

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146856.10 + phase: 0
(1260 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
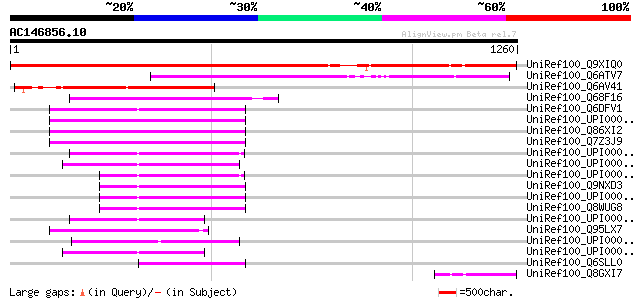
UniRef100_Q9XIQ0 F13O11.26 protein [Arabidopsis thaliana] 1147 0.0
UniRef100_Q6ATV7 Hypothetical protein OJ1285_H07.1 [Oryza sativa] 568 e-160
UniRef100_Q6AV41 Hypothetical protein OSJNBa0063J18.1 [Oryza sat... 387 e-106
UniRef100_Q68F16 LOC446284 protein [Xenopus laevis] 261 1e-67
UniRef100_Q6DFV1 5830426I05Rik protein [Mus musculus] 258 1e-66
UniRef100_UPI000036E07D UPI000036E07D UniRef100 entry 253 3e-65
UniRef100_Q86XI2 MTB protein [Homo sapiens] 253 3e-65
UniRef100_Q7Z3J9 Hypothetical protein DKFZp686G2253 [Homo sapiens] 253 3e-65
UniRef100_UPI00003AB8A8 UPI00003AB8A8 UniRef100 entry 251 1e-64
UniRef100_UPI000033730A UPI000033730A UniRef100 entry 242 6e-62
UniRef100_UPI00003AB8A9 UPI00003AB8A9 UniRef100 entry 213 4e-53
UniRef100_Q9NXD3 Hypothetical protein FLJ20311 [Homo sapiens] 206 4e-51
UniRef100_UPI000036E07C UPI000036E07C UniRef100 entry 206 5e-51
UniRef100_Q8WUG8 MTB protein [Homo sapiens] 206 5e-51
UniRef100_UPI00003AB8A7 UPI00003AB8A7 UniRef100 entry 202 7e-50
UniRef100_Q95LX7 Hypothetical protein [Macaca fascicularis] 192 7e-47
UniRef100_UPI00004392E3 UPI00004392E3 UniRef100 entry 188 8e-46
UniRef100_UPI0000364124 UPI0000364124 UniRef100 entry 187 1e-45
UniRef100_Q6SLL0 More than blood [Mus musculus] 169 6e-40
UniRef100_Q8GXI7 Hypothetical protein At1g64960/F13O11_26 [Arabi... 159 5e-37
>UniRef100_Q9XIQ0 F13O11.26 protein [Arabidopsis thaliana]
Length = 1227
Score = 1147 bits (2966), Expect = 0.0
Identities = 628/1273 (49%), Positives = 840/1273 (65%), Gaps = 64/1273 (5%)
Query: 1 MDKKLCSSLNSSPQEFISLATTLTLKSSKSSLKTLIHSIKPSSNLISSLPPSLYESITTT 60
M+K+L SSL +S +EF+S A LTLKSSK SLKT+I+++KPSS+L SSLP +L+ SI
Sbjct: 1 MEKRLRSSLKTSSEEFLSSAVKLTLKSSKPSLKTIINAVKPSSDLSSSLPLALHNSILHH 60
Query: 61 IQSFLNLLELNNSENPQTPPTATLRRSSRKNTTATTEPSSQSDE-KHKLLEKLQILAHIL 119
+SF LL+ N+ N P + + R T T P S D+ KH++L LQIL+++L
Sbjct: 61 TESFQKLLDEVNNNNTYIPSPSNSPPTKRHRGTTGTSPDSDLDQRKHQILASLQILSYVL 120
Query: 120 FLCVSHPRKVFDFSDLLPGVQALHDNLIVLEADSILSSGIETICEEWWKENLPERESLIS 179
LC+ +P+ F SDLLP QALH+NL + E+DS+L I +CE WWKE L RESLIS
Sbjct: 121 HLCLLNPKNAFPTSDLLPAAQALHNNLRLFESDSVLCLEIAGVCECWWKEGLVGRESLIS 180
Query: 180 QTLPFVLSRSLTLKKKVDVHRVYMFREAFALFDFEDESIEDLKLLLNRCVISPLYLKTED 239
Q+LPF+LSRSLTLKKKVDVHRVYM REAF LFDFEDESIEDL++LL RCV+SPLY+KTED
Sbjct: 181 QSLPFLLSRSLTLKKKVDVHRVYMLREAFTLFDFEDESIEDLRMLLMRCVVSPLYVKTED 240
Query: 240 GRKFLAFLFGLSDQLGKELLAMIRSQIPFGRKSMLEAYGDILFRAWKAAPEDSRSGIEDG 299
G++F++F FGLS QL K LA++++QIP G KS+LE +G ILFRAWK D + IEDG
Sbjct: 241 GQRFVSFAFGLSRQLMKSGLAVVKAQIPLGSKSVLEGFGGILFRAWKEVELDLKGEIEDG 300
Query: 300 FLQDLIEGSIHASSGVFALYIRRVLGGFINQRTVDGVEKLLYRLAEPVIFRSLQAANSNV 359
FLQ +I+ +IHASS FA +RRVLGGFI+QRT GVEKLL+ LAEP+IFRSLQ ANSNV
Sbjct: 301 FLQGIIDSAIHASSSAFAASLRRVLGGFISQRTSQGVEKLLFTLAEPMIFRSLQVANSNV 360
Query: 360 RQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLLEKLLVDDCPEVRTIAVEGSCRVLHL 419
R NALHLLLD+FP+EDPDA+KE KD LLDKQF+LLEKLL D+CP+VR++AVEG RV +L
Sbjct: 361 RLNALHLLLDLFPMEDPDATKEAKDTLLDKQFYLLEKLLSDECPDVRSVAVEGLSRVFYL 420
Query: 420 FWEIIPSPIITKMLTKVISDMSHDVCNEVRLSTLSGIIYLLDNPHSHEVLKVLCPRLGHL 479
FWE+IPS ITK+LTK+ DMSH+ C+EVRLST++GI YLL NP SH +LKV+ PRLGHL
Sbjct: 421 FWEVIPSATITKVLTKIFDDMSHESCSEVRLSTVNGITYLLANPQSHGILKVILPRLGHL 480
Query: 480 MLDNVLTVQVAMAELLLHLNDVQNFQFNKVVVLDVLMSVLASDQPPVAKKLTKLLIPSYF 539
MLD+V +V+VAM +LLL + DV+ FQFN VV LDVL+SVLASDQ VAK + +LLIPSYF
Sbjct: 481 MLDSVTSVRVAMVDLLLLIRDVRAFQFNTVVSLDVLLSVLASDQTHVAKGIARLLIPSYF 540
Query: 540 PSIVPIEEACNRCITLVKRSPMAGAIFCKYAVSEGASKTHLMELVKVFLSLVLSQDKLNA 599
PS EEAC RC TL+ R+P AGA FC++ VS GA+ ++ LV FL+ VLS DKL
Sbjct: 541 PSRKRAEEACQRCRTLINRNPKAGARFCEFLVSLGATVKSVLHLVGFFLNSVLSGDKLLE 600
Query: 600 DQIEGFLIAASYLCDNLVSELCYMDALKELLTAEKVEGLLTGASTEQAQSSLFNIFSTVC 659
+Q EG L AA YLC +LV++ M +LKELL EK++ LL A T QAQSS+ +I + V
Sbjct: 601 NQTEGLLRAAYYLCKDLVADSGCMASLKELLPGEKLKSLLAFAPTAQAQSSVIDIITMVS 660
Query: 660 PDNVAGLLEECMSVVTNCHGLPEDVDRQSKIRSAHKLLLSLGGFDDMFEALTTLLHKAAY 719
PD V+ +LE+CM++V NC GLP D RQ+++RS HKLLLS F D+ T+++ K AY
Sbjct: 661 PDIVSEVLEDCMNLVVNCGGLPSDAGRQTELRSVHKLLLSSNAFCDLIGTFTSIMQKTAY 720
Query: 720 RCHIKFGADMPSHSVTATKRKKSKSSGKFSIKSKIINRKQ--SFEDDYSVAVGVAWQVRD 777
RC I FG ++ ++ + KRKKSKSSGK S++ K ++ K SFE+DY VAVG+AWQ++D
Sbjct: 721 RCQINFGYEVERKNLQSMKRKKSKSSGKSSVRWKHVSGKNAISFEEDYLVAVGIAWQIKD 780
Query: 778 LLQHEDTRKAIFRSQPLEMLFFSLKMVSEVSIEHCGQYEYIDISPVLAYMALALQMTVDN 837
LL ED RK+I S +E L SLK+VS SI EY+D++P
Sbjct: 781 LLTTEDARKSILESD-IEELLLSLKVVSHTSILQATCCEYMDVNP--------------- 824
Query: 838 VGTSSEKNGDSKGKKTKIDSSTLLSEAILDLTIEHVLNCLEKLFGSDD------TVQDHN 891
+ I+D T++H+L C ++LF + D T + N
Sbjct: 825 -------------------------QDIMDQTMDHILVCTDELFQAGDIGTPGTTSPEAN 859
Query: 892 VDSHNLESTTGKNRSSKKRKRTGKNQN----STKRRRLSLTNAGCPSNEGSVHNEPQQVL 947
