein 2a [Zea mays] 291 3e-77
UniRef100_Q7DLV4 Rb1 protein [Zea mays] 288 3e-76
UniRef100_O22344 Retinoblastoma-related protein 1 [Zea mays] 288 3e-76
UniRef100_Q9LKX9 Retinoblastoma related protein RBR1 [Zea mays] 288 3e-76
UniRef100_Q41763 Retinoblastoma-like protein [Zea mays] 286 1e-75
UniRef100_Q8GU40 Putative retinoblastoma protein [Physcomitrella... 204 6e-51
UniRef100_Q5SCB2 Retinoblastoma protein [Ostreococcus tauri] 123 2e-26
>UniRef100_Q9SLZ4 Retinoblastoma-related protein [Pisum sativum]
Length = 1026
Score = 507 bits (1305), Expect = e-142
Identities = 247/263 (93%), Positives = 253/263 (95%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ
Sbjct: 707 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 766
Query: 378 QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC 437
QIRENVY LFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC
Sbjct: 767 QIRENVYCLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC 826
Query: 438 KPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRS 497
KP+VFRSVFVDWSSARRNG + RTGQEHIDIISFYNEVFIPSVKPLLVE+GPGGAT R+
Sbjct: 827 KPEVFRSVFVDWSSARRNGSCKQRTGQEHIDIISFYNEVFIPSVKPLLVEIGPGGATTRN 886
Query: 498 DQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHS 557
D++PEANNKNDGHL Q PGSPRISPFPSLPDMSPKKVSA HNVYVSPLRSSKMDALISHS
Sbjct: 887 DRIPEANNKNDGHLAQCPGSPRISPFPSLPDMSPKKVSATHNVYVSPLRSSKMDALISHS 946
Query: 558 SKSYYACXGESTHAYQSPXKDLT 580
SKSYYAC GESTHAYQSP KDLT
Sbjct: 947 SKSYYACVGESTHAYQSPSKDLT 969
Score = 45.4 bits (106), Expect = 0.005
Identities = 21/22 (95%), Positives = 21/22 (95%)
Query: 66 PVSTAMTTAKWLRTVISPLPIK 87
PVSTAMTTAKWLRTVISPLP K
Sbjct: 417 PVSTAMTTAKWLRTVISPLPSK 438
>UniRef100_Q9M7J0 Retinoblastoma-related protein 1 [Populus tremula x Populus
tremuloides]
Length = 1035
Score = 413 bits (1061), Expect = e-114
Identities = 206/263 (78%), Positives = 228/263 (86%), Gaps = 8/263 (3%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
LKSKL PPPLQSAFASPT+PNPGGGGETCAETGI+VFF+KI KL AVRI+GM+E+LQ SQ
Sbjct: 723 LKSKLPPPPLQSAFASPTRPNPGGGGETCAETGINVFFTKINKLAAVRINGMIEKLQPSQ 782
Query: 378 Q-IRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQ 436
Q IRENVY LFQ IL+ TSLFFNRHIDQIILCCFYGVAKIS+LNLTFREIIYNYR+QP
Sbjct: 783 QHIRENVYRLFQLILSHQTSLFFNRHIDQIILCCFYGVAKISKLNLTFREIIYNYRRQPH 842
Query: 437 CKPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVR 496
CK VFRSVFVDWSSAR NG RTGQ+H+DII+FYNE+FIP+ KPLLV++G G TV+
Sbjct: 843 CKTLVFRSVFVDWSSARHNG----RTGQDHVDIITFYNEIFIPAAKPLLVDVGSAGTTVK 898
Query: 497 SDQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISH 556
+ VPE N DG Q P SP++SPFPSLPDMSPKKVS+ HNVYVSPLRSSKMDALIS+
Sbjct: 899 ASNVPEVGNNKDG---QCPASPKVSPFPSLPDMSPKKVSSVHNVYVSPLRSSKMDALISN 955
Query: 557 SSKSYYACXGESTHAYQSPXKDL 579
SSKSYYAC GESTHAYQSP KDL
Sbjct: 956 SSKSYYACVGESTHAYQSPSKDL 978
Score = 45.4 bits (106), Expect = 0.005
Identities = 20/27 (74%), Positives = 23/27 (85%)
Query: 61 RLLPPPVSTAMTTAKWLRTVISPLPIK 87
++ PVSTAMTTAKWLRT+ISPLP K
Sbjct: 427 KMAATPVSTAMTTAKWLRTIISPLPSK 453
>UniRef100_Q9M5G5 Retinoblastoma-like protein [Euphorbia esula]
Length = 801
Score = 411 bits (1057), Expect = e-113
Identities = 207/264 (78%), Positives = 229/264 (86%), Gaps = 9/264 (3%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
