

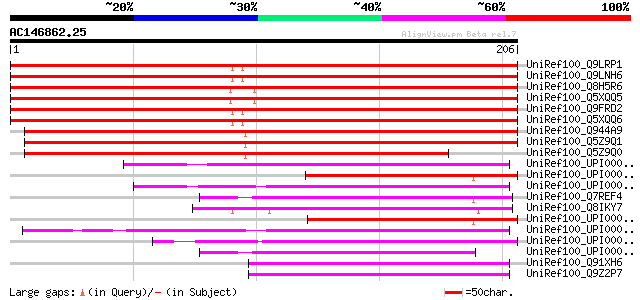
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.25 + phase: 0
(206 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LRP1 Novel plant SNARE 13 [Arabidopsis thaliana] 315 4e-85
UniRef100_Q9LNH6 Novel plant SNARE 12 [Arabidopsis thaliana] 313 2e-84
UniRef100_Q8H5R6 Vesicle soluble NSF attachment protein receptor... 278 5e-74
UniRef100_Q5XQQ5 SNARE 13 [Oryza sativa] 277 1e-73
UniRef100_Q9FRD2 Putative vesicle soluble NSF attachment protein... 270 1e-71
UniRef100_Q5XQQ6 SNARE 12 [Oryza sativa] 269 4e-71
UniRef100_Q944A9 Novel plant SNARE 11 [Arabidopsis thaliana] 253 3e-66
UniRef100_Q5Z9Q1 Putative NPSN12 [Oryza sativa] 247 2e-64
UniRef100_Q5Z9Q0 Putative NPSN12 [Oryza sativa] 204 2e-51
UniRef100_UPI00003ADE2B UPI00003ADE2B UniRef100 entry 59 8e-08
UniRef100_UPI0000430153 UPI0000430153 UniRef100 entry 59 1e-07
UniRef100_UPI00002F9D07 UPI00002F9D07 UniRef100 entry 58 2e-07
UniRef100_Q7REF4 Arabidopsis thaliana At2g35190/T4C15.14, putati... 57 2e-07
UniRef100_Q8IKY7 Hypothetical protein [Plasmodium falciparum] 56 5e-07
UniRef100_UPI00004528FD UPI00004528FD UniRef100 entry 55 1e-06
UniRef100_UPI00003623E6 UPI00003623E6 UniRef100 entry 53 4e-06
UniRef100_UPI0000499EF7 UPI0000499EF7 UniRef100 entry 51 2e-05
UniRef100_UPI0000466BB3 UPI0000466BB3 UniRef100 entry 50 3e-05
UniRef100_Q91XH6 Vesicle transport through interaction with t-SN... 50 3e-05
UniRef100_Q9Z2P7 GES30 [Mus musculus] 50 3e-05
>UniRef100_Q9LRP1 Novel plant SNARE 13 [Arabidopsis thaliana]
Length = 269
Score = 315 bits (808), Expect = 4e-85
Identities = 172/237 (72%), Positives = 188/237 (78%), Gaps = 31/237 (13%)
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MASNL M+PQLEQIHGEIRD FRALANGFQ+LDKIKDS RQS QLEELT KMR+CKRL+K
Sbjct: 1 MASNLPMSPQLEQIHGEIRDHFRALANGFQRLDKIKDSTRQSKQLEELTDKMRECKRLVK 60
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSMT-EKNS-------------------------- 93
+FDRE+KDE A N EVNKQLNDEKQSM E NS
Sbjct: 61 EFDRELKDEEARNSPEVNKQLNDEKQSMIKELNSYVALRKTYMSTLGNKKVELFDMGAGV 120
Query: 94 ----TAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQ 149
TAE NVQ+AS MSNQELV+AGMK MDETDQAIERSKQVV QT+EVGTQTA+ LKGQ
Sbjct: 121 SGEPTAEENVQVASSMSNQELVDAGMKRMDETDQAIERSKQVVEQTLEVGTQTAANLKGQ 180
Query: 150 TEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
T+QMGR+VN LD+IQFSIKKASQLVKEIGRQVATDKCIM FLFLIVCGV+AII+VK+
Sbjct: 181 TDQMGRVVNHLDTIQFSIKKASQLVKEIGRQVATDKCIMGFLFLIVCGVVAIIIVKI 237
>UniRef100_Q9LNH6 Novel plant SNARE 12 [Arabidopsis thaliana]
Length = 265
Score = 313 bits (801), Expect = 2e-84