+ + TT K+R R ++ + ++ + S ++ VL
Sbjct: 860 LSK---KPTTSNGNQPKRRNRNARDDALLVVTVPNEAFKFSDIVFMLHAASEGSKEGGVL 916
Query: 948 CKVKMITAVLKFMADATAMCFAPHNNGSFLNNTSKCIQHILSSLNQLYHHKIEFEEEDKK 1007
KVKM+TA+ KF ++T M A + S +S +++++S N K+EFE+ D K
Sbjct: 917 NKVKMLTAIFKFFVESTEMGLASNFQASMFKFSSAYLKYVISIFNDHSTGKLEFEDADMK 976
Query: 1008 NTIFCLKSSFSYAAKILNVILTDSGGSSIMTSKAFTLANNLLDLIVSTESCLGSAYASRL 1067
I C KSS SYA K +N+++ + +S ++F LAN+LLDL E LGSAYASR+
Sbjct: 977 EMILCTKSSTSYAGKFINLVMRHATEASRPLFESFDLANDLLDLFTMVEKSLGSAYASRI 1036
Query: 1068 VAAAKPWLPDVVLALGSESVLQHTESGSEHLFASEQMKLQFPKWPFIVAKTVLSAVNEGE 1127
V+A PW+PD+VLALG + +E S + + +KL FP W AK L +N+
Sbjct: 1037 VSALNPWIPDLVLALGPCFINNDSEE-SSYTSSFNHIKLCFPSWLLTCAKIELHEINK-- 1093
Query: 1128 GDGECSQADKFSTFNKLTAMLIILLKKNKSIMDAVGDIFLVCSLIGLEQKDFELAAGLLQ 1187
+ ++ F KL + L+K N ++DA+G + L+C + +E++D+ A GLL
Sbjct: 1094 --EDVTETSGFLALKKLRNTIFTLVKGNTKVLDAIGYVLLLCLAVCIEKRDYSTALGLLH 1151
Query: 1188 FVCSKLFNRDDRDWGDL--MLSSLEEIYPKIERQITEASDNDELEKLMHAKELIEPLWTY 1245
VC KL +DR+W +L ML L IYP IER+I E D DE++ L A+EL++P+W Y
Sbjct: 1152 LVCVKLVGSEDREWKELDTMLVLLPRIYPIIEREIGEGRDEDEVKTLEAARELLQPVWMY 1211
Query: 1246 HLYETGKVNMTDE 1258
H+YETG+ +M DE
Sbjct: 1212 HVYETGRFHMMDE 1224
>UniRef100_Q6ATV7 Hypothetical protein OJ1285_H07.1 [Oryza sativa]
Length = 858
Score = 568 bits (1464), Expect = e-160
Identities = 334/898 (37%), Positives = 521/898 (57%), Gaps = 53/898 (5%)
Query: 351 SLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLLEKLLVDDCPEVRTIAV 410
S ANSNVR N+LHLLLD+FPLEDPD +K+ D LL+KQ+FLL+KLL+DDCPE+RT+A+
Sbjct: 4 SQSVANSNVRHNSLHLLLDLFPLEDPDVTKDVNDPLLEKQYFLLDKLLMDDCPEIRTVAI 63
Query: 411 EGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCNEVRLSTLSGIIYLLDNPHSHEVLK 470
EG CR+L+ FWE+IPS I+K L+K++ D+S D C EVR+ST++G+IYLLDNP SHE+LK
Sbjct: 64 EGLCRILNQFWEVIPSLTISKFLSKIVDDISKDSCTEVRVSTINGLIYLLDNPQSHEILK 123
Query: 471 VLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNFQFNKVVVLDVLMSVLASDQPPVAKKL 530
VL PRL ++ D L+V+ + +LLL + D+++FQFNKVV L L+S L++D P VA+K+
Sbjct: 124 VLLPRLSDMVSDPALSVRSSAVDLLLAIRDLRSFQFNKVVGLGTLLSSLSNDHPRVAQKI 183
Query: 531 TKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGAIFCKYAVSEGASKTHLMELVKVFLSL 590
TKLLIPSYFP+ +P++EAC RCI L+KRSP AGA FC++A+SEG+ L+EL+KV ++L
Sbjct: 184 TKLLIPSYFPTKLPLKEACARCIALIKRSPTAGARFCEFALSEGSPPRSLVELIKVSITL 243
Query: 591 VLSQDKLNADQIEGFLIAASYLCDNLVSELCYMDALKELLTAEKVEGLLTGASTEQAQSS 650
L+ +N++Q +G +IA++ L +L E + +L+E K++ L + A+S+
Sbjct: 244 ALAPTGMNSEQTDGLVIASANLIKSLSEERSSLASLREFFANAKLKLLFKTEISVGARSA 303
Query: 651 LFNIFSTVCPDNVAGLLEECMSVVTNCHGLPEDVDRQSKIRSAHKLLLSLGGFDDMFEAL 710
L ++ V PD+++ L +ECM+VV N G+ Q + +AHKL+ S G D+MFEAL
Sbjct: 304 LLSMAPVVSPDDLSALHDECMNVVMNAAGVSTQQGCQEAVLAAHKLVFSSGWSDEMFEAL 363
Query: 711 TTLLHKAAYRCHIKFGADMPSHSVTATKRKKSKSSGKFSIKSKIINRKQSFEDDYSVAVG 770
T +L + + P V +KRKK KS K KS S +D+ + G
Sbjct: 364 TNILQSKVSCFAEIYDIEPPICPVATSKRKKGKSLKKTPAKSGHDIGNGSSSEDFDIVAG 423
Query: 771 VAWQVRDLLQHEDTRKAIFRSQPLEMLFFSLKMVSEVSIEHCGQYEYIDISPVLAYMALA 830
+WQ+ D+L+ E+ R A +S ++ F SLK++ +V IE C Q++ ++ +P+LAY++LA
Sbjct: 424 ASWQINDILKDEEKRVAFLQSSYSDVAFSSLKVICQVYIEQCLQFDSLNATPLLAYLSLA 483
Query: 831 LQMTVDNVGTSSEKNGDSKGKKTKIDSSTLLSEAILDLTIEHVLNCLEKLFGSDDTVQDH 890
+ ++ ++ S + T I+ S ++H+LNC +KL
Sbjct: 484 THSALQDI----DQTDISTSESTTINHS-----------LDHLLNCFDKLLN-------- 520
Query: 891 NVDSHNLESTTGKNRSSKKRKRTGKNQNSTKRRRLSLTNAGCPSNEGSVHNEPQQVLCKV 950
ES TG S K + +N+ S +++ + G P N + +
Sbjct: 521 -------ESVTGSTNSLKLK----QNKKSARQKH----HHGVPEG-----NALRGTVNVY 560
Query: 951 KMITAVLKFMADATAMCFAPHNNGSFLNNTSKCIQHILSSLNQLYHHKIEFEEEDKKNTI 1010
+ T++LKF+ D + N LN ++ S++ F+ D K+ +
Sbjct: 561 MLGTSILKFIVDTITIKLISDNKVGCLNFALSFTKYASSAIKMHQEQSSSFKGNDLKDIL 620
Query: 1011 FCLKSSFSYAAKILNVILTDSGGSSIMTSKAFTLANNLLDLIVSTESCLGSAYASRLVAA 1070
++SSF+YAAK+L+++L +S S +AF LANNLLDL+ S ES GS +A LV+
Sbjct: 621 MLIRSSFTYAAKLLHLVLANSIESQSPPEEAFFLANNLLDLVPSVESAAGSKFALSLVSV 680
Query: 1071 AKPWLPDVVLALGSESVLQHTESGSEHLFASEQMKLQFPKWPFIVAKTVLSAVNEGEGDG 1130
K WLP V++ LG ++ G+ F + P W +AK L + D
Sbjct: 681 VKQWLPVVIMGLGCRWLIGPQAEGNMCDFGGSCL----PLWVVALAKNELLDDEKPRDDD 736
Query: 1131 ECSQADKFS-TFNKLTAMLIILLKK-NKSIMDAVGDIFLVCSLIGLEQKDFELAAGLLQF 1188
+ QA + S + KL M++ILLKK + I+D+V +FL + L++ ++ + GL +F
Sbjct: 737 QSEQASEDSQSSRKLAEMMVILLKKGSPKILDSVAGVFLSTLKLALQRAEYGVVLGLTRF 796
Query: 1189 VCSKLFNRDDRDWGDLMLS--SLEEIYPKIERQITE--ASDNDELEKLMHAKELIEPL 1242
VC +L D L L+ SL E + +I++ + + + ++L AK LI +
Sbjct: 797 VCVRLLGSDSSASEKLHLAHDSLRENFFEIDKHVMDDLVDSEESRQQLESAKALIRSI 854
>UniRef100_Q6AV41 Hypothetical protein OSJNBa0063J18.1 [Oryza sativa]
Length = 496
Score = 387 bits (995), Expect = e-106
Identities = 217/512 (42%), Positives = 318/512 (61%), Gaps = 59/512 (11%)
Query: 11 SSPQEFISLATTLTLKSSKSSL------KTLIHSIKPSSNLISSLPPSLYESITTTIQSF 64
SS + ++LA TL ++ + L K L+HS+ S ++ SLP +L
Sbjct: 21 SSAADLLALAATLFPAAADAPLRDPPHLKHLVHSLPDSHPVLLSLPGALAPP-------- 72