LKSK+ PPPLQSAFASPT+PNPGGGGETCAET IS+FF KI KL AVRI+GM+ERLQ S
Sbjct: 489 LKSKI-PPPLQSAFASPTRPNPGGGGETCAETAISIFFCKINKLAAVRINGMIERLQQSH 547
Query: 378 Q-IRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQ 436
IRENVY LFQ+IL+ TSLFFNRHIDQIILCCFYGVAKIS++NLTFREII NYRKQPQ
Sbjct: 548 HHIRENVYRLFQQILSHRTSLFFNRHIDQIILCCFYGVAKISKVNLTFREIICNYRKQPQ 607
Query: 437 CKPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVR 496
CKPQVFRSVFVDWSS+R+NG RTGQ+H+DII+FYNE+FIP+ KPLLVE+G G +
Sbjct: 608 CKPQVFRSVFVDWSSSRQNG----RTGQDHVDIITFYNEIFIPAAKPLLVEVGSSGTITK 663
Query: 497 SDQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISH 556
S QVPE N DG Q P SP++SPFPSLPDMSPKKVSA+HNVYVSPLR+SKMDALISH
Sbjct: 664 STQVPEVINSKDG---QCPQSPKVSPFPSLPDMSPKKVSASHNVYVSPLRTSKMDALISH 720
Query: 557 SSKSYYACXGESTHAYQSPXKDLT 580
SSKSYYAC GESTHAYQSP KDLT
Sbjct: 721 SSKSYYACVGESTHAYQSPSKDLT 744
Score = 45.1 bits (105), Expect = 0.006
Identities = 21/36 (58%), Positives = 26/36 (71%)
Query: 61 RLLPPPVSTAMTTAKWLRTVISPLPIKAVTRARAIL 96
++ PV+TAMTTAKWLRTVISPLP K ++ L
Sbjct: 193 KIADTPVTTAMTTAKWLRTVISPLPSKPSSQLERFL 228
>UniRef100_Q9LHH8 Retinoblastoma-related protein [Arabidopsis thaliana]
Length = 1021
Score = 402 bits (1032), Expect = e-110
Identities = 201/263 (76%), Positives = 227/263 (85%), Gaps = 11/263 (4%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
+KSK+LPPPLQSAFASPT+PNPGGGGETCAETGI++FF+KI KL AVRI+GMVERLQLSQ
Sbjct: 705 VKSKMLPPPLQSAFASPTRPNPGGGGETCAETGINIFFTKINKLAAVRINGMVERLQLSQ 764
Query: 378 QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC 437
QIRE+VY FQ +L Q TSL F+RHIDQIILCCFYGVAKISQ++LTFREIIYNYRKQPQC
Sbjct: 765 QIRESVYCFFQHVLAQRTSLLFSRHIDQIILCCFYGVAKISQMSLTFREIIYNYRKQPQC 824
Query: 438 KPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRS 497
KP VFRSV+VD RR G R G +H+DII+FYNE+FIP+VKPLLVELGP VR+
Sbjct: 825 KPLVFRSVYVDALQCRRQG----RIGPDHVDIITFYNEIFIPAVKPLLVELGP----VRN 876
Query: 498 DQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHS 557
D+ EANNK +G Q PGSP++S FPS+PDMSPKKVSA HNVYVSPLR SKMDALISHS
Sbjct: 877 DRAVEANNKPEG---QCPGSPKVSVFPSVPDMSPKKVSAVHNVYVSPLRGSKMDALISHS 933
Query: 558 SKSYYACXGESTHAYQSPXKDLT 580
+KSYYAC GESTHAYQSP KDL+
Sbjct: 934 TKSYYACVGESTHAYQSPSKDLS 956
Score = 42.7 bits (99), Expect = 0.032
Identities = 20/24 (83%), Positives = 21/24 (87%)
Query: 61 RLLPPPVSTAMTTAKWLRTVISPL 84
+L PVSTAMTTAKWLRTVISPL
Sbjct: 410 KLAATPVSTAMTTAKWLRTVISPL 433
>UniRef100_Q9C7C9 Retinoblastoma-related protein, putative; 44014-38352 [Arabidopsis
thaliana]
Length = 1088
Score = 402 bits (1032), Expect = e-110
Identities = 201/263 (76%), Positives = 227/263 (85%), Gaps = 11/263 (4%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
+KSK+LPPPLQSAFASPT+PNPGGGGETCAETGI++FF+KI KL AVRI+GMVERLQLSQ
Sbjct: 773 VKSKMLPPPLQSAFASPTRPNPGGGGETCAETGINIFFTKINKLAAVRINGMVERLQLSQ 832
Query: 378 QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC 437
QIRE+VY FQ +L Q TSL F+RHIDQIILCCFYGVAKISQ++LTFREIIYNYRKQPQC
Sbjct: 833 QIRESVYCFFQHVLAQRTSLLFSRHIDQIILCCFYGVAKISQMSLTFREIIYNYRKQPQC 892
Query: 438 KPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRS 497
KP VFRSV+VD RR G R G +H+DII+FYNE+FIP+VKPLLVELGP VR+
Sbjct: 893 KPLVFRSVYVDALQCRRQG----RIGPDHVDIITFYNEIFIPAVKPLLVELGP----VRN 944
Query: 498 DQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHS 557
