

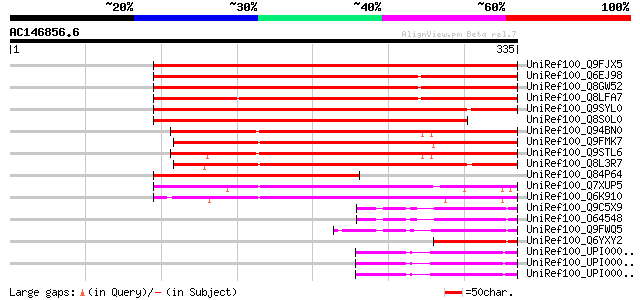
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146856.6 - phase: 0 /pseudo
(335 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FJX5 Arabidopsis thaliana genomic DNA, chromosome 5,... 372 e-101
UniRef100_Q6EJ98 BTB and TAZ domain protein 5 [Arabidopsis thali... 326 6e-88
UniRef100_Q8GW52 Hypothetical protein At4g37610/F19F18_100 [Arab... 324 2e-87
UniRef100_Q8LFA7 Hypothetical protein [Arabidopsis thaliana] 324 2e-87
UniRef100_Q9SYL0 F3F20.14 protein [Arabidopsis thaliana] 285 9e-76
UniRef100_Q8S0L0 B1078G07.24 protein [Oryza sativa] 254 2e-66
UniRef100_Q94BN0 Hypothetical protein At3g48360 [Arabidopsis tha... 216 9e-55
UniRef100_Q9FMK7 Similarity to unknown protein [Arabidopsis thal... 215 1e-54
UniRef100_Q9STL6 Hypothetical protein T29H11_120 [Arabidopsis th... 214 3e-54
UniRef100_Q8L3R7 P0497A05.15 protein [Oryza sativa] 179 1e-43
UniRef100_Q84P64 Hypothetical protein [Oryza sativa] 157 4e-37
UniRef100_Q7XUP5 OSJNBb0011N17.18 protein [Oryza sativa] 152 9e-36
UniRef100_Q6K910 Putative speckle-type POZ [Oryza sativa] 130 5e-29
UniRef100_Q9C5X9 P300/CBP acetyltransferase-related protein 2 [A... 67 7e-10
UniRef100_O64548 YUP8H12R.38 protein [Arabidopsis thaliana] 67 7e-10
UniRef100_Q9FWQ5 F17F16.8 protein [Arabidopsis thaliana] 65 3e-09
UniRef100_Q6YXY2 Putative HAC5 [Oryza sativa] 62 2e-08
UniRef100_UPI00003AAEA7 UPI00003AAEA7 UniRef100 entry 59 2e-07
UniRef100_UPI00003AAEA6 UPI00003AAEA6 UniRef100 entry 59 2e-07
UniRef100_UPI00003AAEA5 UPI00003AAEA5 UniRef100 entry 59 2e-07
>UniRef100_Q9FJX5 Arabidopsis thaliana genomic DNA, chromosome 5, TAC clone:K9I9
[Arabidopsis thaliana]
Length = 372
Score = 372 bits (954), Expect = e-101
Identities = 174/241 (72%), Positives = 201/241 (83%), Gaps = 1/241 (0%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
MLKQA R +W +ISI GVPHDAVRVFIR+LYSSCYEKEEM EF++HLL+LSH Y VP L
Sbjct: 89 MLKQAKRHGKWHTISIRGVPHDAVRVFIRFLYSSCYEKEEMNEFIMHLLLLSHAYVVPQL 148
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KR CE LE GLLT +N+VDVFQLALLCD PRLSLI HR I+K+F +S +E W AMK+S
Sbjct: 149 KRVCEWHLEHGLLTTENVVDVFQLALLCDFPRLSLISHRMIMKHFNELSATEAWTAMKKS 208
Query: 216 HPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDF 275
HP LEKE+ +S+I E N++KER+RK N++ +Y QLY+AMEALVHICRDGC+TIGPHDKDF
Sbjct: 209 HPFLEKEVRDSVIIEANTRKERMRKRNDQRIYSQLYEAMEALVHICRDGCKTIGPHDKDF 268
Query: 276 KANQ-PCRYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPL 334
K N C Y +CKGLE L+RHFAGCKLRVPGGC HCKRMWQLLELHSR+CA D CRVPL
Sbjct: 269 KPNHATCNYEACKGLESLIRHFAGCKLRVPGGCVHCKRMWQLLELHSRVCAGSDQCRVPL 328
Query: 335 C 335
C
Sbjct: 329 C 329
>UniRef100_Q6EJ98 BTB and TAZ domain protein 5 [Arabidopsis thaliana]
Length = 368
Score = 326 bits (835), Expect = 6e-88