Query: 65 LNLLELNNSENPQTPPTATLRRSSRKNTTATTEPSSQSDEKHKLLEKLQILAHILFLCVS 124
L++ P PP A +L H+L S
Sbjct: 73 -----LSDGGAPGYPPRAAA-----------------------------VLLHLLLTHPS 98
Query: 125 HPRKVFDFSDLLPGVQALHDNLIVLEADSILSSGIETICEEW-WKENLPERESLISQTLP 183
HP + + DLLP + LHD L L D + + C E W+ P RE++++QTLP
Sbjct: 99 HPPR---WGDLLPPLARLHDRLAQLATDDPPLAALAVACFELAWRAAAPGREAVVAQTLP 155
Query: 184 FVLSRSLTLKKKVD---VHRVYMFREAFALFDFEDE-SIEDLKLLLNRCVISPLYLKTED 239
++ + +L+ + R+ R+A AL D++D+ SI D K+LL RC +SPL+LK E+
Sbjct: 156 YLFAEALSCGSATARPVLRRLLALRDALALLDYDDDDSISDFKMLLLRCFVSPLFLKAEE 215
Query: 240 GRKFLAFLFGLSDQLGKELLAMIRSQI--PFGRKSMLEAYGDILFRAWKAAPEDSRSGIE 297
GRK L+ + G+S+ L +E L +IR+Q+ P +++ L AYG+++FRAWK R +
Sbjct: 216 GRKLLSLVLGVSEGLAREGLELIRAQVGMPGVKRAALVAYGEVVFRAWKDGGW-VRGEVG 274
Query: 298 DGFLQDLIEGSIHASSGVFALYIRRVLGGFINQRTVDGVEKLLYRLAEPVIFRSLQAANS 357
+ FLQ ++EG++HA S A R++L F+ QR V GVEKL+++LAEPV+FRSLQ ANS
Sbjct: 275 EAFLQGMLEGAVHARSKELAKAARKLLSAFVEQRMVAGVEKLIFQLAEPVLFRSLQVANS 334
Query: 358 NVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLLEKLLVDDCPEVRTIAVEGSCRVL 417
NVR N+LHLLLD+FPLEDPD +K+ D LL+KQ+FLL+KLL+DDCPE+RT+A+EG CR+L
Sbjct: 335 NVRHNSLHLLLDLFPLEDPDVTKDVNDPLLEKQYFLLDKLLMDDCPEIRTVAIEGLCRIL 394
Query: 418 HLFWEIIPSPIITKMLTKVISDMSHDVCNEVRLSTLSGIIYLLDNPHSHEVLKVLCPRLG 477
+ FWE+IPS I+K L+K++ D+S D C EVR+ST++G+IYLLDNP SHE+LKVL PRL
Sbjct: 395 NQFWEVIPSLTISKFLSKIVDDISKDSCTEVRVSTINGLIYLLDNPQSHEILKVLLPRLS 454
Query: 478 HLMLDNVLTVQVAMAELLLHLNDVQNFQFNKV 509
++ D L+V+ + +LLL + D+++FQFNKV
Sbjct: 455 DMVSDPALSVRSSAVDLLLAIRDLRSFQFNKV 486
>UniRef100_Q68F16 LOC446284 protein [Xenopus laevis]
Length = 1108
Score = 261 bits (667), Expect = 1e-67
Identities = 168/524 (32%), Positives = 274/524 (52%), Gaps = 29/524 (5%)
Query: 150 EADSILSSGIETICEEWWKENLPERESLISQTLPFVLSRSLTLKKKV-DVHRVYMFREAF 208
+++S ++ I +CE WW++ L +E +L++SL +K V D+ R++ +A
Sbjct: 87 KSESHITLAIRHLCEAWWEKGLQGKEEFGKTAFLLLLAKSLEVKCVVADIGRLWHLHQAL 146
Query: 209 ALFDFEDESIEDLKLLLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPF 268
FDF E ++K LL +C +S ++K E+GR+FL+FLF K + I++Q+
Sbjct: 147 LSFDFNSEESHNVKDLLLQCFLSINHIKKEEGRRFLSFLFSWDVNFIKMIHGTIKNQLQC 206
Query: 269 GRKSMLEAYGDILFRAWKAAPEDSRSGIEDGFLQDLIEGSIHAS-SGVFALYIRRVLGGF 327
KS++ DI FRAWK A D IE +QD + +H + +R VL F
Sbjct: 207 LPKSLMTHIADIYFRAWKKASGDVLQTIEQSCIQDFMHHGVHLPRNSPLHPKVREVLSYF 266
Query: 328 INQRTVDGVEKLLYRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLL 387
Q+ GVE++LY L +P+I+R L+A NS VR NA L ++ FP+ DP+ + ED D +
Sbjct: 267 HQQKLRQGVEEMLYNLYQPIIWRGLKARNSEVRSNAALLFVEAFPIRDPNLNHEDMDNEI 326
Query: 388 DKQFFLLEKLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCN- 446
KQF L LL D P VR+ V G C++ +WE+IP I+T +L K++ D++ DV +
Sbjct: 327 QKQFEELFNLLEDPQPLVRSTGVLGVCKITAKYWEMIPPAILTDLLRKILGDLAADVSSA 386
Query: 447 EVRLSTLSGIIYLLDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNFQF 506
+VR S + LLDN SH +L+ + P L + DN V+VA ++LL + V+ +F
Sbjct: 387 DVRCSVFKCLPILLDNKLSHPLLENMLPALKFCLHDNSEKVRVAFVDMLLKIKAVRAAKF 446
Query: 507 NKVVVLDVLMSVLASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGAIF 566
K+ ++ +++ L D PV++++ LL S+FP E C RC+ L++ +P A F
Sbjct: 447 WKICPMEQILARLEVDSRPVSRRIVNLLFNSFFPVNQQEEVWCERCVALIQMNPAAARKF 506
Query: 567 CKYAVSEGA--SKTHLMELVKVFLSLVLSQDKLNADQIEGFLIAASYLCDNLVSELCYMD 624
+YA A + LM ++ L+ +++ N D +
Sbjct: 507 YQYAYEHTAPTNIAKLMLTIRRCLNACINRTVPNEDTED--------------------- 545
Query: 625 ALKELLTAEKVEGLLTGASTEQAQSSLFNIFSTVCPDNVAGLLE 668
E EG++ G S ++ +S L N+ ST ++A LLE
Sbjct: 546 ---EDDDEGDGEGIVRGDSEKENKSVLENVLSTEDSASMASLLE 586
>UniRef100_Q6DFV1 5830426I05Rik protein [Mus musculus]
Length = 1138
Score = 258 bits (658), Expect = 1e-66
Identities = 158/492 (32%), Positives = 266/492 (53%), Gaps = 6/492 (1%)
Query: 99 SSQSDEKHKLLEKLQILAHILFLCVSHPRKVFDFSDLLPGVQALHDNLIVL-EADSILSS 157
S S + K ++ + + ++ VS + ++ LL L+ L L E++ L +
Sbjct: 83 SEHSPKMRKSIKIICAIVTVILASVSIINEHENYGALLECAVILNGILYALPESEQKLQN 142
Query: 158 GIETICEEWWKENLPERESLISQTLPFVLSRSLTLKKKVDVHRVYMFREAFALFDFEDES 217
I+ +C +WW+ LP +E + +L RSL K DV R++ +A FD++ E
Sbjct: 143 SIQDLCVKWWERGLPAKEDMGKTAFIMLLRRSLETKSGADVCRLWRIHQALYCFDYDWEE 202
Query: 218 IEDLKLLLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPFGRKSMLEAY 277
++K +L C I+ Y+K E+GR+FL+FLF + K + I++Q+ +KS++
Sbjct: 203 SREIKDMLLECFINVNYIKKEEGRRFLSFLFSWNVDFIKMIHETIKNQLAGLQKSLMVHI 262
Query: 278 GDILFRAWKAAPEDSRSGIEDGFLQDLIEGSIHA--SSGVFALYIRRVLGGFINQRTVDG 335
+I FRAWK A IE +QD + IH S V + +R VL F Q+ G
Sbjct: 263 AEIYFRAWKKASGKMLETIEYDCIQDFMFHGIHLLRRSPVHSK-VREVLSYFHQQKVRQG 321
Query: 336 VEKLLYRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLLE 395
VE++LYRL +P+++R L+A NS VR NA L ++ FP+ DP+ + + D + KQF L
Sbjct: 322 VEEMLYRLYKPILWRGLKARNSEVRSNAALLFVEAFPIRDPNFTATEMDNEIQKQFEELY 381
Query: 396 KLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCN-EVRLSTLS 454
L+ D P VR+ + G C++ +WE++P I+ L KV +++ D+ + +VR S