Identities = 170/237 (71%), Positives = 187/237 (78%), Gaps = 31/237 (13%)
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MAS L M+P LEQIHGEIRD FRALANGFQ+LDKIKDS+RQS QLEEL KMRDCKRL+K
Sbjct: 1 MASELPMSPHLEQIHGEIRDHFRALANGFQRLDKIKDSSRQSKQLEELAEKMRDCKRLVK 60
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSMT-EKNS-------------------------- 93
+FDRE+KD A N +VNKQLNDEKQSM E NS
Sbjct: 61 EFDRELKDGEARNSPQVNKQLNDEKQSMIKELNSYVALRKTYLNTLGNKKVELFDTGAGV 120
Query: 94 ----TAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQ 149
TAE NVQ+AS MSNQELV+AGMK MDETDQAIERSKQVVHQT+EVGTQTAS LKGQ
Sbjct: 121 SGEPTAEENVQMASTMSNQELVDAGMKRMDETDQAIERSKQVVHQTLEVGTQTASNLKGQ 180
Query: 150 TEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
T+QMGR+VN+LD+IQFS+KKASQLVKEIGRQVATDKCIM FLFLIVCGVIAII+VK+
Sbjct: 181 TDQMGRVVNDLDTIQFSLKKASQLVKEIGRQVATDKCIMAFLFLIVCGVIAIIIVKI 237
>UniRef100_Q8H5R6 Vesicle soluble NSF attachment protein receptor-like protein [Oryza
sativa]
Length = 266
Score = 278 bits (712), Expect = 5e-74
Identities = 150/234 (64%), Positives = 179/234 (76%), Gaps = 28/234 (11%)
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MAS++ M+P+LEQ+ GEI+DIFRAL NGFQK+DKIKDSNRQS QLE+LTGKMR+CKRLIK
Sbjct: 1 MASDVPMSPELEQVDGEIQDIFRALQNGFQKMDKIKDSNRQSKQLEDLTGKMRECKRLIK 60
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSM---------------------------TEKNS 93
+FDR +K++ N +VNKQLND+KQ M T +
Sbjct: 61 EFDRILKEDEKKNSADVNKQLNDKKQLMIKELNSYVTLRKTYQSSLGNKRIELFDTGNDQ 120
Query: 94 TAEGN-VQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQ 152
AE N VQ+ASEMSNQ+L++AG K MD+TDQ IERSK+VV QT+EVG+QTA+ L QTEQ
Sbjct: 121 VAEDNTVQMASEMSNQQLMDAGRKQMDQTDQVIERSKKVVAQTVEVGSQTAAALSQQTEQ 180
Query: 153 MGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
M RI NELDS+ FS+KKASQ+VKEIGRQVATDKCIM FLFLIVCGVIAIIVVK+
Sbjct: 181 MKRIGNELDSVHFSLKKASQMVKEIGRQVATDKCIMAFLFLIVCGVIAIIVVKI 234
>UniRef100_Q5XQQ5 SNARE 13 [Oryza sativa]
Length = 266
Score = 277 bits (709), Expect = 1e-73
Identities = 150/234 (64%), Positives = 178/234 (75%), Gaps = 28/234 (11%)
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MAS++ M P+LEQ+ GEI+DIFRAL NGFQK+DKIKDSNRQS QLE+LTGKMR+CKRLIK
Sbjct: 1 MASDVPMGPELEQVDGEIQDIFRALQNGFQKMDKIKDSNRQSKQLEDLTGKMRECKRLIK 60
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSM---------------------------TEKNS 93
+FDR +K++ N +VNKQLND+KQ M T +
Sbjct: 61 EFDRILKEDEKKNSADVNKQLNDKKQLMIKELNSYVTLRKTYQSSLGNKRIELFDTGNDQ 120
Query: 94 TAEGN-VQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQ 152
AE N VQ+ASEMSNQ+L++AG K MD+TDQ IERSK+VV QT+EVG+QTA+ L QTEQ