D+ EANNK +G Q PGSP++S FPS+PDMSPKKVSA HNVYVSPLR SKMDALISHS
Sbjct: 945 DRAVEANNKPEG---QCPGSPKVSVFPSVPDMSPKKVSAVHNVYVSPLRGSKMDALISHS 1001
Query: 558 SKSYYACXGESTHAYQSPXKDLT 580
+KSYYAC GESTHAYQSP KDL+
Sbjct: 1002 TKSYYACVGESTHAYQSPSKDLS 1024
Score = 42.7 bits (99), Expect = 0.032
Identities = 20/24 (83%), Positives = 21/24 (87%)
Query: 61 RLLPPPVSTAMTTAKWLRTVISPL 84
+L PVSTAMTTAKWLRTVISPL
Sbjct: 478 KLAATPVSTAMTTAKWLRTVISPL 501
>UniRef100_Q93ZS5 Putative retinoblastoma-related protein [Arabidopsis thaliana]
Length = 1013
Score = 402 bits (1032), Expect = e-110
Identities = 201/263 (76%), Positives = 227/263 (85%), Gaps = 11/263 (4%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
+KSK+LPPPLQSAFASPT+PNPGGGGETCAETGI++FF+KI KL AVRI+GMVERLQLSQ
Sbjct: 697 VKSKMLPPPLQSAFASPTRPNPGGGGETCAETGINIFFTKINKLAAVRINGMVERLQLSQ 756
Query: 378 QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC 437
QIRE+VY FQ +L Q TSL F+RHIDQIILCCFYGVAKISQ++LTFREIIYNYRKQPQC
Sbjct: 757 QIRESVYCFFQHVLAQRTSLLFSRHIDQIILCCFYGVAKISQMSLTFREIIYNYRKQPQC 816
Query: 438 KPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRS 497
KP VFRSV+VD RR G R G +H+DII+FYNE+FIP+VKPLLVELGP VR+
Sbjct: 817 KPLVFRSVYVDALQCRRQG----RIGPDHVDIITFYNEIFIPAVKPLLVELGP----VRN 868
Query: 498 DQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHS 557
D+ EANNK +G Q PGSP++S FPS+PDMSPKKVSA HNVYVSPLR SKMDALISHS
Sbjct: 869 DRAVEANNKPEG---QCPGSPKVSVFPSVPDMSPKKVSAVHNVYVSPLRGSKMDALISHS 925
Query: 558 SKSYYACXGESTHAYQSPXKDLT 580
+KSYYAC GESTHAYQSP KDL+
Sbjct: 926 TKSYYACVGESTHAYQSPSKDLS 948
Score = 42.7 bits (99), Expect = 0.032
Identities = 20/24 (83%), Positives = 21/24 (87%)
Query: 61 RLLPPPVSTAMTTAKWLRTVISPL 84
+L PVSTAMTTAKWLRTVISPL
Sbjct: 402 KLAATPVSTAMTTAKWLRTVISPL 425
>UniRef100_Q66WV0 Retinoblastoma protein [Nicotiana benthamiana]
Length = 1003
Score = 398 bits (1022), Expect = e-109
Identities = 203/262 (77%), Positives = 219/262 (83%), Gaps = 13/262 (4%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
+KSK PP L SAFASPT+PNPGGGGETCAET I+VFF KIVKL AVRI+GM+ERLQLSQ
Sbjct: 699 IKSKFPPPALHSAFASPTRPNPGGGGETCAETAINVFFGKIVKLAAVRINGMIERLQLSQ 758
Query: 378 QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC 437
QIRE VY LFQ+IL+Q TSLFFNRHIDQIILC FYGVAKISQLNLTF+EII NYRKQPQC
Sbjct: 759 QIRETVYCLFQKILSQRTSLFFNRHIDQIILCSFYGVAKISQLNLTFKEIICNYRKQPQC 818
Query: 438 KPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRS 497
KPQVFRSVFVDW+ AR N RTG +H+DII+FYNE+FIPSVKPLLVEL P G
Sbjct: 819 KPQVFRSVFVDWTLARHN----VRTGADHVDIITFYNEMFIPSVKPLLVELAPAGNN--- 871
Query: 498 DQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHS 557
E N+ NDG Q P SP+ SPFP LPDMSPKKVSA HNVYVSPLR+SKMDALISHS
Sbjct: 872 ---SEKNDHNDG---QGPASPKPSPFPKLPDMSPKKVSAVHNVYVSPLRASKMDALISHS 925
Query: 558 SKSYYACXGESTHAYQSPXKDL 579
SKSYYAC GESTHAYQSP KDL
Sbjct: 926 SKSYYACVGESTHAYQSPSKDL 947
Score = 39.7 bits (91), Expect = 0.27
Identities = 18/22 (81%), Positives = 20/22 (90%)
Query: 66 PVSTAMTTAKWLRTVISPLPIK 87
PVSTAMTTA+WLRTVI+PL K
Sbjct: 406 PVSTAMTTARWLRTVIAPLQPK 427
>UniRef100_Q8H252 Retinoblastoma-like protein [Cocos nucifera]
Length = 1011
Score = 396 bits (1018), Expect = e-109
Identities = 202/263 (76%), Positives = 221/263 (83%), Gaps = 9/263 (3%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