Identities = 154/241 (63%), Positives = 186/241 (76%), Gaps = 2/241 (0%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
M+KQ R + +SISI GVPH A+RVFIR+LYSSCYEK++M++F +HLLVLSHVY VPHL
Sbjct: 84 MMKQHKRKSHRKSISILGVPHHALRVFIRFLYSSCYEKQDMEDFAIHLLVLSHVYVVPHL 143
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KR CE + E LL +N++DVFQLALLCDAPRL L+CHR IL NF VS SEGW+AMK+S
Sbjct: 144 KRVCESEFESSLLNKENVIDVFQLALLCDAPRLGLLCHRMILNNFEEVSTSEGWQAMKES 203
Query: 216 HPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDF 275
HP L+KE+L S+ E NS K+R RK E + Y QLY+AMEA VHICRDGCR IGP K
Sbjct: 204 HPRLQKELLRSVAYELNSLKQRNRKQKEIQTYTQLYEAMEAFVHICRDGCREIGP-TKTE 262
Query: 276 KANQPCRYTSCKGLELLVRHFAGCKLR-VPGGCGHCKRMWQLLELHSRLCADPDFCRVPL 334
+ C + +C GLE L++H AGCKLR +PGGC CKRMWQLLELHSR+C D + C+VPL
Sbjct: 263 TPHMSCGFQACNGLEQLLKHLAGCKLRSIPGGCSRCKRMWQLLELHSRICVDSEQCKVPL 322
Query: 335 C 335
C
Sbjct: 323 C 323
>UniRef100_Q8GW52 Hypothetical protein At4g37610/F19F18_100 [Arabidopsis thaliana]
Length = 368
Score = 324 bits (831), Expect = 2e-87
Identities = 153/241 (63%), Positives = 185/241 (76%), Gaps = 2/241 (0%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
M+KQ R + +SISI GVPH A+RVFIR+LYSSCYEK++M++F +HLLVLSHVY VPHL
Sbjct: 84 MMKQHKRKSHRKSISILGVPHHALRVFIRFLYSSCYEKQDMEDFAIHLLVLSHVYVVPHL 143
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KR CE + E LL +N++DVFQLALLCDAPRL L+CHR IL NF VS SEGW+ MK+S
Sbjct: 144 KRVCESEFESSLLNKENVIDVFQLALLCDAPRLGLLCHRMILNNFEEVSTSEGWQTMKES 203
Query: 216 HPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDF 275
HP L+KE+L S+ E NS K+R RK E + Y QLY+AMEA VHICRDGCR IGP K
Sbjct: 204 HPRLQKELLRSVAYELNSLKQRNRKQKEIQTYTQLYEAMEAFVHICRDGCREIGP-TKTE 262
Query: 276 KANQPCRYTSCKGLELLVRHFAGCKLR-VPGGCGHCKRMWQLLELHSRLCADPDFCRVPL 334
+ C + +C GLE L++H AGCKLR +PGGC CKRMWQLLELHSR+C D + C+VPL
Sbjct: 263 TPHMSCGFQACNGLEQLLKHLAGCKLRSIPGGCSRCKRMWQLLELHSRICVDSEQCKVPL 322
Query: 335 C 335
C
Sbjct: 323 C 323
>UniRef100_Q8LFA7 Hypothetical protein [Arabidopsis thaliana]
Length = 367
Score = 324 bits (831), Expect = 2e-87
Identities = 156/241 (64%), Positives = 187/241 (76%), Gaps = 3/241 (1%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
M+KQ R + +SISI GVPH AVRVFIR+LYSSCYEK++M++F +HLLVLSHVY VPHL
Sbjct: 84 MMKQHKRKSHRKSISILGVPHHAVRVFIRFLYSSCYEKQDMEDFAIHLLVLSHVY-VPHL 142
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KR CE + E LL +N++DVFQLALLCDAPRL L+CHR ILKNF VS SEGW+AMK+S
Sbjct: 143 KRVCESEFESSLLNKENVIDVFQLALLCDAPRLGLLCHRMILKNFEEVSTSEGWQAMKES 202
Query: 216 HPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDF 275
HP L+KE+L S+ E NS K+R RK E + Y QLY+AMEA VHICRDGCR IGP K
Sbjct: 203 HPRLQKELLRSVAYELNSLKQRNRKQKEIQTYTQLYEAMEAFVHICRDGCREIGP-TKTE 261