Sbjct: 121 VAEDNTVQMASEMSNQQLMDAGRKQMDQTDQVIERSKKVVAQTVEVGSQTAAALSQQTEQ 180
Query: 153 MGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
M RI NELDS+ FS+KKASQ+VKEIGRQVATDKCIM FLFLIVCGVIAIIVVK+
Sbjct: 181 MKRIGNELDSVHFSLKKASQMVKEIGRQVATDKCIMAFLFLIVCGVIAIIVVKI 234
>UniRef100_Q9FRD2 Putative vesicle soluble NSF attachment protein receptor [Oryza
sativa]
Length = 275
Score = 270 bits (691), Expect = 1e-71
Identities = 147/238 (61%), Positives = 176/238 (73%), Gaps = 32/238 (13%)
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MAS++ M+P+LEQI GE++DIFRAL NGFQK+DKIKDS+RQ+ QLE+LT KM++CKRLIK
Sbjct: 1 MASDVPMSPELEQIDGEVQDIFRALQNGFQKMDKIKDSSRQAKQLEDLTAKMKECKRLIK 60
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSMT-EKNS-------------------------- 93
+FDR +KDE + NP EV+KQLND KQ M E NS
Sbjct: 61 EFDRILKDEESNNPPEVHKQLNDRKQYMIKELNSYVTLRKTYQSSLGNNNKRVELFDMGA 120
Query: 94 -----TAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKG 148
AE N+Q+AS M+NQ+L++AG + M +TDQAI+RSK VV QTIE GTQTAS L
Sbjct: 121 GSSEPAAEDNIQIASAMTNQQLMDAGREQMTQTDQAIDRSKMVVAQTIETGTQTASALSQ 180
Query: 149 QTEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
QTEQM RI NELD++ FS+KKASQLVKEIGRQVATDKCIM LFLIVCGVIAIIVVK+
Sbjct: 181 QTEQMKRIGNELDTVHFSLKKASQLVKEIGRQVATDKCIMALLFLIVCGVIAIIVVKI 238
>UniRef100_Q5XQQ6 SNARE 12 [Oryza sativa]
Length = 275
Score = 269 bits (687), Expect = 4e-71
Identities = 146/238 (61%), Positives = 176/238 (73%), Gaps = 32/238 (13%)
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MAS++ M+P+LEQI GE++DIFRAL NGFQK+DKIKDS+RQ+ QLE+LT KM++CKRLIK
Sbjct: 1 MASDVPMSPELEQIDGEVQDIFRALQNGFQKMDKIKDSSRQAKQLEDLTAKMKECKRLIK 60
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSMT-EKNS-------------------------- 93
+FDR +KDE + NP EV+KQLND KQ M E NS
Sbjct: 61 EFDRILKDEESNNPPEVHKQLNDRKQYMIKELNSYVTLRKTYQSSLGNNNKRVELFDMGA 120
Query: 94 -----TAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKG 148
AE N+Q+AS M+NQ+L++AG + M +TDQAI+RSK VV QTIE GTQTAS L
Sbjct: 121 GSSEPAAEDNIQIASAMTNQQLMDAGREQMTQTDQAIDRSKMVVAQTIETGTQTASALSQ 180
Query: 149 QTEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
QTE+M RI NELD++ FS+KKASQLVKEIGRQVATDKCIM LFLIVCGVIAIIVVK+
Sbjct: 181 QTERMKRIGNELDTVHFSLKKASQLVKEIGRQVATDKCIMALLFLIVCGVIAIIVVKI 238
>UniRef100_Q944A9 Novel plant SNARE 11 [Arabidopsis thaliana]
Length = 265
Score = 253 bits (645), Expect = 3e-66
Identities = 136/229 (59%), Positives = 164/229 (71%), Gaps = 29/229 (12%)
Query: 7 MTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREI 66
++ +L +I G+I DIFRAL+NGFQKL+KIKD+NRQS QLEELT KMRDCK LIKDFDREI