LKSKL PPLQSAFASPT+PNP GGGETCAETGI++FF+KIVKL A+RI + ERLQL Q
Sbjct: 701 LKSKL--PPLQSAFASPTRPNPAGGGETCAETGINIFFNKIVKLAAIRIRSLCERLQLPQ 758
Query: 378 QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC 437
QI E VY L Q+IL+Q T+LFFNRHIDQIILC FYGVAKISQ+ LTF+EIIYNYRKQPQC
Sbjct: 759 QILEWVYCLIQQILSQRTALFFNRHIDQIILCSFYGVAKISQMALTFKEIIYNYRKQPQC 818
Query: 438 KPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRS 497
KPQVFRSVFV W S NG +TGQEH+DII+FYNEVFIPSVKPLLVELGP G +S
Sbjct: 819 KPQVFRSVFVHWPSTSHNG----KTGQEHVDIITFYNEVFIPSVKPLLVELGPAGVAQKS 874
Query: 498 DQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHS 557
PE +N D Q PGSPR+SPFP+LPDMSPKKVSAAHNVYVSPLRSSKMDAL+S S
Sbjct: 875 KSSPEDSNNADS---QIPGSPRLSPFPNLPDMSPKKVSAAHNVYVSPLRSSKMDALLSPS 931
Query: 558 SKSYYACXGESTHAYQSPXKDLT 580
SKSYYAC GESTHAYQSP KDLT
Sbjct: 932 SKSYYACVGESTHAYQSPSKDLT 954
Score = 42.7 bits (99), Expect = 0.032
Identities = 19/25 (76%), Positives = 21/25 (84%)
Query: 66 PVSTAMTTAKWLRTVISPLPIKAVT 90
PVSTAMTTAKWLR +ISPLP + T
Sbjct: 412 PVSTAMTTAKWLRNIISPLPSRPST 436
>UniRef100_O82677 Retinoblastoma-related protein [Chenopodium rubrum]
Length = 1012
Score = 392 bits (1007), Expect = e-107
Identities = 204/273 (74%), Positives = 225/273 (81%), Gaps = 20/273 (7%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERL---- 373
LK KL PP LQSAFASPT+ +PGGGGETCAETGI++FFSKIVKL AVRI+GM+ERL
Sbjct: 693 LKQKLTPPALQSAFASPTRLSPGGGGETCAETGINIFFSKIVKLAAVRINGMIERLEKLE 752
Query: 374 ------QLSQQIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREI 427
QL +Q+RENVY F ILNQ TSLFFNRHIDQIILC FYGVAKISQ LTF+EI
Sbjct: 753 KQSQTQQLREQLRENVYCRFHIILNQRTSLFFNRHIDQIILCAFYGVAKISQYELTFKEI 812
Query: 428 IYNYRKQPQCKPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVE 487
I NYRKQPQCKPQVFRSVFVDWS +RRN R GQ+H+DII+FYNEVFIPSVKPLL E
Sbjct: 813 ILNYRKQPQCKPQVFRSVFVDWSFSRRN-----RQGQDHVDIITFYNEVFIPSVKPLLGE 867
Query: 488 LGPGGATVRSDQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRS 547
LGP GA V++D +PE+NN DG Q PGSP++S FPSLPDMSPKKV A NVYVSPLRS
Sbjct: 868 LGPQGACVKTDLIPESNNDKDG---QCPGSPKLSTFPSLPDMSPKKV--AKNVYVSPLRS 922
Query: 548 SKMDALISHSSKSYYACXGESTHAYQSPXKDLT 580
SKMDALIS+SSKSYYAC GESTHA+QSP KDLT
Sbjct: 923 SKMDALISNSSKSYYACVGESTHAFQSPSKDLT 955
Score = 46.2 bits (108), Expect = 0.003
Identities = 21/27 (77%), Positives = 23/27 (84%)
Query: 61 RLLPPPVSTAMTTAKWLRTVISPLPIK 87
R++ PVSTAMTTAKWLRT ISPLP K
Sbjct: 399 RMISTPVSTAMTTAKWLRTYISPLPSK 425
>UniRef100_Q84QM3 Putative Retinoblastoma protein [Oryza sativa]
Length = 1010
Score = 353 bits (907), Expect = 6e-96
Identities = 184/265 (69%), Positives = 208/265 (78%), Gaps = 18/265 (6%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
LKSKL PPLQSAF SPT+PNP GGE CAETGI VF SKI KL A+RI G+ ERLQLSQ
Sbjct: 702 LKSKL--PPLQSAFLSPTRPNPAAGGELCAETGIGVFLSKIAKLAAIRIRGLCERLQLSQ 759
Query: 378 QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC 437
Q+ E VYSL Q+I+ Q T+LFFNRHIDQIILC YGVAKISQL LTF+EII+ YRKQ QC
Sbjct: 760 QVLERVYSLVQQIIIQQTALFFNRHIDQIILCSIYGVAKISQLALTFKEIIFGYRKQSQC 819
Query: 438 KPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRS 497
KPQVFRSV+V W+S RNG +TG++H+DII+FYNEVFIP+VKPLLVELG G
Sbjct: 820 KPQVFRSVYVHWASRSRNG----KTGEDHVDIITFYNEVFIPTVKPLLVELGSG------ 869
Query: 498 DQVPEANNKNDGHLVQN---PGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALI 554