Query: 276 KANQPCRYTSCKGLELLVRHFAGCKLR-VPGGCGHCKRMWQLLELHSRLCADPDFCRVPL 334
+ C + +C GLE L++H AGCKLR +PGGC CKRMWQLLELHSR+C D + C+VPL
Sbjct: 262 TPHMSCGFQACNGLEQLLKHLAGCKLRSIPGGCSRCKRMWQLLELHSRICVDSEQCKVPL 321
Query: 335 C 335
C
Sbjct: 322 C 322
>UniRef100_Q9SYL0 F3F20.14 protein [Arabidopsis thaliana]
Length = 322
Score = 285 bits (730), Expect = 9e-76
Identities = 134/241 (55%), Positives = 181/241 (74%), Gaps = 3/241 (1%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
+L Q+ N + I GVP +AV +FIR+LYSSCYE+EEMK+FVLHLLVLSH Y+VP L
Sbjct: 83 LLNQSRDKNGNTYLKIHGVPCEAVYMFIRFLYSSCYEEEEMKKFVLHLLVLSHCYSVPSL 142
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KR C + L+ G + +N++DV QLA CD R+ +C ++K+F++VS +EGWK MK+S
Sbjct: 143 KRLCVEILDQGWINKENVIDVLQLARNCDVTRICFVCLSMVIKDFKSVSSTEGWKVMKRS 202
Query: 216 HPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDF 275
+P+LE+E++E++I+ ++ K+ER RK+ EREVYLQLY+AMEALVHICR+GC TIGP DK
Sbjct: 203 NPLLEQELIEAVIESDSRKQERRRKLEEREVYLQLYEAMEALVHICREGCGTIGPRDKAL 262
Query: 276 KANQP-CRYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPL 334
K + C++ +CKGLE +RHF GCK R C HCKRMWQLL+LHS +C D + C+V L
Sbjct: 263 KGSHTVCKFPACKGLEGALRHFLGCKSR--ASCSHCKRMWQLLQLHSCICDDSNSCKVSL 320
Query: 335 C 335
C
Sbjct: 321 C 321
>UniRef100_Q8S0L0 B1078G07.24 protein [Oryza sativa]
Length = 587
Score = 254 bits (650), Expect = 2e-66
Identities = 117/208 (56%), Positives = 167/208 (80%), Gaps = 1/208 (0%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
ML++A R I I GVP +AV VFIR+LYSS +E+ +MK +VLHLLVLSHV++VP L
Sbjct: 307 MLEEAKVQGGIRHILIPGVPSEAVHVFIRFLYSSRFEQYQMKRYVLHLLVLSHVFSVPSL 366
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KR C +LE LL+ +N+VD+ QLA LCDAPRLSL+C R I+ +F+ ++++EGW+ M+Q+
Sbjct: 367 KRVCINQLETSLLSPENVVDILQLARLCDAPRLSLVCTRMIIGDFKAITQTEGWRVMRQA 426
Query: 216 HPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDF 275
+P LE+E+LES+++E+ ++ER R++ E +VYLQL++AMEAL+HICRDGCRTIGP D+
Sbjct: 427 NPSLEQELLESLVEEDTKRQERARRLEENKVYLQLHEAMEALIHICRDGCRTIGPRDQTL 486
Query: 276 KANQP-CRYTSCKGLELLVRHFAGCKLR 302
K++Q CR+ +CKG+ELL+RHF+ CK+R
Sbjct: 487 KSSQAVCRFPACKGIELLLRHFSACKMR 514
>UniRef100_Q94BN0 Hypothetical protein At3g48360 [Arabidopsis thaliana]
Length = 364
Score = 216 bits (549), Expect = 9e-55
Identities = 115/239 (48%), Positives = 157/239 (65%), Gaps = 11/239 (4%)
Query: 107 RSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKLELG 166
R I I GVP DAV VFI++LYSS ++EM+ + +HLL LSHVY V LK+ C + + +
Sbjct: 78 RVIKILGVPCDAVSVFIKFLYSSSLTEDEMERYGIHLLALSHVYMVTQLKQRCSKGV-VQ 136
Query: 167 LLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILES 226
LT +N+VDV QLA LCDAP + L R I F+TV ++EGWK +++ P LE +IL+
Sbjct: 137 RLTTENVVDVLQLARLCDAPDVCLRSMRLIHSQFKTVEQTEGWKFIQEHDPFLELDILQF 196