Sbjct: 7 VSEELAEIEGQINDIFRALSNGFQKLEKIKDANRQSRQLEELTDKMRDCKSLIKDFDREI 66
Query: 67 KDEGAGNPEEVNKQLNDEKQSMTEKNST-----------------------------AEG 97
K +GN N+ LND +QSM ++ ++ E
Sbjct: 67 KSLESGNDASTNRMLNDRRQSMVKELNSYVALKKKYSSNLASNNKRVDLFDGPGEEHMEE 126
Query: 98 NVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIV 157
NV LAS MSNQEL++ G MD+TDQAIER K++V +TI VGT T++ LK QTEQM R+V
Sbjct: 127 NVLLASNMSNQELMDKGNSMMDDTDQAIERGKKIVQETINVGTDTSAALKPQTEQMSRVV 186
Query: 158 NELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
NELDSI FS+KKAS+LVKEIGRQVATDKCIM FLFLIV GVIAII+VK+
Sbjct: 187 NELDSIHFSLKKASKLVKEIGRQVATDKCIMAFLFLIVIGVIAIIIVKI 235
>UniRef100_Q5Z9Q1 Putative NPSN12 [Oryza sativa]
Length = 261
Score = 247 bits (630), Expect = 2e-64
Identities = 135/227 (59%), Positives = 160/227 (70%), Gaps = 27/227 (11%)
Query: 7 MTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREI 66
+ P+L +I G+I DI RAL NGFQKLDKIKD+NR+S QLEELT KMRDCKRLIKDF+R +
Sbjct: 6 VNPELAEIDGQIGDILRALQNGFQKLDKIKDANRRSRQLEELTDKMRDCKRLIKDFERVV 65
Query: 67 KDEGAGNPEEVNKQLNDEKQSMTEKNST---------------------------AEGNV 99
KD E + L+D KQSM ++ ++ E NV
Sbjct: 66 KDMAGSTDPETARMLHDRKQSMIKELNSYVALKKQYASENKRVDLFDGPSVEDGFGEENV 125
Query: 100 QLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNE 159
LAS M+NQ+L++ G + MDETDQAI RSKQ V +TI VGT+TA+ LK QTEQM RIVNE
Sbjct: 126 LLASNMTNQQLMDQGNQLMDETDQAIARSKQTVQETINVGTETAAALKSQTEQMSRIVNE 185
Query: 160 LDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
LDSI FSIKKASQ+VKEIGRQVATD+CIM LFLIV GVIAIIVVK+
Sbjct: 186 LDSIHFSIKKASQMVKEIGRQVATDRCIMALLFLIVAGVIAIIVVKI 232
>UniRef100_Q5Z9Q0 Putative NPSN12 [Oryza sativa]
Length = 206
Score = 204 bits (518), Expect = 2e-51
Identities = 112/199 (56%), Positives = 135/199 (67%), Gaps = 27/199 (13%)
Query: 7 MTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREI 66
+ P+L +I G+I DI RAL NGFQKLDKIKD+NR+S QLEELT KMRDCKRLIKDF+R +
Sbjct: 6 VNPELAEIDGQIGDILRALQNGFQKLDKIKDANRRSRQLEELTDKMRDCKRLIKDFERVV 65
Query: 67 KDEGAGNPEEVNKQLNDEKQSMTEKNST---------------------------AEGNV 99
KD E + L+D KQSM ++ ++ E NV
Sbjct: 66 KDMAGSTDPETARMLHDRKQSMIKELNSYVALKKQYASENKRVDLFDGPSVEDGFGEENV 125
Query: 100 QLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNE 159
LAS M+NQ+L++ G + MDETDQAI RSKQ V +TI VGT+TA+ LK QTEQM RIVNE
Sbjct: 126 LLASNMTNQQLMDQGNQLMDETDQAIARSKQTVQETINVGTETAAALKSQTEQMSRIVNE 185
Query: 160 LDSIQFSIKKASQLVKEIG 178
LDSI FSIKKASQ+VKEIG
Sbjct: 186 LDSIHFSIKKASQMVKEIG 204
>UniRef100_UPI00003ADE2B UPI00003ADE2B UniRef100 entry
Length = 199