N KN+ + P SPR+S FP+LPDMSPKKVSAAHNVYVSPLR+SKMD L+
Sbjct: 870 ---TSPNKKNEEKCAADGPYPESPRLSRFPNLPDMSPKKVSAAHNVYVSPLRTSKMDTLL 926
Query: 555 SHSSKSYYACXGESTHAYQSPXKDL 579
S SSKSYYAC GESTHA+QSP KDL
Sbjct: 927 SPSSKSYYACVGESTHAFQSPSKDL 951
Score = 42.4 bits (98), Expect = 0.041
Identities = 19/22 (86%), Positives = 20/22 (90%)
Query: 66 PVSTAMTTAKWLRTVISPLPIK 87
PVSTAMTTAKWLR+ ISPLP K
Sbjct: 420 PVSTAMTTAKWLRSTISPLPSK 441
>UniRef100_Q9SXN6 NtRb1 protein [Nicotiana tabacum]
Length = 961
Score = 318 bits (814), Expect = 4e-85
Identities = 171/262 (65%), Positives = 184/262 (69%), Gaps = 54/262 (20%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
+KSK PP LQSAFASPT+PNPGGGGETCAET I+VFF KIVKL AVRI+GM+ERLQLSQ
Sbjct: 698 IKSKFPPPALQSAFASPTRPNPGGGGETCAETAINVFFGKIVKLAAVRINGMIERLQLSQ 757
Query: 378 QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC 437
QIRE VY LFQ+IL+Q TSLFFNRHIDQIILC FYGVAK
Sbjct: 758 QIRETVYCLFQKILSQRTSLFFNRHIDQIILCSFYGVAK--------------------- 796
Query: 438 KPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRS 497
RTG +H+DII+FYNE+FIPSVKPLLVEL P G
Sbjct: 797 -----------------------RTGADHVDIITFYNEMFIPSVKPLLVELAPAGNA--- 830
Query: 498 DQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHS 557
E NN NDG Q P SP+ SPFP LPDMSPKKVSA HNVYVSPLR+SKMDALISHS
Sbjct: 831 ----EKNNHNDG---QGPASPKPSPFPKLPDMSPKKVSAVHNVYVSPLRASKMDALISHS 883
Query: 558 SKSYYACXGESTHAYQSPXKDL 579
SKSYYAC GESTHAYQSP KDL
Sbjct: 884 SKSYYACVGESTHAYQSPSKDL 905
Score = 39.7 bits (91), Expect = 0.27
Identities = 18/22 (81%), Positives = 20/22 (90%)
Query: 66 PVSTAMTTAKWLRTVISPLPIK 87
PVSTAMTTA+WLRTVI+PL K
Sbjct: 405 PVSTAMTTARWLRTVIAPLQPK 426
>UniRef100_Q8H0J6 Retinoblastoma-related protein [Zea mays]
Length = 866
Score = 295 bits (756), Expect = 2e-78
Identities = 150/257 (58%), Positives = 187/257 (72%), Gaps = 22/257 (8%)
Query: 326 PLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYS 385
PLQS FASPT NP GG E CA+ I +FFSKI+KL A+RI + ER+Q +Q E VY+
Sbjct: 571 PLQSTFASPTVSNPVGGNEKCADVTIQIFFSKILKLAAIRIRNLCERIQYMEQT-ERVYN 629
Query: 386 LFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSV 445
+F++IL+Q T+LFFNRHIDQ+ILCC YGVAK+ QL L+FREI+ NY+K+ QCKP+VF S+
Sbjct: 630 VFKQILDQQTTLFFNRHIDQLILCCLYGVAKVCQLELSFREILNNYKKEAQCKPEVFLSI 689
Query: 446 FVDWSSARRNGGSRHRTG---QEHIDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPE 502
++ GSR+ G H+DII+FYNEVF+P+ KP LV L + S PE
Sbjct: 690 YI---------GSRNHNGVLISRHVDIITFYNEVFVPAAKPFLVSL------ISSGTRPE 734
Query: 503 ANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHSSKSYY 562
G Q PGSP++SPFP+LPDMSPKKVSA+HNVYVSPLR +KMD L+S SS+S+Y
Sbjct: 735 DKKNASG---QVPGSPKLSPFPNLPDMSPKKVSASHNVYVSPLRQTKMDLLLSPSSRSFY 791
Query: 563 ACXGESTHAYQSPXKDL 579
AC GE THAYQSP KDL
Sbjct: 792 ACIGEGTHAYQSPSKDL 808
Score = 37.7 bits (86), Expect = 1.0
Identities = 16/31 (51%), Positives = 24/31 (76%)
Query: 66 PVSTAMTTAKWLRTVISPLPIKAVTRARAIL 96
P+++AMTTAKWLR VIS LP K ++ + ++
Sbjct: 275 PITSAMTTAKWLREVISSLPEKPSSKLQQLM 305
>UniRef100_O22346 Retinoblastoma-related protein 2b [Zea mays]
Length = 584
Score = 291 bits (746), Expect = 3e-77
Identities = 148/257 (57%), Positives = 186/257 (71%), Gaps = 22/257 (8%)
Query: 326 PLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYS 385
