

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146856.2 + phase: 0
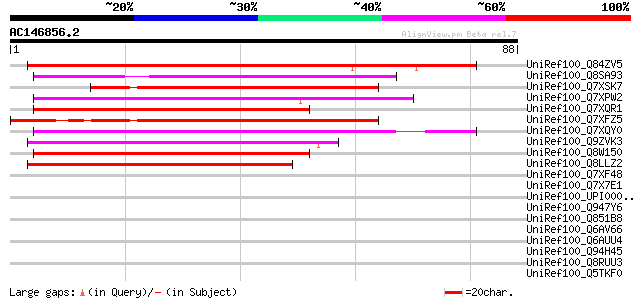
(88 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84ZV5 Polyprotein [Glycine max] 55 3e-07
UniRef100_Q8SA93 Putative polyprotein [Zea mays] 53 2e-06
UniRef100_Q7XSK7 OSJNBa0059D20.8 protein [Oryza sativa] 48 7e-05
UniRef100_Q7XPW2 OSJNBa0032F06.19 protein [Oryza sativa] 47 1e-04
UniRef100_Q7XQR1 OSJNBa0091D06.3 protein [Oryza sativa] 46 2e-04
UniRef100_Q7XFZ5 Putative retroelement [Oryza sativa] 45 3e-04
UniRef100_Q7XQY0 OSJNBb0108J11.11 protein [Oryza sativa] 45 6e-04
UniRef100_Q9ZVK3 Putative retroelement pol polyprotein [Arabidop... 44 8e-04
UniRef100_Q8W150 Polyprotein [Oryza sativa] 44 8e-04
UniRef100_Q8LLZ2 Putative retroelement [Oryza sativa] 44 8e-04
UniRef100_Q7XF48 Similar to 22 kDa kafirin cluster;Ty3-Gypsytype... 44 0.001
UniRef100_Q7X7E1 OSJNBa0006M15.7 protein [Oryza sativa] 44 0.001
UniRef100_UPI000027A252 UPI000027A252 UniRef100 entry 43 0.002
UniRef100_Q947Y6 Putative retroelement [Oryza sativa] 43 0.002
UniRef100_Q851B8 Hypothetical protein OSJNBa0023A13.1 [Oryza sat... 43 0.002
UniRef100_Q6AV66 Hypothetical protein OSJNBa0035N15.2 [Oryza sat... 43 0.002
UniRef100_Q6AUU4 Putative pol polyprotein [Oryza sativa] 43 0.002
UniRef100_Q94H45 Putative polyprotein [Oryza sativa] 43 0.002
UniRef100_Q8RUU3 Putative gag-pol polyprotein [Oryza sativa] 43 0.002
UniRef100_Q5TKF0 Hypothetical protein OSJNBa0030I14.16 [Oryza sa... 43 0.002
>UniRef100_Q84ZV5 Polyprotein [Glycine max]
Length = 1552
Score = 55.5 bits (132), Expect = 3e-07
Identities = 31/87 (35%), Positives = 53/87 (60%), Gaps = 9/87 (10%)
Query: 4 QPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWPSPILEDKVV-EGDG 62
QP+K+L +R + Q+LVQW+ +EATWED ++++ S+P+ LEDKVV +G+G
Sbjct: 1442 QPVKILASRIIIRGHNQIEQILVQWENGLQDEATWEDIEDIKASYPTFNLEDKVVFKGEG 1501
Query: 63 NDTYSLD--------SKHATEPGIRNE 81
N T + ++ ++E G+ N+
Sbjct: 1502 NVTNGMSRGEKVNNTAESSSERGLHNK 1528
>UniRef100_Q8SA93 Putative polyprotein [Zea mays]
Length = 2749
Score = 52.8 bits (125), Expect = 2e-06
Identities = 30/63 (47%), Positives = 37/63 (58%), Gaps = 4/63 (6%)
Query: 5 PLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWPSPILEDKVVEGDGND 64
PLKVL + R I LVQW+GLSPEEATWE D+ R +P LED++ G D
Sbjct: 1793 PLKVLRAQQRRGTWHI----LVQWQGLSPEEATWEPLDDFRGLYPDFQLEDELFAHAGRD 1848
Query: 65 TYS 67
+S
Sbjct: 1849 AWS 1851
>UniRef100_Q7XSK7 OSJNBa0059D20.8 protein [Oryza sativa]
Length = 1463
Score = 47.8 bits (112), Expect = 7e-05
Identities = 23/50 (46%), Positives = 33/50 (66%), Gaps = 1/50 (2%)
Query: 15 RTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWPSPILEDKVVEGDGND 64
R +G++ Q LVQWKG SP ATWED + +R +P+ LED++ +G D
Sbjct: 1365 RLARGVR-QALVQWKGESPASATWEDIEVLRAKYPALQLEDELSLEEGGD 1413
>UniRef100_Q7XPW2 OSJNBa0032F06.19 protein [Oryza sativa]
Length = 1575
Score = 47.0 bits (110), Expect = 1e-04
Identities = 24/67 (35%), Positives = 37/67 (54%), Gaps = 1/67 (1%)
Query: 5 PLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWP-SPILEDKVVEGDGN 63
P+K+L RS+R + Q+L++W G+S TWE DEM+ +P +P +G N
Sbjct: 1478 PIKILDHRSIRRGSKMIKQVLIRWSGVSSSADTWESLDEMQAKFPAAPAWGQAETQGGRN 1537
Query: 64 DTYSLDS 70
T S+ S
Sbjct: 1538 VTASIQS 1544