Score = 58.9 bits (141), Expect = 8e-08
Identities = 35/157 (22%), Positives = 79/157 (50%), Gaps = 8/157 (5%)
Query: 47 ELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMS 106
++ K+R +R + F RE+K+ G L Q + + N Q + S
Sbjct: 45 QMMSKIRAYRRDLSMFQREMKNTDLG--------LGPGSQGDMKYGIFSTENEQSTNLQS 96
Query: 107 NQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFS 166
+ L+ G ++++ Q+IERS ++ +T ++GT L Q EQ+ R + L + +
Sbjct: 97 QRVLLLQGTESLNRASQSIERSHRIAAETDQIGTDIIEELGEQREQLERTKSRLVNTSEN 156
Query: 167 IKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIV 203
+ K+ ++++ + R++AT+K ++ + ++ ++ +V
Sbjct: 157 LSKSRKILRSMSRRIATNKLLLSIIIILELAILGGVV 193
>UniRef100_UPI0000430153 UPI0000430153 UniRef100 entry
Length = 222
Score = 58.5 bits (140), Expect = 1e-07
Identities = 34/87 (39%), Positives = 55/87 (63%), Gaps = 1/87 (1%)
Query: 121 TDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGRQ 180
TD++I R KQ + T ++G + + + Q EQ+ RI NELD+++ +I KA+ +K I R
Sbjct: 126 TDESIIRMKQSLTYTDKLGDDSLNKMNTQKEQLDRIRNELDNVKDNIYKANISLKAIARN 185
Query: 181 VATDKCI-MLFLFLIVCGVIAIIVVKV 206
ATD C+ ML L +C +I I+++ V
Sbjct: 186 TATDFCVQMLCGILSICLLIIIVLLVV 212
>UniRef100_UPI00002F9D07 UPI00002F9D07 UniRef100 entry
Length = 224
Score = 57.8 bits (138), Expect = 2e-07
Identities = 33/153 (21%), Positives = 80/153 (51%), Gaps = 8/153 (5%)
Query: 51 KMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQEL 110
K+R +R + R++K +G + K+ +E ++ T++G++ S + L
Sbjct: 74 KLRMYRRDLAKVQRDMKSSNSG----LAKRTAEEGTRAGRQHKTSKGHIS----RSQRAL 125
Query: 111 VNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKA 170
+ G + + Q+IERS+++ ++T ++GT L Q EQ+ R L + ++ ++
Sbjct: 126 LLQGTEARNNASQSIERSQRIANETEQIGTDIIEELGEQREQLDRTRGRLVNTGENLSRS 185
Query: 171 SQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIV 203
++++ I R++ T+K ++ + L+ ++ +V
Sbjct: 186 RKILRSISRRLVTNKLLLAVIILMELAILGAVV 218
>UniRef100_Q7REF4 Arabidopsis thaliana At2g35190/T4C15.14, putative [Plasmodium
yoelii yoelii]
Length = 224
Score = 57.4 bits (137), Expect = 2e-07
Identities = 34/128 (26%), Positives = 70/128 (54%), Gaps = 6/128 (4%)
Query: 78 NKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIE 137
NK + + KQ + N + ++ +EL G D+T+Q+I R K +V ++ +
Sbjct: 97 NKLIYENKQDENKNNE-----INKLKYITPKELEIRGNLIQDQTEQSIFRMKNLVDESEQ 151
Query: 138 VGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCI-MLFLFLIVC 196
+ A L Q E++ ++ +++D + ++ A + +KEI ++ A+D+ I +LF+ +I+
Sbjct: 152 ITRIAAVKLNEQNEKLKKVKDKVDDVDINVSTAKETLKEIMKEAASDRFIRLLFIMIIIV 211
Query: 197 GVIAIIVV 204
VI I V+
Sbjct: 212 LVILISVI 219
>UniRef100_Q8IKY7 Hypothetical protein [Plasmodium falciparum]