Sbjct: 382 NLIEDPYPRVRSTGILGVCKISSKYWEMMPPNILVDFLKKVTGELAFDISSADVRCSVFK 441
Query: 455 GIIYLLDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNFQFNKVVVLDV 514
+ +LDN SH +L+ L P L + + DN V+VA +LLL + V+ +F K+ ++
Sbjct: 442 CLPIILDNKLSHPLLEQLLPTLRYSLHDNSEKVRVAFVDLLLKIKAVRAAKFWKICPMED 501
Query: 515 LMSVLASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGAIFCKYAVSEG 574
++ L D PV+++L L+ S+ P P E C RC+TL++ + A F +YA E
Sbjct: 502 ILVRLEMDSRPVSRRLVSLIFNSFLPVNQPEEVWCERCVTLIQMNRAAARRFYQYA-HEH 560
Query: 575 ASKTHLMELVKV 586
+ T++ +L+ V
Sbjct: 561 TASTNIAKLIHV 572
>UniRef100_UPI000036E07D UPI000036E07D UniRef100 entry
Length = 1156
Score = 253 bits (646), Expect = 3e-65
Identities = 160/493 (32%), Positives = 267/493 (53%), Gaps = 7/493 (1%)
Query: 99 SSQSDEKHKLLEKLQILAHILFLCVSHPRKVFDFSDLLPGVQALHDNLIVL-EADSILSS 157
+ + K +E + + ++ VS + ++ LL V L+ L L E++ L S
Sbjct: 83 TEHGSKMRKSIEIIYAITSVILASVSVINESENYEALLECVIILNGILYALPESERKLQS 142
Query: 158 GIETICEEWWKENLPERESLISQTLPFVLSRSLTLKKKVDVHRVYMFREAFALFDFEDES 217
I+ +C WW++ LP +E +L RSL K DV R++ +A FD++ E
Sbjct: 143 SIQDLCVTWWEKGLPAKEDTGKTAFVMLLRRSLETKTGADVCRLWRIHQALYCFDYDLEE 202
Query: 218 IEDLKLLLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPFGRKSMLEAY 277
++K +L C I+ Y+K E+GR+FL+ LF + K + I++Q+ +KS++
Sbjct: 203 SGEIKDMLLECFININYIKKEEGRRFLSCLFNWNINFIKMIHGTIKNQLQGLQKSLMVYI 262
Query: 278 GDILFRAWKAAPEDSRSGIEDGFLQDLIEGSIH--ASSGVFALYIRRVLGGFINQRTV-D 334
+I FRAWK A IE+ +QD + IH S V + +R VL F +Q+ V
Sbjct: 263 AEIYFRAWKKASGKILEAIENDCIQDFMFHGIHLPRRSPVHS-KVREVLSYFHHQKKVRQ 321
Query: 335 GVEKLLYRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLL 394
GVE++LYRL +P+++R L+A NS VR NA L ++ FP+ DP+ + D + KQF L
Sbjct: 322 GVEEMLYRLYKPILWRGLKARNSEVRSNAALLFVEAFPIRDPNLHAIEMDSEIQKQFEEL 381
Query: 395 EKLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCN-EVRLSTL 453
LL D P VR+ + G C++ +WE++P I+ +L KV +++ D + +VR S
Sbjct: 382 YSLLEDPYPMVRSTGILGVCKITSKYWEMMPPTILIDLLKKVTGELAFDTSSADVRCSVF 441
Query: 454 SGIIYLLDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNFQFNKVVVLD 513
+ +LDN SH +L+ L P L + + DN V+VA ++LL + V+ +F K+ ++
Sbjct: 442 KCLPMILDNKLSHPLLEQLLPALRYSLHDNSEKVRVAFVDMLLKIKAVRAAKFWKICPME 501
Query: 514 VLMSVLASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGAIFCKYAVSE 573
++ L +D PV+++L L+ S+ P P E C RC+TLV+ + A F +YA E
Sbjct: 502 HILVRLETDSRPVSRRLVSLIFNSFLPVNQPEEVWCERCVTLVQMNHAAARRFYQYA-HE 560
Query: 574 GASKTHLMELVKV 586
+ T++ +L+ V
Sbjct: 561 HTACTNIAKLIHV 573
>UniRef100_Q86XI2 MTB protein [Homo sapiens]
Length = 1143
Score = 253 bits (646), Expect = 3e-65
Identities = 160/493 (32%), Positives = 267/493 (53%), Gaps = 7/493 (1%)
Query: 99 SSQSDEKHKLLEKLQILAHILFLCVSHPRKVFDFSDLLPGVQALHDNLIVL-EADSILSS 157
+ + K +E + + ++ VS + ++ LL V L+ L L E++ L S
Sbjct: 83 TEHGSKMRKSIEIIYAITSVILASVSVINESENYEALLECVIILNGILYALPESERKLQS 142
Query: 158 GIETICEEWWKENLPERESLISQTLPFVLSRSLTLKKKVDVHRVYMFREAFALFDFEDES 217
I+ +C WW++ LP +E +L RSL K DV R++ +A FD++ E
Sbjct: 143 SIQDLCVTWWEKGLPAKEDTGKTAFVMLLRRSLETKTGADVCRLWRIHQALYCFDYDLEE 202
Query: 218 IEDLKLLLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPFGRKSMLEAY 277
++K +L C I+ Y+K E+GR+FL+ LF + K + I++Q+ +KS++
Sbjct: 203 SGEIKDMLLECFININYIKKEEGRRFLSCLFNWNINFIKMIHGTIKNQLQGLQKSLMVYI 262
Query: 278 GDILFRAWKAAPEDSRSGIEDGFLQDLIEGSIH--ASSGVFALYIRRVLGGFINQRTV-D 334
+I FRAWK A IE+ +QD + IH S V + +R VL F +Q+ V
Sbjct: 263 AEIYFRAWKKASGKILEAIENDCIQDFMFHGIHLPRRSPVHS-KVREVLSYFHHQKKVRQ 321
Query: 335 GVEKLLYRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLL 394
GVE++LYRL +P+++R L+A NS VR NA L ++ FP+ DP+ + D + KQF L
Sbjct: 322 GVEEMLYRLYKPILWRGLKARNSEVRSNAALLFVEAFPIRDPNLHAIEMDSEIQKQFEEL 381
Query: 395 EKLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCN-EVRLSTL 453
LL D P VR+ + G C++ +WE++P I+ +L KV +++ D + +VR S
Sbjct: 382 YSLLEDPYPMVRSTGILGVCKITSKYWEMMPPTILIDLLKKVTGELAFDTSSADVRCSVF 441
Query: 454 SGIIYLLDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNFQFNKVVVLD 513
+ +LDN SH +L+ L P L + + DN V+VA ++LL + V+ +F K+ ++
Sbjct: 442 KCLPMILDNKLSHPLLEQLLPALRYSLHDNSEKVRVAFVDMLLKIKAVRAAKFWKICPME 501
Query: 514 VLMSVLASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGAIFCKYAVSE 573
++ L +D PV+++L L+ S+ P P E C RC+TLV+ + A F +YA E
Sbjct: 502 HILVRLETDSRPVSRRLVSLIFNSFLPVNQPEEVWCERCVTLVQMNHAAARRFYQYA-HE 560
Query: 574 GASKTHLMELVKV 586
+ T++ +L+ V
Sbjct: 561 HTACTNIAKLIHV 573
>UniRef100_Q7Z3J9 Hypothetical protein DKFZp686G2253 [Homo sapiens]
Length = 1156
Score = 253 bits (646), Expect = 3e-65
Identities = 160/493 (32%), Positives = 267/493 (53%), Gaps = 7/493 (1%)
Query: 99 SSQSDEKHKLLEKLQILAHILFLCVSHPRKVFDFSDLLPGVQALHDNLIVL-EADSILSS 157
+ + K +E + + ++ VS + ++ LL V L+ L L E++ L S
Sbjct: 83 TEHGSKMRKSIEIIYAITSVILASVSVINESENYEALLECVIILNGILYALPESERKLQS 142
Query: 158 GIETICEEWWKENLPERESLISQTLPFVLSRSLTLKKKVDVHRVYMFREAFALFDFEDES 217