PLQS FASPT NP GG E CA+ I +FFSKI+KL A+RI + ER+Q +Q E VY+
Sbjct: 316 PLQSTFASPTVSNPVGGNEKCADVTIQIFFSKILKLAAIRIRNLCERIQYMEQT-ERVYN 374
Query: 386 LFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSV 445
+F++IL+Q T+LFFNRH+ Q+ILCC YGVAK+ QL L+FREI+ NY+K+ QCKP+VF S+
Sbjct: 375 VFKQILDQQTTLFFNRHMHQLILCCLYGVAKVCQLELSFREILNNYKKEAQCKPEVFLSI 434
Query: 446 FVDWSSARRNGGSRHRTG---QEHIDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPE 502
++ GSR+ G H+DII+FYNEVF+P+ KP LV L + S PE
Sbjct: 435 YI---------GSRNHNGVLISRHVDIITFYNEVFVPAAKPFLVSL------ISSGTRPE 479
Query: 503 ANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHSSKSYY 562
G Q PGSP++SPFP+LPDMSPKKVSA+HNVYVSPLR +KMD L+S SS+S+Y
Sbjct: 480 DKKNASG---QVPGSPKLSPFPNLPDMSPKKVSASHNVYVSPLRQTKMDLLLSPSSRSFY 536
Query: 563 ACXGESTHAYQSPXKDL 579
AC GE THAYQSP KDL
Sbjct: 537 ACIGEGTHAYQSPSKDL 553
Score = 37.7 bits (86), Expect = 1.0
Identities = 16/31 (51%), Positives = 24/31 (76%)
Query: 66 PVSTAMTTAKWLRTVISPLPIKAVTRARAIL 96
P+++AMTTAKWLR VIS LP K ++ + ++
Sbjct: 20 PITSAMTTAKWLREVISSLPEKPSSKLQQLM 50
>UniRef100_O22345 Retinoblastoma-related protein 2a [Zea mays]
Length = 420
Score = 291 bits (746), Expect = 3e-77
Identities = 148/257 (57%), Positives = 186/257 (71%), Gaps = 22/257 (8%)
Query: 326 PLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYS 385
PLQS FASPT NP GG E CA+ I +FFSKI+KL A+RI + ER+Q +Q E VY+
Sbjct: 125 PLQSTFASPTVSNPVGGNEKCADVTIQIFFSKILKLAAIRIRNLCERIQYMEQT-ERVYN 183
Query: 386 LFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSV 445
+F++IL+Q T+LFFNRH+ Q+ILCC YGVAK+ QL L+FREI+ NY+K+ QCKP+VF S+
Sbjct: 184 VFKQILDQQTTLFFNRHMHQLILCCLYGVAKVCQLELSFREILNNYKKEAQCKPEVFLSI 243
Query: 446 FVDWSSARRNGGSRHRTG---QEHIDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPE 502
++ GSR+ G H+DII+FYNEVF+P+ KP LV L + S PE
Sbjct: 244 YI---------GSRNHNGVLISRHVDIITFYNEVFVPAAKPFLVSL------ISSGTRPE 288
Query: 503 ANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHSSKSYY 562
G Q PGSP++SPFP+LPDMSPKKVSA+HNVYVSPLR +KMD L+S SS+S+Y
Sbjct: 289 DKKNASG---QVPGSPKLSPFPNLPDMSPKKVSASHNVYVSPLRQTKMDLLLSPSSRSFY 345
Query: 563 ACXGESTHAYQSPXKDL 579
AC GE THAYQSP KDL
Sbjct: 346 ACIGEGTHAYQSPSKDL 362
>UniRef100_Q7DLV4 Rb1 protein [Zea mays]
Length = 683
Score = 288 bits (738), Expect = 3e-76
Identities = 148/254 (58%), Positives = 184/254 (72%), Gaps = 16/254 (6%)
Query: 326 PLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYS 385
PLQS FASPT NP GG E CA+ I +FFSKI+KL A+RI + ER+Q +Q E VY+
Sbjct: 388 PLQSTFASPTVCNPVGGNEKCADVTIHIFFSKILKLAAIRIRNLCERVQCVEQT-ERVYN 446
Query: 386 LFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSV 445
+F++IL Q T+LFFNRHIDQ+ILCC YGVAK+ QL LTFREI+ NY+++ QCKP+VF S+
Sbjct: 447 VFKQILEQQTTLFFNRHIDQLILCCLYGVAKVCQLELTFREILNNYKREAQCKPEVFSSI 506
Query: 446 FVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANN 505
++ S RNG R H+ II+FYNEVF+P+ KP LV L + S PE
Sbjct: 507 YI--GSTNRNGVLVSR----HVGIITFYNEVFVPAAKPFLVSL------ISSGTHPEDKK 554
Query: 506 KNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHSSKSYYACX 565
G + PGSP+ SPFP+LPDMSPKKVSA+HNVYVSPLR +K+D L+S SS+S+YAC
Sbjct: 555 NASGQI---PGSPKPSPFPNLPDMSPKKVSASHNVYVSPLRQTKLDLLLSPSSRSFYACI 611
Query: 566 GESTHAYQSPXKDL 579
GE THAYQSP KDL
Sbjct: 612 GEGTHAYQSPSKDL 625
Score = 38.1 bits (87), Expect = 0.78