Length = 253
Score = 56.2 bits (134), Expect = 5e-07
Identities = 34/136 (25%), Positives = 73/136 (53%), Gaps = 6/136 (4%)
Query: 75 EEVNKQLNDEKQSMT-EKNSTAEGNVQLASE--MSNQELVNAGMKTMDETDQAIERSKQV 131
E +N Q + +K + E++ A+ N +L +S +++ G D+TD AI R K +
Sbjct: 83 ENLNIQFDFKKSKLIYEEDKQAKENKKLLKVRYVSAKDIETKGDIIQDQTDDAIFRMKMM 142
Query: 132 VHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIML-- 189
V ++ + A L Q E++ + ++++ + ++ A Q +KEI ++ TD+ + L
Sbjct: 143 VDESENITKDAADKLNIQNEKLQKARDKIEDVDINVYSAKQTLKEIAKEAVTDRFVRLMS 202
Query: 190 -FLFLIVCGVIAIIVV 204
+F++V +I +I++
Sbjct: 203 ILIFIVVSVLITVIII 218
>UniRef100_UPI00004528FD UPI00004528FD UniRef100 entry
Length = 222
Score = 55.1 bits (131), Expect = 1e-06
Identities = 32/86 (37%), Positives = 54/86 (62%), Gaps = 1/86 (1%)
Query: 122 DQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGRQV 181
D++I R K+ + T ++G + + + Q EQ+ RI NELD+++ +I KA+ +K I R
Sbjct: 127 DESIIRMKKSLTYTEKLGDDSLNKMNTQKEQLDRIRNELDNVKDNIYKANISLKAIARNT 186
Query: 182 ATDKCI-MLFLFLIVCGVIAIIVVKV 206
ATD C+ ML L +C +I I+++ V
Sbjct: 187 ATDFCVQMLCGILSICLLIIIVLLVV 212
>UniRef100_UPI00003623E6 UPI00003623E6 UniRef100 entry
Length = 224
Score = 53.1 bits (126), Expect = 4e-06
Identities = 36/198 (18%), Positives = 96/198 (48%), Gaps = 16/198 (8%)
Query: 6 KMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDRE 65
++ ++ GE ++ +A+ ++L S R + ++ K+R +R + R+
Sbjct: 37 RLVRAFDERQGEAEEVLQAME---EELRAAPPSYRNT-----MSTKLRVYRRDLAKLQRD 88
Query: 66 IKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAI 125
+K +G ++ S ++ST + S + L+ G ++++ Q+I
Sbjct: 89 MKSSASGFSHFPSEGSRHGIYSSQNQHSTYQ--------QSQRALLLQGSESLNTASQSI 140
Query: 126 ERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDK 185
ERS+++ +T ++G L GQ EQ+ R + L + ++ ++ ++++ + R++ T+K
Sbjct: 141 ERSQRIATETEQIGNDIIEELGGQREQLDRTRDRLVNTGENLSRSRKILRSMSRRLVTNK 200
Query: 186 CIMLFLFLIVCGVIAIIV 203
++ + ++ ++ +V
Sbjct: 201 LLLAVIIVMELAILGAVV 218
>UniRef100_UPI0000499EF7 UPI0000499EF7 UniRef100 entry
Length = 214
Score = 51.2 bits (121), Expect = 2e-05
Identities = 32/148 (21%), Positives = 74/148 (49%), Gaps = 9/148 (6%)
Query: 59 IKDFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTM 118
+ DF+ E+K ++ +LN Q +T++ ST + V + +++ E + ++
Sbjct: 73 LADFNAEVK--------QLEGELNAATQKLTKELSTDKKEVNV-DDLNADEALGLAVQIQ 123
Query: 119 DETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIG 178
+E ++ + ++ ++G + L Q EQ+ + +D I ++ A++ + I
Sbjct: 124 EEDKNHLDNAINQINDAKQIGGTISLQLDQQNEQLDHCIELVDEIDSTLDIANKELNGIA 183
Query: 179 RQVATDKCIMLFLFLIVCGVIAIIVVKV 206