I+ +C WW++ LP +E +L RSL K DV R++ +A FD++ E
Sbjct: 143 SIQDLCVTWWEKGLPAKEDTGKTAFVMLLRRSLETKTGADVCRLWRIHQALYCFDYDLEE 202
Query: 218 IEDLKLLLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPFGRKSMLEAY 277
++K +L C I+ Y+K E+GR+FL+ LF + K + I++Q+ +KS++
Sbjct: 203 SGEIKDMLLECFININYIKKEEGRRFLSCLFNWNINFIKMIHGTIKNQLQGLQKSLMVYI 262
Query: 278 GDILFRAWKAAPEDSRSGIEDGFLQDLIEGSIH--ASSGVFALYIRRVLGGFINQRTV-D 334
+I FRAWK A IE+ +QD + IH S V + +R VL F +Q+ V
Sbjct: 263 AEIYFRAWKKASGKILEAIENDCIQDFMFHGIHLPRRSPVHS-KVREVLSYFHHQKKVRQ 321
Query: 335 GVEKLLYRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLL 394
GVE++LYRL +P+++R L+A NS VR NA L ++ FP+ DP+ + D + KQF L
Sbjct: 322 GVEEMLYRLYKPILWRGLKARNSEVRSNAALLFVEAFPIRDPNLHAIEMDSEIQKQFEEL 381
Query: 395 EKLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCN-EVRLSTL 453
LL D P VR+ + G C++ +WE++P I+ +L KV +++ D + +VR S
Sbjct: 382 YSLLEDPYPMVRSTGILGVCKITSKYWEMMPPTILIDLLKKVTGELAFDTSSADVRCSVF 441
Query: 454 SGIIYLLDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNFQFNKVVVLD 513
+ +LDN SH +L+ L P L + + DN V+VA ++LL + V+ +F K+ ++
Sbjct: 442 KCLPMILDNKLSHPLLEQLLPALRYSLHDNSEKVRVAFVDMLLKIKAVRAAKFWKICPME 501
Query: 514 VLMSVLASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGAIFCKYAVSE 573
++ L +D PV+++L L+ S+ P P E C RC+TLV+ + A F +YA E
Sbjct: 502 HILVRLETDSRPVSRRLVSLIFNSFLPVNQPEEVWCERCVTLVQMNHAAARRFYQYA-HE 560
Query: 574 GASKTHLMELVKV 586
+ T++ +L+ V
Sbjct: 561 HTACTNIAKLIHV 573
>UniRef100_UPI00003AB8A8 UPI00003AB8A8 UniRef100 entry
Length = 1142
Score = 251 bits (640), Expect = 1e-64
Identities = 146/440 (33%), Positives = 249/440 (56%), Gaps = 7/440 (1%)
Query: 150 EADSILSSGIETICEEWWKENLPERESLISQTLPFVLSRSLTLKKK-VDVHRVYMFREAF 208
E+++IL S I +CE WW++ L +E L +L +SL+ VD+ ++ +
Sbjct: 134 ESENILRSAIRRVCEMWWEKGLEGKEQLGKTAFIMLLKKSLSKAATGVDIVCLWKLHQTL 193
Query: 209 ALFDFEDESIEDLKLLLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPF 268
FD++ E ++K LL +C +S ++K E+GR+ L+F F + K + +++Q+ F
Sbjct: 194 LCFDYDSEDSNEIKDLLLQCFMSVKHIKKEEGRRILSFFFSWNVNFIKMIHGTVKNQLQF 253
Query: 269 GRKSMLEAYGDILFRAWKAAPEDSRSGIEDGFLQDLIEGSIHA--SSGVFALYIRRVLGG 326
+S++E +I FRAWK ++ +E +QD + IH SS V++ +R +L
Sbjct: 254 FPRSLVEYISEIYFRAWKKVSGETLEILEHNCIQDFMHHGIHLPRSSSVYSK-VREMLSY 312
Query: 327 FINQRTV-DGVEKLLYRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDK 385
F Q V GVE++LYRL +P+++R+L+A NS VR NA L +D FP+ DP + E+ D
Sbjct: 313 FHKQSKVRQGVEEMLYRLYKPILWRALKARNSEVRSNAAFLFIDAFPIRDPSFNTEEMDA 372
Query: 386 LLDKQFFLLEKLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVC 445
+ KQF L LL D P VR+ + G ++ +WE+IP +I +L K+I +++ DV
Sbjct: 373 EIQKQFEELFSLLEDPHPVVRSTGILGVSQITSKYWEMIPPTVIADLLKKLIEELACDVT 432
Query: 446 N-EVRLSTLSGIIYLLDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNF 504
+ +VR S + LDN SH +L+ L P + H + DN V+VA ++LL + +
Sbjct: 433 SADVRCSVFKCLPITLDNKLSHPLLEQLLPTVKHSLHDNSEKVRVAFVDMLLKVKATKAA 492
Query: 505 QFNKVVVLDVLMSVLASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGA 564
+F K+ ++ L++ L D PV++++ LL S+ P P + C RC+TL++ +P A
Sbjct: 493 KFWKICPMEHLLARLEVDSRPVSRRIVNLLFNSFLPVNQPEDVWCERCVTLIQMNPAAAR 552
Query: 565 IFCKYAVSEGASKTHLMELV 584
F +YA E + T++ +L+
Sbjct: 553 KFYQYAY-EYTAPTNIAKLM 571
>UniRef100_UPI000033730A UPI000033730A UniRef100 entry
Length = 1181
Score = 242 bits (617), Expect = 6e-62
Identities = 147/444 (33%), Positives = 240/444 (53%), Gaps = 6/444 (1%)
Query: 132 FSDLLPGVQALHDNLI-VLEADSILSSGIETICEEWWKENLPERESLISQTLPFVLSRSL 190
++ LL LHD L E D L I +CE WWK++L E++ L +SL
Sbjct: 109 YTALLQVALRLHDVLTSTSEQDGPLQHHIHALCEAWWKKDLKEKDKFGRTAFLLSLKKSL 168
Query: 191 TLKKKV-DVHRVYMFREAFALFDFEDESIEDLKLLLNRCVISPLYLKTEDGRKFLAFLFG 249
KK V ++ RV+ D+ E +++ LL +C P +LK +DG++FL FLF
Sbjct: 169 ASKKPVTEIQRVWSLHNVLLTLDYTSEDNKEITDLLLQCFNRPAFLKNDDGKRFLVFLFS 228
Query: 250 LSDQLGKELLAMIRSQIPFGRKSMLEAYGDILFRAWKAAPEDSRSGIEDGFLQDLIEGSI 309
+ + I++Q+ F K+M +I FRAWK A D + IE +QD ++ +I
Sbjct: 229 WNVSFIWLIHGTIKNQLEFYSKTMTSHMTEIYFRAWKKATRDVQEEIESSCVQDFMQNAI 288
Query: 310 --HASSGVFALYIRRVLGGFINQRTVDGVEKLLYRLAEPVIFRSLQAANSNVRQNALHLL 367
+S V A +R+++ F +++ V+K+L L +P+++++L A N VR NA L
Sbjct: 289 FLRRTSPVHAK-VRQIVSYFHSRKGCHKVDKMLSLLYKPILWKALSAPNFEVRANATLLF 347
Query: 368 LDIFPLEDPDASKEDKDKLLDKQFFLLEKLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSP 427
+ FP+ DPD + E D+ + +Q + LL D P VR+ AV G C++L WE++P P
Sbjct: 348 TEAFPVHDPDQNNESTDEAIQRQLDTVMTLLDDPHPTVRSTAVLGVCKILGQCWELLPPP 407
Query: 428 IITKMLTKVISDMSHDVCN-EVRLSTLSGIIYLLDNPHSHEVLKVLCPRLGHLMLDNVLT 486
IIT L K++ +++ D + +VR S + +LDN SH VL+ L P L + + DN
Sbjct: 408 IITDFLKKLVMELATDCSSPDVRCSVFKCLALVLDNALSHPVLEKLLPTLKYSLHDNSEK 467
Query: 487 VQVAMAELLLHLNDVQNFQFNKVVVLDVLMSVLASDQPPVAKKLTKLLIPSYFPSIVPIE 546
V+ A ++L+ + V+ +F V +D L++ LA D V++++ LL S+FP
Sbjct: 468 VRTAFLDMLIKVKAVRAAKFWDVCNMDHLLARLAIDSQSVSRRIVDLLFKSFFPVNESER 527
Query: 547 EACNRCITLVKRSPMAGAIFCKYA 570
E C RC+TL++ +P A F ++A