>UniRef100_Q7XQR1 OSJNBa0091D06.3 protein [Oryza sativa]
Length = 753
Score = 46.2 bits (108), Expect = 2e-04
Identities = 18/48 (37%), Positives = 30/48 (62%)
Query: 5 PLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWPSPI 52
PL++L R + Q+L+QW PE+ATWED D++R+ +P+ +
Sbjct: 395 PLRILDRRLTKRGNSAVAQVLIQWSASVPEDATWEDLDDLRSRFPAAL 442
>UniRef100_Q7XFZ5 Putative retroelement [Oryza sativa]
Length = 1476
Score = 45.4 bits (106), Expect = 3e-04
Identities = 27/64 (42%), Positives = 40/64 (62%), Gaps = 4/64 (6%)
Query: 1 MDHQPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWPSPILEDKVVEG 60
+D +P +V TRS R +G++ Q+LV WKG S ATWED D + +P+ LED++
Sbjct: 1375 IDPEPERV--TRS-RLARGVR-QVLVHWKGESAASATWEDLDTFKERYPAFQLEDELALE 1430
Query: 61 DGND 64
+G D
Sbjct: 1431 EGRD 1434
>UniRef100_Q7XQY0 OSJNBb0108J11.11 protein [Oryza sativa]
Length = 815
Score = 44.7 bits (104), Expect = 6e-04
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 5/77 (6%)
Query: 5 PLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWPSPILEDKVVEGDGND 64
PL+VL R + + Q+L++W G P TWE+E E+R +P+ +V G D
Sbjct: 731 PLQVLDRRFVHKGSKMVEQVLIKWSGGEPAAVTWENEQELRRRFPTTTTWGQVATQGGGD 790
Query: 65 TYSLDSKHATEPGIRNE 81
+ A+E G++ E
Sbjct: 791 VTA-----ASEQGVQPE 802
>UniRef100_Q9ZVK3 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 945
Score = 44.3 bits (103), Expect = 8e-04
Identities = 24/56 (42%), Positives = 33/56 (58%), Gaps = 2/56 (3%)
Query: 4 QPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWPSPI--LEDKV 57
QP VL R + +LVQW+GL E+ATWE+ D++ S+P + LEDKV
Sbjct: 888 QPTAVLEVRWRSQDKKRVADLLVQWEGLHIEDATWEEYDQLAASFPEFVLNLEDKV 943
>UniRef100_Q8W150 Polyprotein [Oryza sativa]
Length = 933
Score = 44.3 bits (103), Expect = 8e-04
Identities = 19/48 (39%), Positives = 30/48 (61%)
Query: 5 PLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWPSPI 52
P+++L TR + + Q+LV W L EATWEDE+E++ +PS +
Sbjct: 828 PVQILDTRIQKKGNKVVRQILVCWSNLPAVEATWEDEEELKQRFPSAL 875
>UniRef100_Q8LLZ2 Putative retroelement [Oryza sativa]
Length = 1786
Score = 44.3 bits (103), Expect = 8e-04
Identities = 20/46 (43%), Positives = 28/46 (60%)
Query: 4 QPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWP 49
+P+K+L TR RT + VQW + EEATWE EDE++ + P
Sbjct: 1726 RPVKILDTRERRTRNRVIRFCKVQWSNHAEEEATWEREDELKAAHP 1771
>UniRef100_Q7XF48 Similar to 22 kDa kafirin cluster;Ty3-Gypsytype [Oryza sativa]
Length = 169
Score = 43.9 bits (102), Expect = 0.001
Identities = 20/46 (43%), Positives = 28/46 (60%)
Query: 4 QPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWP 49
+P+K+L T RT + VQW + EEATWE EDE++T+ P
Sbjct: 109 RPVKILDTMERRTRNRVIRFCKVQWSNHAEEEATWEREDELKTAHP 154
>UniRef100_Q7X7E1 OSJNBa0006M15.7 protein [Oryza sativa]
Length = 1807
Score = 43.5 bits (101), Expect = 0.001
Identities = 19/46 (41%), Positives = 28/46 (60%)
Query: 4 QPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWP 49
+P+K+L T RT + VQW + EEATWE EDE++ ++P
Sbjct: 1747 RPVKILDTMERRTRNRVIRFCKVQWSNHAEEEATWEREDELKAAYP 1792
>UniRef100_UPI000027A252 UPI000027A252 UniRef100 entry
Length = 69
Score = 43.1 bits (100), Expect = 0.002
Identities = 18/48 (37%), Positives = 29/48 (59%)
Query: 2 DHQPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWP 49
+ +P+K+L T RT + VQW + EEATWE +DE++ ++P
Sbjct: 7 EERPVKILDTMERRTRNRVIRFCKVQWSNHAEEEATWERQDELKAAYP 54