Query: 227 MIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPH---DKDFKA-----N 278
+ D E+ KK R R E+++Y+QL +AME + HIC GC +GP D + K+ +
Sbjct: 197 IDDAESRKKRRRRHRKEQDLYMQLSEAMECIEHICTQGCTLVGPSNVVDNNKKSMTAEKS 256
Query: 279 QPCR-YTSCKGLELLVRHFAGCKLR-VPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
+PC+ +++C GL+LL+RHFA CK R GC CKRM QL LHS +C PD CRVPLC
Sbjct: 257 EPCKAFSTCYGLQLLIRHFAVCKRRNNDKGCLRCKRMLQLFRLHSLICDQPDSCRVPLC 315
>UniRef100_Q9FMK7 Similarity to unknown protein [Arabidopsis thaliana]
Length = 365
Score = 215 bits (548), Expect = 1e-54
Identities = 115/235 (48%), Positives = 153/235 (64%), Gaps = 9/235 (3%)
Query: 109 ISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKLELGLL 168
I I GVP DAV VF+R+LYS + EM+++ +HLL LSHVY V LK+ C + + +
Sbjct: 70 IKILGVPCDAVSVFVRFLYSPSVTENEMEKYGIHLLALSHVYMVTQLKQRCTKGVG-ERV 128
Query: 169 TMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILESMI 228
T +N+VD+ QLA LCDAP L L C R I F+TV ++EGWK +++ P LE +IL+ +
Sbjct: 129 TAENVVDILQLARLCDAPDLCLKCMRFIHYKFKTVEQTEGWKFLQEHDPFLELDILQFID 188
Query: 229 DEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDK-DFKAN-----QPC- 281
D E+ KK R R E+ +YLQL +AME + HIC +GC +GP D K+ PC
Sbjct: 189 DAESRKKRRRRHRREQNLYLQLSEAMECIEHICTEGCTLVGPSSNLDNKSTCQAKPGPCS 248
Query: 282 RYTSCKGLELLVRHFAGCKLRVPG-GCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
+++C GL+LL+RHFA CK RV G GC CKRM QLL LHS +C + CRVPLC
Sbjct: 249 AFSTCYGLQLLIRHFAVCKKRVDGKGCVRCKRMIQLLRLHSSICDQSESCRVPLC 303
>UniRef100_Q9STL6 Hypothetical protein T29H11_120 [Arabidopsis thaliana]
Length = 367
Score = 214 bits (545), Expect = 3e-54
Identities = 116/242 (47%), Positives = 158/242 (64%), Gaps = 14/242 (5%)
Query: 107 RSISIFGVPHDAVRVFIRYLYSS---CYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKL 163
R I I GVP DAV VFI++LYSS C ++EM+ + +HLL LSHVY V LK+ C + +
Sbjct: 78 RVIKILGVPCDAVSVFIKFLYSSRLVCLTEDEMERYGIHLLALSHVYMVTQLKQRCSKGV 137
Query: 164 ELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEI 223
+ LT +N+VDV QLA LCDAP + L R I F+TV ++EGWK +++ P LE +I
Sbjct: 138 -VQRLTTENVVDVLQLARLCDAPDVCLRSMRLIHSQFKTVEQTEGWKFIQEHDPFLELDI 196
Query: 224 LESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPH---DKDFKA--- 277
L+ + D E+ KK R R E+++Y+QL +AME + HIC GC +GP D + K+
Sbjct: 197 LQFIDDAESRKKRRRRHRKEQDLYMQLSEAMECIEHICTQGCTLVGPSNVVDNNKKSMTA 256
Query: 278 --NQPCR-YTSCKGLELLVRHFAGCKLR-VPGGCGHCKRMWQLLELHSRLCADPDFCRVP 333
++PC+ +++C GL+LL+RHFA CK R GC CKRM QL LHS +C PD CRVP
Sbjct: 257 EKSEPCKAFSTCYGLQLLIRHFAVCKRRNNDKGCLRCKRMLQLFRLHSLICDQPDSCRVP 316
Query: 334 LC 335
LC
Sbjct: 317 LC 318
>UniRef100_Q8L3R7 P0497A05.15 protein [Oryza sativa]
Length = 347
Score = 179 bits (453), Expect = 1e-43
Identities = 98/231 (42%), Positives = 141/231 (60%), Gaps = 8/231 (3%)