Sbjct: 528 EWCCRCVTLIQMNPTAARKFYQFA 551
>UniRef100_UPI00003AB8A9 UPI00003AB8A9 UniRef100 entry
Length = 886
Score = 213 bits (541), Expect = 4e-53
Identities = 125/365 (34%), Positives = 210/365 (57%), Gaps = 6/365 (1%)
Query: 224 LLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPFGRKSMLEAYGDILFR 283
LL +C +S ++K E+GR+ L+F F + K + +++Q+ F +S++E +I FR
Sbjct: 1 LLLQCFMSVKHIKKEEGRRILSFFFSWNVNFIKMIHGTVKNQLQFFPRSLVEYISEIYFR 60
Query: 284 AWKAAPEDSRSGIEDGFLQDLIEGSIHA--SSGVFALYIRRVLGGFINQRTV-DGVEKLL 340
AWK ++ +E +QD + IH SS V++ +R +L F Q V GVE++L
Sbjct: 61 AWKKVSGETLEILEHNCIQDFMHHGIHLPRSSSVYSK-VREMLSYFHKQSKVRQGVEEML 119
Query: 341 YRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLLEKLLVD 400
YRL +P+++R+L+A NS VR NA L +D FP+ DP + E+ D + KQF L LL D
Sbjct: 120 YRLYKPILWRALKARNSEVRSNAAFLFIDAFPIRDPSFNTEEMDAEIQKQFEELFSLLED 179
Query: 401 DCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCN-EVRLSTLSGIIYL 459
P VR+ + G ++ +WE+IP +I +L K+I +++ DV + +VR S +
Sbjct: 180 PHPVVRSTGILGVSQITSKYWEMIPPTVIADLLKKLIEELACDVTSADVRCSVFKCLPIT 239
Query: 460 LDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNFQFNKVVVLDVLMSVL 519
LDN SH +L+ L P + H + DN V+VA ++LL + + +F K+ ++ L++ L
Sbjct: 240 LDNKLSHPLLEQLLPTVKHSLHDNSEKVRVAFVDMLLKVKATKAAKFWKICPMEHLLARL 299
Query: 520 ASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGAIFCKYAVSEGASKTH 579
D PV++++ LL S+ P P + C RC+TL++ +P A F +YA E + T+
Sbjct: 300 EVDSRPVSRRIVNLLFNSFLPVNQPEDVWCERCVTLIQMNPAAARKFYQYAY-EYTAPTN 358
Query: 580 LMELV 584
+ +L+
Sbjct: 359 IAKLM 363
>UniRef100_Q9NXD3 Hypothetical protein FLJ20311 [Homo sapiens]
Length = 888
Score = 206 bits (524), Expect = 4e-51
Identities = 127/367 (34%), Positives = 208/367 (56%), Gaps = 6/367 (1%)
Query: 224 LLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPFGRKSMLEAYGDILFR 283
+L C I+ Y+K E+GR+FL+ LF + K + I++Q+ +KS++ +I FR
Sbjct: 1 MLLECFININYIKKEEGRRFLSCLFNWNINFIKMIHGTIKNQLQGLQKSLMVYIAEIYFR 60
Query: 284 AWKAAPEDSRSGIEDGFLQDLIEGSIHAS--SGVFALYIRRVLGGFINQRTV-DGVEKLL 340
AWK A IE+ +QD + IH S V + +R VL F +Q+ V GVE++L
Sbjct: 61 AWKKASGKILEAIENDCIQDFMFHGIHLPRRSPVHSK-VREVLSYFHHQKKVRQGVEEML 119
Query: 341 YRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLLEKLLVD 400
YRL +P+++R L+A NS VR NA L ++ FP+ DP+ + D + KQF L LL D
Sbjct: 120 YRLYKPILWRGLKARNSEVRSNAALLFVEAFPIRDPNLHAIEMDSKIQKQFEELYSLLED 179
Query: 401 DCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCN-EVRLSTLSGIIYL 459
P VR+ + G C++ +WE++P I+ +L KV +++ D + +VR S + +
Sbjct: 180 PYPMVRSTGILGVCKITSKYWEMMPPTILIDLLKKVTGELAFDTSSADVRCSVFKCLPMI 239
Query: 460 LDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNFQFNKVVVLDVLMSVL 519
LDN SH +L+ L P L + + DN V+VA ++LL + V+ +F K+ ++ ++ L
Sbjct: 240 LDNKLSHPLLEQLLPALRYSLHDNSEKVRVAFVDMLLKIKAVRAAKFWKICPMEHILVRL 299
Query: 520 ASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGAIFCKYAVSEGASKTH 579
+D PV+++L L+ S+ P P E C RC+TLV+ + A F +YA E + T+
Sbjct: 300 ETDSRPVSRRLVSLIFNSFLPVNQPEEVWCERCVTLVQMNHAAARRFYQYA-HEHTACTN 358
Query: 580 LMELVKV 586
+ +L+ V
Sbjct: 359 IAKLIHV 365
>UniRef100_UPI000036E07C UPI000036E07C UniRef100 entry
Length = 888
Score = 206 bits (523), Expect = 5e-51
Identities = 127/367 (34%), Positives = 208/367 (56%), Gaps = 6/367 (1%)
Query: 224 LLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPFGRKSMLEAYGDILFR 283
+L C I+ Y+K E+GR+FL+ LF + K + I++Q+ +KS++ +I FR
Sbjct: 1 MLLECFININYIKKEEGRRFLSCLFNWNINFIKMIHGTIKNQLQGLQKSLMVYIAEIYFR 60
Query: 284 AWKAAPEDSRSGIEDGFLQDLIEGSIHAS--SGVFALYIRRVLGGFINQRTV-DGVEKLL 340
AWK A IE+ +QD + IH S V + +R VL F +Q+ V GVE++L
Sbjct: 61 AWKKASGKILEAIENDCIQDFMFHGIHLPRRSPVHSK-VREVLSYFHHQKKVRQGVEEML 119
Query: 341 YRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLLEKLLVD 400
YRL +P+++R L+A NS VR NA L ++ FP+ DP+ + D + KQF L LL D
Sbjct: 120 YRLYKPILWRGLKARNSEVRSNAALLFVEAFPIRDPNLHAIEMDSEIQKQFEELYSLLED 179
Query: 401 DCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCN-EVRLSTLSGIIYL 459
P VR+ + G C++ +WE++P I+ +L KV +++ D + +VR S + +
Sbjct: 180 PYPMVRSTGILGVCKITSKYWEMMPPTILIDLLKKVTGELAFDTSSADVRCSVFKCLPMI 239
Query: 460 LDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNFQFNKVVVLDVLMSVL 519
LDN SH +L+ L P L + + DN V+VA ++LL + V+ +F K+ ++ ++ L
Sbjct: 240 LDNKLSHPLLEQLLPALRYSLHDNSEKVRVAFVDMLLKIKAVRAAKFWKICPMEHILVRL 299
Query: 520 ASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGAIFCKYAVSEGASKTH 579
+D PV+++L L+ S+ P P E C RC+TLV+ + A F +YA E + T+
Sbjct: 300 ETDSRPVSRRLVSLIFNSFLPVNQPEEVWCERCVTLVQMNHAAARRFYQYA-HEHTACTN 358
Query: 580 LMELVKV 586
+ +L+ V
Sbjct: 359 IAKLIHV 365
>UniRef100_Q8WUG8 MTB protein [Homo sapiens]
Length = 935
Score = 206 bits (523), Expect = 5e-51
Identities = 127/367 (34%), Positives = 208/367 (56%), Gaps = 6/367 (1%)
Query: 224 LLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPFGRKSMLEAYGDILFR 283
+L C I+ Y+K E+GR+FL+ LF + K + I++Q+ +KS++ +I FR