Identities = 18/31 (58%), Positives = 23/31 (74%)
Query: 66 PVSTAMTTAKWLRTVISPLPIKAVTRARAIL 96
PV++AMTTAKWLR VIS LP K ++ + L
Sbjct: 92 PVTSAMTTAKWLREVISSLPDKPSSKLQQFL 122
>UniRef100_O22344 Retinoblastoma-related protein 1 [Zea mays]
Length = 866
Score = 288 bits (738), Expect = 3e-76
Identities = 148/254 (58%), Positives = 184/254 (72%), Gaps = 16/254 (6%)
Query: 326 PLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYS 385
PLQS FASPT NP GG E CA+ I +FFSKI+KL A+RI + ER+Q +Q E VY+
Sbjct: 571 PLQSTFASPTVCNPVGGNEKCADVTIHIFFSKILKLAAIRIRNLCERVQCVEQT-ERVYN 629
Query: 386 LFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSV 445
+F++IL Q T+LFFNRHIDQ+ILCC YGVAK+ QL LTFREI+ NY+++ QCKP+VF S+
Sbjct: 630 VFKQILEQQTTLFFNRHIDQLILCCLYGVAKVCQLELTFREILNNYKREAQCKPEVFSSI 689
Query: 446 FVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANN 505
++ S RNG R H+ II+FYNEVF+P+ KP LV L + S PE
Sbjct: 690 YI--GSTNRNGVLVSR----HVGIITFYNEVFVPAAKPFLVSL------ISSGTHPEDKK 737
Query: 506 KNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHSSKSYYACX 565
G + PGSP+ SPFP+LPDMSPKKVSA+HNVYVSPLR +K+D L+S SS+S+YAC
Sbjct: 738 NASGQI---PGSPKPSPFPNLPDMSPKKVSASHNVYVSPLRQTKLDLLLSPSSRSFYACI 794
Query: 566 GESTHAYQSPXKDL 579
GE THAYQSP KDL
Sbjct: 795 GEGTHAYQSPSKDL 808
Score = 38.1 bits (87), Expect = 0.78
Identities = 18/31 (58%), Positives = 23/31 (74%)
Query: 66 PVSTAMTTAKWLRTVISPLPIKAVTRARAIL 96
PV++AMTTAKWLR VIS LP K ++ + L
Sbjct: 275 PVTSAMTTAKWLREVISSLPDKPSSKLQQFL 305
>UniRef100_Q9LKX9 Retinoblastoma related protein RBR1 [Zea mays]
Length = 867
Score = 288 bits (737), Expect = 3e-76
Identities = 148/254 (58%), Positives = 184/254 (72%), Gaps = 16/254 (6%)
Query: 326 PLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYS 385
PLQS FASPT NP GG E CA+ I +FFSKI+KL A+RI + ER+Q +Q E VY+
Sbjct: 572 PLQSTFASPTVCNPVGGNEKCADLTIHIFFSKILKLAAIRIRNLCERVQCVEQT-ERVYN 630
Query: 386 LFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSV 445
+F++IL Q T+LFFNRHIDQ+ILCC YGVAK+ QL LTFREI+ NY+++ QCKP+VF S+
Sbjct: 631 VFKQILEQQTTLFFNRHIDQLILCCLYGVAKVCQLELTFREILNNYKREAQCKPEVFSSI 690
Query: 446 FVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANN 505
++ S RNG R H+ II+FYNEVF+P+ KP LV L + S PE
Sbjct: 691 YI--GSTNRNGVLVSR----HVGIITFYNEVFVPAAKPFLVSL------ISSGTHPEDKK 738
Query: 506 KNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHSSKSYYACX 565
G + PGSP+ SPFP+LPDMSPKKVSA+HNVYVSPLR +K+D L+S SS+S+YAC
Sbjct: 739 NASGQI---PGSPKPSPFPNLPDMSPKKVSASHNVYVSPLRQTKLDLLLSPSSRSFYACI 795
Query: 566 GESTHAYQSPXKDL 579
GE THAYQSP KDL
Sbjct: 796 GEGTHAYQSPSKDL 809
Score = 38.1 bits (87), Expect = 0.78
Identities = 18/31 (58%), Positives = 23/31 (74%)
Query: 66 PVSTAMTTAKWLRTVISPLPIKAVTRARAIL 96
PV++AMTTAKWLR VIS LP K ++ + L
Sbjct: 276 PVTSAMTTAKWLREVISSLPDKPSSKLQQFL 306
>UniRef100_Q41763 Retinoblastoma-like protein [Zea mays]
Length = 527
Score = 286 bits (732), Expect = 1e-75
Identities = 147/254 (57%), Positives = 183/254 (71%), Gaps = 16/254 (6%)
Query: 326 PLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYS 385
PLQS FASPT NP GG E CA+ I +FFSKI+K A+RI + ER+Q +Q E VY+
Sbjct: 232 PLQSTFASPTVCNPVGGNEKCADVTIHIFFSKILKFPAIRIRNLCERVQCVEQT-ERVYN 290
Query: 386 LFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSV 445
+F++IL Q T+LFFNRHIDQ+ILCC YGVAK+ QL LTFREI+ NY+++ QCKP+VF S+