Query: 109 ISIFGVPHDAVRVFIRYLY--SSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKLELG 166
+ I GV DA F+R LY SS E+E ++ +LVL+H Y VP LKR CE +
Sbjct: 68 VLIRGVTDDAAAAFVRLLYAGSSGDEEEIDEKSAAQMLVLAHAYRVPWLKRRCEGAIG-S 126
Query: 167 LLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILES 226
LT +++VD QLA LCDAP+L L C R + K F+ V ++E W+ ++++ P LE +IL+
Sbjct: 127 RLTAESVVDTMQLAALCDAPQLHLRCTRLLAKEFKAVEKTEAWRFLQENDPWLELDILQR 186
Query: 227 MIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDK-DFKANQPCRYTS 285
+ D + +++ RK E+ VY++L +AM+ L HIC +GC +GP + A P T+
Sbjct: 187 LHDADLRRRKWRRKRAEQGVYVELSEAMDCLSHICTEGCTEVGPVGRAPAAAPCPAYATA 246
Query: 286 CKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPD-FCRVPLC 335
C+GL+LL+RHF+ C C C+RMWQLL LH+ LC PD C PLC
Sbjct: 247 CRGLQLLIRHFSRCHRT---SCPRCQRMWQLLRLHAALCDLPDGHCNTPLC 294
>UniRef100_Q84P64 Hypothetical protein [Oryza sativa]
Length = 270
Score = 157 bits (397), Expect = 4e-37
Identities = 74/136 (54%), Positives = 107/136 (78%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
ML++A R I I GVP +AV VFIR+LYSS +E+ +MK +VLHLLVLSHV++VP L
Sbjct: 127 MLEEAKVQGGIRHILIPGVPSEAVHVFIRFLYSSRFEQYQMKRYVLHLLVLSHVFSVPSL 186
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KR C +LE LL+ +N+VD+ QLA LCDAPRLSL+C R I+ +F+ ++++EGW+ M+Q+
Sbjct: 187 KRVCINQLETSLLSPENVVDILQLARLCDAPRLSLVCTRMIIGDFKAITQTEGWRVMRQA 246
Query: 216 HPVLEKEILESMIDEE 231
+P LE+E+LES+++E+
Sbjct: 247 NPSLEQELLESLVEED 262
>UniRef100_Q7XUP5 OSJNBb0011N17.18 protein [Oryza sativa]
Length = 355
Score = 152 bits (385), Expect = 9e-36
Identities = 100/254 (39%), Positives = 136/254 (53%), Gaps = 19/254 (7%)
Query: 96 MLKQANRS-NRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLH---LLVLSHVYA 151
M+++A R N +I + GV DAV F++ LY+S E+ + H LL LSH Y
Sbjct: 45 MIERARRGWNAECTIRVLGVSSDAVFAFLQLLYASRVTPEDEEVVTAHGPQLLALSHAYR 104
Query: 152 VPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKA 211
+ LKR E + LT ++ VD+ +LA LCDAPRL L C R K+F V SEGW+
Sbjct: 105 IGWLKRAAEASVT-ARLTPEHAVDMLKLARLCDAPRLYLRCARLAAKDFAAVERSEGWRF 163
Query: 212 MKQSHPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPH 271
++ LE EIL+ + D + ++ R+ RE Y QL +AM++L HI D +
Sbjct: 164 ARRHDAALELEILQLLEDADQRRERWARERASREAYRQLGEAMDSLEHIFSDDGCSCADA 223
Query: 272 DKDFKANQPCRYTSCKGLELLVRHFAGC---KLRVPGGCGHCKRMWQLLELHSRLC--AD 326
D D + P C+GL LL+RH+A C K GGC CKRM QL LH+ +C A
Sbjct: 224 DADADTDAP----PCRGLRLLMRHYATCGARKAAPGGGCTRCKRMVQLFRLHASVCDRAA 279
Query: 327 PDF-----CRVPLC 335
P CRVPLC
Sbjct: 280 PHDDGDRPCRVPLC 293
>UniRef100_Q6K910 Putative speckle-type POZ [Oryza sativa]
Length = 361
Score = 130 bits (327), Expect = 5e-29
Identities = 94/260 (36%), Positives = 129/260 (49%), Gaps = 24/260 (9%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSC------------YEKEEMKEFVLHL 143