>UniRef100_Q947Y6 Putative retroelement [Oryza sativa]
Length = 1461
Score = 43.1 bits (100), Expect = 0.002
Identities = 24/61 (39%), Positives = 35/61 (57%), Gaps = 4/61 (6%)
Query: 4 QPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWPSPILEDKVVEGDGN 63
+P +V+ R R G++ Q+LVQWKG S ATWED + +P+ LED++ G
Sbjct: 1364 EPERVIKARLAR---GVR-QVLVQWKGTSAASATWEDREPFFARYPALQLEDELPLDGGG 1419
Query: 64 D 64
D
Sbjct: 1420 D 1420
>UniRef100_Q851B8 Hypothetical protein OSJNBa0023A13.1 [Oryza sativa]
Length = 264
Score = 43.1 bits (100), Expect = 0.002
Identities = 20/46 (43%), Positives = 27/46 (58%)
Query: 4 QPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWP 49
+P+K+L T RT + VQW + EEATWE EDEM+ + P
Sbjct: 204 RPVKILDTMERRTRNQVIRFCKVQWSNHAEEEATWEREDEMKAAHP 249
>UniRef100_Q6AV66 Hypothetical protein OSJNBa0035N15.2 [Oryza sativa]
Length = 1169
Score = 43.1 bits (100), Expect = 0.002
Identities = 18/48 (37%), Positives = 29/48 (59%)
Query: 2 DHQPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWP 49
+ +P+K+L T RT + VQW + EEATWE +DE++ ++P
Sbjct: 1107 EERPVKILDTMERRTRNRVIRFCKVQWSNHAEEEATWERQDELKAAYP 1154
>UniRef100_Q6AUU4 Putative pol polyprotein [Oryza sativa]
Length = 922
Score = 43.1 bits (100), Expect = 0.002
Identities = 18/48 (37%), Positives = 29/48 (59%)
Query: 2 DHQPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWP 49
+ +P+K+L T RT + VQW + EEATWE +DE++ ++P
Sbjct: 860 EERPVKILDTMERRTRNRVIRFCKVQWSNHAEEEATWERQDELKAAYP 907
>UniRef100_Q94H45 Putative polyprotein [Oryza sativa]
Length = 1475
Score = 43.1 bits (100), Expect = 0.002
Identities = 20/46 (43%), Positives = 27/46 (58%)
Query: 4 QPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWP 49
+P+K+L T RT + VQW + EEATWE EDEM+ + P
Sbjct: 1415 RPVKILDTMERRTRNQVIRFCKVQWSNHAEEEATWEREDEMKAAHP 1460
>UniRef100_Q8RUU3 Putative gag-pol polyprotein [Oryza sativa]
Length = 1338
Score = 42.7 bits (99), Expect = 0.002
Identities = 24/60 (40%), Positives = 35/60 (58%), Gaps = 4/60 (6%)
Query: 5 PLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWPSPILEDKVVEGDGND 64
P +VL R R G++ Q+LVQW+GL +WED D+ R +PS L D+++ G D
Sbjct: 1254 PRQVLRARLAR---GVR-QLLVQWEGLPASATSWEDLDDFRNRYPSFQLADELLIEGGRD 1309
>UniRef100_Q5TKF0 Hypothetical protein OSJNBa0030I14.16 [Oryza sativa]
Length = 1253
Score = 42.7 bits (99), Expect = 0.002
Identities = 20/46 (43%), Positives = 27/46 (58%)
Query: 4 QPLKVLGTRSLRTPQGIKTQMLVQWKGLSPEEATWEDEDEMRTSWP 49
+P ++L T RT + VQW LS EEATWE EDE++ + P
Sbjct: 1193 KPTRILETSERRTRNKVIRFCKVQWSHLSKEEATWEREDELKAAHP 1238
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.307 0.128 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 163,206,696
Number of Sequences: 2790947
Number of extensions: 6501065
Number of successful extensions: 10805
Number of sequences better than 10.0: 400
Number of HSP's better than 10.0 without gapping: 385
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 10412
Number of HSP's gapped (non-prelim): 408
length of query: 88
length of database: 848,049,833
effective HSP length: 64
effective length of query: 24
effective length of database: 669,429,225
effective search space: 16066301400
effective search space used: 16066301400
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146856.2