R++ATDK IM+ + + V +I V+ +
Sbjct: 184 RRLATDKIIMVLIIVCVLVIIGCSVLYI 211
>UniRef100_UPI0000466BB3 UPI0000466BB3 UniRef100 entry
Length = 190
Score = 50.4 bits (119), Expect = 3e-05
Identities = 28/112 (25%), Positives = 59/112 (52%), Gaps = 5/112 (4%)
Query: 78 NKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIE 137
NK + + KQ + N + ++ +EL G D+T+Q+I R K +V ++ +
Sbjct: 82 NKLIYENKQDENKNNE-----INRLKYVTPKELEIRGNLIQDQTEQSIFRMKNMVDESEQ 136
Query: 138 VGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIML 189
+ A L Q E++ ++ +++D + ++ A + +KEI ++ A+D+ I L
Sbjct: 137 ITRIAAVKLNEQNEKLRKVKDKVDDVDINVSSAKETLKEIMKEAASDRFIRL 188
>UniRef100_Q91XH6 Vesicle transport through interaction with t-SNAREs 1B homolog [Mus
musculus]
Length = 232
Score = 50.4 bits (119), Expect = 3e-05
Identities = 25/106 (23%), Positives = 59/106 (55%)
Query: 98 NVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIV 157
N L S + L+ G ++++ Q+IERS ++ +T ++GT+ L Q +Q+ R
Sbjct: 119 NEHLNRLQSQRALLLQGTESLNRATQSIERSHRIATETDQIGTEIIEELGEQRDQLERTK 178
Query: 158 NELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIV 203
+ L + ++ K+ ++++ + R+V T+K ++ + L+ ++ +V
Sbjct: 179 SRLVNTNENLSKSRKILRSMSRKVITNKLLLSVIILLELAILVGLV 224
>UniRef100_Q9Z2P7 GES30 [Mus musculus]
Length = 231
Score = 50.4 bits (119), Expect = 3e-05
Identities = 25/106 (23%), Positives = 59/106 (55%)
Query: 98 NVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIV 157
N L S + L+ G ++++ Q+IERS ++ +T ++GT+ L Q +Q+ R
Sbjct: 118 NEHLNRLQSQRALLLQGTESLNRATQSIERSHRIATETDQIGTEIIEELGEQRDQLERTK 177
Query: 158 NELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIV 203
+ L + ++ K+ ++++ + R+V T+K ++ + L+ ++ +V
Sbjct: 178 SRLVNTNENLSKSRKILRSMSRKVITNKLLLSVIILLELAILVGLV 223
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.130 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 284,366,889
Number of Sequences: 2790947
Number of extensions: 10661380
Number of successful extensions: 53236
Number of sequences better than 10.0: 1444
Number of HSP's better than 10.0 without gapping: 183
Number of HSP's successfully gapped in prelim test: 1289
Number of HSP's that attempted gapping in prelim test: 51759
Number of HSP's gapped (non-prelim): 2508
length of query: 206
length of database: 848,049,833
effective HSP length: 121
effective length of query: 85
effective length of database: 510,345,246
effective search space: 43379345910
effective search space used: 43379345910
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 72 (32.3 bits)
Medicago: description of AC146862.25