M++QA R + I G AV VF+R+LY++ +E+ + E L
Sbjct: 44 MIEQAPRGG---VVPIAGASTGAVVVFLRFLYAASVRGAAAAAAAAEWEEAALAEHGAAL 100
Query: 144 LVLSHVYAVPH-LKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRT 202
+ L+H Y V LKR E+ + + + VD +LA LCDAPRL L C R +
Sbjct: 101 MALAHAYRVAGPLKRRAEEAVA-ARVAAEGAVDAMKLAALCDAPRLYLWCARLAGRELAA 159
Query: 203 VSESEGWKAMKQSHPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICR 262
V ESEGW+ + L ++L+ + D + K+ R+ + VYLQL DAM AL +
Sbjct: 160 VRESEGWRFAARHDAALRADLLQLIRDADQRKERWGRERGSQGVYLQLSDAMAALERVFA 219
Query: 263 DGCRTIGPHDKDFKANQPCRYTS-C---KGLELLVRH-FAGCKLRVPGGCGHCKRMWQLL 317
P + Q CR S C +GL L RH FAGC RV GGC C+R + LL
Sbjct: 220 RAAHGSPPPLPPPRTGQCCRMASPCAHRRGLLQLARHFFAGCGRRVAGGCTPCRRFFLLL 279
Query: 318 ELHSRLC--ADPDFCRVPLC 335
LHS +C +D D C VPLC
Sbjct: 280 RLHSSVCDKSDDDSCGVPLC 299
>UniRef100_Q9C5X9 P300/CBP acetyltransferase-related protein 2 [Arabidopsis thaliana]
Length = 1654
Score = 67.0 bits (162), Expect = 7e-10
Identities = 36/106 (33%), Positives = 58/106 (53%), Gaps = 18/106 (16%)
Query: 230 EENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDFKANQPCRYTSCKGL 289
++N++ + R++ LQL ++ LVH + CR+ C+Y +C+ +
Sbjct: 1566 DQNAQNKEARQLR----VLQLRKMLDLLVHASQ--CRSAH-----------CQYPNCRKV 1608
Query: 290 ELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
+ L RH CK+R GGC CK+MW LL+LH+R C + + C VP C
Sbjct: 1609 KGLFRHGINCKVRASGGCVLCKKMWYLLQLHARACKESE-CHVPRC 1653
>UniRef100_O64548 YUP8H12R.38 protein [Arabidopsis thaliana]
Length = 1516
Score = 67.0 bits (162), Expect = 7e-10
Identities = 36/106 (33%), Positives = 58/106 (53%), Gaps = 18/106 (16%)
Query: 230 EENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDFKANQPCRYTSCKGL 289
++N++ + R++ LQL ++ LVH + CR+ C+Y +C+ +
Sbjct: 1428 DQNAQNKEARQLR----VLQLRKMLDLLVHASQ--CRSAH-----------CQYPNCRKV 1470
Query: 290 ELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
+ L RH CK+R GGC CK+MW LL+LH+R C + + C VP C
Sbjct: 1471 KGLFRHGINCKVRASGGCVLCKKMWYLLQLHARACKESE-CHVPRC 1515
>UniRef100_Q9FWQ5 F17F16.8 protein [Arabidopsis thaliana]
Length = 1498
Score = 64.7 bits (156), Expect = 3e-09
Identities = 39/121 (32%), Positives = 62/121 (51%), Gaps = 20/121 (16%)
Query: 215 SHPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKD 274
+HP K + ++N++ + R++ LQL ++ LVH + CR+
Sbjct: 1360 NHP--HKLTTHPSLADQNAQNKEARQLR----VLQLRKMLDLLVHASQ--CRS------- 1404
Query: 275 FKANQPCRYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPL 334
C Y +C+ ++ L RH CK+R GGC CK+MW LL+LH+R C + + C VP
Sbjct: 1405 ----PVCLYPNCRKVKGLFRHGLRCKVRASGGCVLCKKMWYLLQLHARACKESE-CDVPR 1459
Query: 335 C 335
C
Sbjct: 1460 C 1460
>UniRef100_Q6YXY2 Putative HAC5 [Oryza sativa]
Length = 1623
Score = 62.0 bits (149), Expect = 2e-08