Sbjct: 291 VFKQILEQQTTLFFNRHIDQLILCCLYGVAKVCQLELTFREILNNYKREAQCKPEVFSSI 350
Query: 446 FVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANN 505
++ S RNG R H+ II+FYNEVF+P+ KP LV L + S PE
Sbjct: 351 YI--GSTNRNGVLVSR----HVGIITFYNEVFVPAAKPFLVSL------ISSGTHPEDKK 398
Query: 506 KNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHSSKSYYACX 565
G + PGSP+ SPFP+LPDMSPKKVSA+HNVYVSPLR +K+D L+S SS+S+YAC
Sbjct: 399 NASGQI---PGSPKPSPFPNLPDMSPKKVSASHNVYVSPLRQTKLDLLLSPSSRSFYACI 455
Query: 566 GESTHAYQSPXKDL 579
GE THAYQSP KDL
Sbjct: 456 GEGTHAYQSPSKDL 469
>UniRef100_Q8GU40 Putative retinoblastoma protein [Physcomitrella patens]
Length = 597
Score = 204 bits (519), Expect = 6e-51
Identities = 101/165 (61%), Positives = 122/165 (73%), Gaps = 4/165 (2%)
Query: 325 PPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVY 384
PPL AFASP +PNP GGETCAET I+VFF K++KL AVRI + ERLQ SQQ+ E V
Sbjct: 422 PPLLLAFASPQRPNPQAGGETCAETCINVFFQKVLKLAAVRIRNLCERLQQSQQVVERVN 481
Query: 385 SLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRS 444
LN T LFFNRHIDQIILCC YG+ K+S++N+TFR+IIY+YRKQPQCKP VFR+
Sbjct: 482 RALHHALNHETGLFFNRHIDQIILCCVYGICKVSKVNVTFRDIIYHYRKQPQCKPYVFRN 541
Query: 445 VFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELG 489
V +D S R+G + GQE DII FYNEVF+PS K L+++G
Sbjct: 542 VLLDGSVVWRSG----KAGQETGDIIRFYNEVFVPSTKSFLLQVG 582
Score = 38.9 bits (89), Expect = 0.46
Identities = 17/21 (80%), Positives = 18/21 (84%)
Query: 64 PPPVSTAMTTAKWLRTVISPL 84
P PVS MTTAKWLRTVI+PL
Sbjct: 115 PTPVSITMTTAKWLRTVIAPL 135
>UniRef100_Q5SCB2 Retinoblastoma protein [Ostreococcus tauri]
Length = 987
Score = 123 bits (308), Expect = 2e-26
Identities = 77/251 (30%), Positives = 128/251 (50%), Gaps = 32/251 (12%)
Query: 342 GGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYSLFQRILNQWTSLFFNR 401
G + CA + +FF+K+++L A R+ + RL+LS ++ +VY+L + ++ + T+L +NR
Sbjct: 726 GCDVCAFKALQIFFAKVMQLAARRLGDLASRLKLSPEVTRDVYALIEHVIYEQTNLVYNR 785
Query: 402 HIDQIILCCFYGVAKIS---QLNLTFREIIYNYRKQPQCKPQVFRSVFVDWSSARRNGGS 458
H+DQIIL YGV K++ + F++IIY Y KQPQC ++F +V ++ + +
Sbjct: 786 HVDQIILASVYGVCKVNGGCGGAVQFKDIIYQYSKQPQCTEEIFWTVVIEQTDPELEVST 845
Query: 459 RHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANNKNDGHLVQNPGSP 518
R DIISFYN+VF+ V+ L+ L + A DG V
Sbjct: 846 RG-------DIISFYNKVFVSRVRTFLLALRERAEAL------AAQKTTDGEKVSVD--- 889
Query: 519 RISPFPSLPDMSPKKVSAAH-NVYVSPLRSSKMDALISHS----------SKSYYACXGE 567
PFP +++ + N+YVSP+R + A++ + ++S +A GE
Sbjct: 890 --EPFPFGISSPRRRLPVENQNIYVSPMRPEREAAMLQDAAVQGEPSTPRTRSLFATIGE 947
Query: 568 STHAYQSPXKD 578
S H S D
Sbjct: 948 SIHGDPSSAND 958
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.342 0.148 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 881,992,339
Number of Sequences: 2790947
Number of extensions: 33311891
Number of successful extensions: 110447
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 110148
Number of HSP's gapped (non-prelim): 147
length of query: 624
length of database: 848,049,833
effective HSP length: 134
effective length of query: 490
effective length of database: 474,062,935
effective search space: 232290838150
effective search space used: 232290838150
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 78 (34.7 bits)
Medicago: description of AC146862.5