Sbjct: 1 MLLECFININYIKKEEGRRFLSCLFNWNINFIKMIHGTIKNQLQGLQKSLMVYIAEIYFR 60
Query: 284 AWKAAPEDSRSGIEDGFLQDLIEGSIHAS--SGVFALYIRRVLGGFINQRTV-DGVEKLL 340
AWK A IE+ +QD + IH S V + +R VL F +Q+ V GVE++L
Sbjct: 61 AWKKASGKILEAIENDCIQDFMFHGIHLPRRSPVHSK-VREVLSYFHHQKKVRQGVEEML 119
Query: 341 YRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLLEKLLVD 400
YRL +P+++R L+A NS VR NA L ++ FP+ DP+ + D + KQF L LL D
Sbjct: 120 YRLYKPILWRGLKARNSEVRSNAALLFVEAFPIRDPNLHAIEMDSEIQKQFEELYSLLED 179
Query: 401 DCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCN-EVRLSTLSGIIYL 459
P VR+ + G C++ +WE++P I+ +L KV +++ D + +VR S + +
Sbjct: 180 PYPMVRSTGILGVCKITSKYWEMMPPTILIDLLKKVTGELAFDTSSADVRCSVFKCLPMI 239
Query: 460 LDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNFQFNKVVVLDVLMSVL 519
LDN SH +L+ L P L + + DN V+VA ++LL + V+ +F K+ ++ ++ L
Sbjct: 240 LDNKLSHPLLEQLLPALRYSLHDNSEKVRVAFVDMLLKIKAVRAAKFWKICPMEHILVRL 299
Query: 520 ASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGAIFCKYAVSEGASKTH 579
+D PV+++L L+ S+ P P E C RC+TLV+ + A F +YA E + T+
Sbjct: 300 ETDSRPVSRRLVSLIFNSFLPVNQPEEVWCERCVTLVQMNHAAARRFYQYA-HEHTACTN 358
Query: 580 LMELVKV 586
+ +L+ V
Sbjct: 359 IAKLIHV 365
>UniRef100_UPI00003AB8A7 UPI00003AB8A7 UniRef100 entry
Length = 879
Score = 202 bits (513), Expect = 7e-50
Identities = 116/339 (34%), Positives = 192/339 (56%), Gaps = 6/339 (1%)
Query: 150 EADSILSSGIETICEEWWKENLPERESLISQTLPFVLSRSLTLKKK-VDVHRVYMFREAF 208
E+++IL S I +CE WW++ L +E L +L +SL+ VD+ ++ +
Sbjct: 134 ESENILRSAIRRVCEMWWEKGLEGKEQLGKTAFIMLLKKSLSKAATGVDIVCLWKLHQTL 193
Query: 209 ALFDFEDESIEDLKLLLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPF 268
FD++ E ++K LL +C +S ++K E+GR+ L+F F + K + +++Q+ F
Sbjct: 194 LCFDYDSEDSNEIKDLLLQCFMSVKHIKKEEGRRILSFFFSWNVNFIKMIHGTVKNQLQF 253
Query: 269 GRKSMLEAYGDILFRAWKAAPEDSRSGIEDGFLQDLIEGSIHA--SSGVFALYIRRVLGG 326
+S++E +I FRAWK ++ +E +QD + IH SS V++ +R +L
Sbjct: 254 FPRSLVEYISEIYFRAWKKVSGETLEILEHNCIQDFMHHGIHLPRSSSVYSK-VREMLSY 312
Query: 327 FINQRTV-DGVEKLLYRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDK 385
F Q V GVE++LYRL +P+++R+L+A NS VR NA L +D FP+ DP + E+ D
Sbjct: 313 FHKQSKVRQGVEEMLYRLYKPILWRALKARNSEVRSNAAFLFIDAFPIRDPSFNTEEMDA 372
Query: 386 LLDKQFFLLEKLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVC 445
+ KQF L LL D P VR+ + G ++ +WE+IP +I +L K+I +++ DV
Sbjct: 373 EIQKQFEELFSLLEDPHPVVRSTGILGVSQITSKYWEMIPPTVIADLLKKLIEELACDVT 432
Query: 446 N-EVRLSTLSGIIYLLDNPHSHEVLKVLCPRLGHLMLDN 483
+ +VR S + LDN SH +L+ L P + H + DN
Sbjct: 433 SADVRCSVFKCLPITLDNKLSHPLLEQLLPTVKHSLHDN 471
>UniRef100_Q95LX7 Hypothetical protein [Macaca fascicularis]
Length = 877
Score = 192 bits (487), Expect = 7e-47
Identities = 125/400 (31%), Positives = 210/400 (52%), Gaps = 15/400 (3%)
Query: 99 SSQSDEKHKLLEKLQILAHILFLCVSHPRKVFDFSDLLPGVQALHDNLIVL-EADSILSS 157
+ + K ++ + + ++ VS + ++ LL V L+ L L E++ L S
Sbjct: 83 TEHGSKMRKSIDIIYAITSVVLASVSVINESENYEALLECVIILNGILYALPESERKLQS 142
Query: 158 GIETICEEWWKENLPERESLISQTLPFVLSRSLTLKKKVDVHRVYMFREAFALFDFEDES 217
I+ +C WW++ LP +E +L RSL K D+ R++ +A FD++ E
Sbjct: 143 SIQDLCVTWWEKGLPAKEDTGKTAFVMLLRRSLETKTGADICRLWRIHQALYCFDYDLEE 202
Query: 218 IEDLKLLLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPFGRKSMLEAY 277
++K +L C IS Y+K E+GR+FL+ LF + + + I++Q+ +KS++
Sbjct: 203 SGEIKDMLLECFISINYIKKEEGRRFLSSLFNWNINFVRMIHGTIKNQLQGLQKSLMVYI 262
Query: 278 GDILFRAWKAAPEDSRSGIEDGFLQDLIEGSIH--ASSGVFALYIRRVLGGFINQRTV-D 334
+I FRAWK A IE+ +QD + IH S V + +R VL F +Q+ V
Sbjct: 263 AEIYFRAWKKASGKILETIENDCIQDFMFHGIHLPRRSPVHS-KVREVLSYFHHQKKVRQ 321
Query: 335 GVEKLLYRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFFLL 394
GVE++LYRL +P+++ L+A NS VR NA L ++ FP+ DP+ + D + KQF L
Sbjct: 322 GVEEMLYRLYKPILWGGLKARNSEVRSNAALLFVEAFPIRDPNLHAIEMDSEIQKQFEEL 381
Query: 395 EKLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCN-EVRLSTL 453
LL D P VR+ + G C++ +WE++P I+ +L KV +++ D + +VR S
Sbjct: 382 YSLLEDPYPMVRSTGILGVCKITSKYWEMMPPTILIDLLKKVTGELAFDTSSADVRCSVF 441
Query: 454 SGIIYLLDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAE 493
+ +LDN SH +L+ ++DN L + A A+
Sbjct: 442 KCLPMILDNKLSHPLLE---------LVDNWLPTEHAQAK 472
>UniRef100_UPI00004392E3 UPI00004392E3 UniRef100 entry
Length = 987
Score = 188 bits (478), Expect = 8e-46
Identities = 134/423 (31%), Positives = 219/423 (51%), Gaps = 14/423 (3%)
Query: 155 LSSGIETICEEWWKENLPERESLISQTLPFVLSRSLTLKKKVDVHRVYMFREAFALFDFE 214
L I + E WW+ +L +E L L ++TL K V V R+ RE DF
Sbjct: 50 LQQAIHWLFECWWRRDLQGKEELGWTAFLVCLENTVTLDKPVLVVRLCSLREVLLSVDFA 109
Query: 215 DESIEDLKLLLNRCVISPLYLKTEDGRKFLAFLFGLSDQLGKELLAMIRSQIPFGRKSML 274
E + + L +C ++K E+G++FLAFLF +D + + I++Q+ F K++
Sbjct: 110 SEKGQQVIDPLLQCFFRASHIKQEEGKRFLAFLFSWNDNFIRMIHETIKNQLQFFPKTLS 169
Query: 275 EAYGDILFRAWKAAPEDSRSGIEDGFLQDLIEGS--IHASSGVFALYIRRVLGGFINQRT 332
+I FRAW+ A IE +QDL++ + +H +S V + +R++L F +
Sbjct: 170 VHVAEIYFRAWRKASGPFLEEIESACIQDLMQHALLLHRTSPVHS-KVRQILTYFHKHKF 228