Identities = 26/55 (47%), Positives = 35/55 (63%), Gaps = 1/55 (1%)
Query: 281 CRYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
C+Y +C+ ++ L RH CK R GGC CK+MW +L+LH+R C D C VP C
Sbjct: 1533 CQYPNCRKVKGLFRHGMQCKTRASGGCVLCKKMWYMLQLHARACRDSG-CNVPRC 1586
>UniRef100_UPI00003AAEA7 UPI00003AAEA7 UniRef100 entry
Length = 1847
Score = 58.9 bits (141), Expect = 2e-07
Identities = 34/107 (31%), Positives = 54/107 (49%), Gaps = 14/107 (13%)
Query: 229 DEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDFKANQPCRYTSCKG 288
DE NS+ E+ K + L + +++LVH C+ CR N C SC+
Sbjct: 1304 DESNSQGEQQSKSPQESRRLSIQRCIQSLVHACQ--CR-----------NANCSLPSCQK 1350
Query: 289 LELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
++ +V+H GCK + GGC CK++ L H++ C + + C VP C
Sbjct: 1351 MKRVVQHTKGCKRKTNGGCPVCKQLIALCCYHAKHCQE-NKCPVPFC 1396
>UniRef100_UPI00003AAEA6 UPI00003AAEA6 UniRef100 entry
Length = 2328
Score = 58.9 bits (141), Expect = 2e-07
Identities = 34/107 (31%), Positives = 54/107 (49%), Gaps = 14/107 (13%)
Query: 229 DEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDFKANQPCRYTSCKG 288
DE NS+ E+ K + L + +++LVH C+ CR N C SC+
Sbjct: 1673 DESNSQGEQQSKSPQESRRLSIQRCIQSLVHACQ--CR-----------NANCSLPSCQK 1719
Query: 289 LELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
++ +V+H GCK + GGC CK++ L H++ C + + C VP C
Sbjct: 1720 MKRVVQHTKGCKRKTNGGCPVCKQLIALCCYHAKHCQE-NKCPVPFC 1765
>UniRef100_UPI00003AAEA5 UPI00003AAEA5 UniRef100 entry
Length = 918
Score = 58.9 bits (141), Expect = 2e-07
Identities = 34/107 (31%), Positives = 54/107 (49%), Gaps = 14/107 (13%)
Query: 229 DEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDFKANQPCRYTSCKG 288
DE NS+ E+ K + L + +++LVH C+ CR N C SC+
Sbjct: 232 DESNSQGEQQSKSPQESRRLSIQRCIQSLVHACQ--CR-----------NANCSLPSCQK 278
Query: 289 LELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
++ +V+H GCK + GGC CK++ L H++ C + + C VP C
Sbjct: 279 MKRVVQHTKGCKRKTNGGCPVCKQLIALCCYHAKHCQE-NKCPVPFC 324
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.342 0.149 0.512
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 510,407,129
Number of Sequences: 2790947
Number of extensions: 19487314
Number of successful extensions: 80922
Number of sequences better than 10.0: 345
Number of HSP's better than 10.0 without gapping: 90
Number of HSP's successfully gapped in prelim test: 255
Number of HSP's that attempted gapping in prelim test: 80628
Number of HSP's gapped (non-prelim): 425
length of query: 335
length of database: 848,049,833
effective HSP length: 128
effective length of query: 207
effective length of database: 490,808,617
effective search space: 101597383719
effective search space used: 101597383719
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146856.6