Query: 333 VDGVEKLLYRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDASKEDKDKLLDKQFF 392
+GV+++L+RL +PV++++L A+ LL + A ++D L +
Sbjct: 229 REGVDEMLHRLYKPVLWKALXASQKISSPAGQAWLL------NSRAVRKDISMLYCCETC 282
Query: 393 L----LEKLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISDMSHDVCN-E 447
L LL D P VR+ AV G C VL WE+IPS +IT +L K+I +++D + +
Sbjct: 283 LCLCVCVALLDDPQPLVRSSAVLGVCSVLARCWEVIPSAVITDLLEKLILQLANDTSSPD 342
Query: 448 VRLSTLSGIIYLLDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHLNDVQNFQFN 507
VR S I +LDN SH +++ L P L + D+ V+VA +LL + + +F
Sbjct: 343 VRCSVFMCISIILDNSLSHPLMEKLLPALKSSLHDSSEKVRVAFVGMLLKIKAARAAKFW 402
Query: 508 KVVVLDVLMSVLASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKRSPMAGAIFC 567
KV L+ L++ L D PV+K++ LL S+FP P C RC+TL++ +P A F
Sbjct: 403 KVCSLEHLLARLEMDSAPVSKRIVNLLFNSFFPVNQPETVWCERCVTLIQTNPGAARKFY 462
Query: 568 KYA 570
++A
Sbjct: 463 QHA 465
>UniRef100_UPI0000364124 UPI0000364124 UniRef100 entry
Length = 805
Score = 187 bits (476), Expect = 1e-45
Identities = 113/357 (31%), Positives = 190/357 (52%), Gaps = 6/357 (1%)
Query: 132 FSDLLPGVQALHDNLI-VLEADSILSSGIETICEEWWKENLPERESLISQTLPFVLSRSL 190
+S LL LHD + E D+ L + I +CE WWK++L E+E L + L
Sbjct: 113 YSALLQVALRLHDVFVSTFERDAPLQNHIHNLCEAWWKKDLKEKEKFGRTAFFVSLKKIL 172
Query: 191 TLKKKV-DVHRVYMFREAFALFDFEDESIEDLKLLLNRCVISPLYLKTEDGRKFLAFLFG 249
KK V ++ RV+ D+ E ++ LL C P+++K ++G++FL FLF
Sbjct: 173 ASKKPVTEIQRVWSLHNVLLTLDYTAEDNNEMIDLLLECFHRPMFIKNDNGKRFLVFLFS 232
Query: 250 LSDQLGKELLAMIRSQIPFGRKSMLEAYGDILFRAWKAAPEDSRSGIEDGFLQDLIEGSI 309
+ L I++Q+ F K+M ++ FRAWK A + + IE +QD ++ +I
Sbjct: 233 WNINFIWVLHGTIKNQLEFYSKTMTAHIAEVYFRAWKKATGEVQEQIESSCIQDFMQNAI 292
Query: 310 --HASSGVFALYIRRVLGGFINQRTVDGVEKLLYRLAEPVIFRSLQAANSNVRQNALHLL 367
+S V A +R+V+ F +++ V V+++L RL +P+++++L N VR NA +
Sbjct: 293 LLRRTSPVHAK-VRQVVSYFHSKKDVSKVDQMLCRLYKPILWKALSVPNFEVRANAALVF 351
Query: 368 LDIFPLEDPDASKEDKDKLLDKQFFLLEKLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSP 427
+ FP+ DPD + + D+ + Q + LL D P VR+ A+ G C++L WE++P
Sbjct: 352 TEAFPVHDPDQNSKSTDEAIQNQLDTVMTLLNDPHPCVRSTAILGVCKILAKCWELLPPS 411
Query: 428 IITKMLTKVISDMSHDVCN-EVRLSTLSGIIYLLDNPHSHEVLKVLCPRLGHLMLDN 483
IIT L K++ +M+ D + +VR S + +LDN SH +L+ P L + + DN
Sbjct: 412 IITDFLKKLVMEMATDCSSPDVRCSVFKCLCIVLDNVFSHPLLEKFLPALKNSLHDN 468
>UniRef100_Q6SLL0 More than blood [Mus musculus]
Length = 849
Score = 169 bits (427), Expect = 6e-40
Identities = 93/268 (34%), Positives = 155/268 (57%), Gaps = 2/268 (0%)
Query: 320 IRRVLGGFINQRTVDGVEKLLYRLAEPVIFRSLQAANSNVRQNALHLLLDIFPLEDPDAS 379
+R VL F Q+ GVE++LYRL +P+++R L+A NS VR NA L ++ FP+ DP+ +
Sbjct: 17 VREVLSYFHQQKVRQGVEEMLYRLYKPILWRGLKARNSEVRSNAALLFVEAFPIRDPNFT 76
Query: 380 KEDKDKLLDKQFFLLEKLLVDDCPEVRTIAVEGSCRVLHLFWEIIPSPIITKMLTKVISD 439
+ D + KQF L L+ D P VR+ + G C++ +WE++P I+ L KV +
Sbjct: 77 ATEMDNEIQKQFEELYNLIEDPYPRVRSTGILGVCKISSKYWEMMPPNILVDFLKKVTGE 136
Query: 440 MSHDVCN-EVRLSTLSGIIYLLDNPHSHEVLKVLCPRLGHLMLDNVLTVQVAMAELLLHL 498
++ D+ + +VR S + +LDN SH +L+ L P L + + DN V+VA +LLL +
Sbjct: 137 LAFDISSADVRCSVFKCLPIILDNKLSHPLLEQLLPTLRYSLHDNSEKVRVAFVDLLLKI 196
Query: 499 NDVQNFQFNKVVVLDVLMSVLASDQPPVAKKLTKLLIPSYFPSIVPIEEACNRCITLVKR 558
V+ +F K+ ++ ++ L D PV+++L L+ S+ P P E C RC+TL++
Sbjct: 197 KAVRAAKFWKICPMEDILVRLEMDSRPVSRRLVSLIFNSFLPVNQPEEVWCERCVTLIQM 256
Query: 559 SPMAGAIFCKYAVSEGASKTHLMELVKV 586
+ A F +YA E + T++ +L+ V
Sbjct: 257 NRAAARRFYQYA-HEHTASTNIAKLIHV 283
>UniRef100_Q8GXI7 Hypothetical protein At1g64960/F13O11_26 [Arabidopsis thaliana]
Length = 205
Score = 159 bits (402), Expect = 5e-37
Identities = 84/205 (40%), Positives = 123/205 (59%), Gaps = 7/205 (3%)
Query: 1056 ESCLGSAYASRLVAAAKPWLPDVVLALGSESVLQHTESGSEHLFASEQMKLQFPKWPFIV 1115
E LGSAYASR+V+A PW+PD+VLALG + +E S + + +KL FP W
Sbjct: 3 EKSLGSAYASRIVSALNPWIPDLVLALGPCFINNDSEESS-YTSSFNHIKLCFPSWLLTC 61
Query: 1116 AKTVLSAVNEGEGDGECSQADKFSTFNKLTAMLIILLKKNKSIMDAVGDIFLVCSLIGLE 1175
AK L +N+ + ++ F KL + L+K N ++DA+G + L+C + +E
Sbjct: 62 AKIELHEINKED----VTETSGFLALKKLRNTIFTLVKGNTKVLDAIGYVLLLCLAVCIE 117
Query: 1176 QKDFELAAGLLQFVCSKLFNRDDRDWGDL--MLSSLEEIYPKIERQITEASDNDELEKLM 1233
++D+ A GLL VC KL +DR+W +L ML IYP IER+I E D DE++ L
Sbjct: 118 KRDYSTALGLLHLVCVKLVGSEDREWKELDTMLVLPPRIYPIIEREIGEGRDEDEVKTLE 177
Query: 1234 HAKELIEPLWTYHLYETGKVNMTDE 1258
A+EL++P+W YH+YETG+ +M DE
Sbjct: 178 AARELLQPVWMYHVYETGRFHMMDE 202
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.134 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,938,648,391
Number of Sequences: 2790947
Number of extensions: 79056778
Number of successful extensions: 237113
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 41
Number of HSP's successfully gapped in prelim test: 45
Number of HSP's that attempted gapping in prelim test: 236798
Number of HSP's gapped (non-prelim): 218
length of query: 1260
length of database: 848,049,833
effective HSP length: 139
effective length of query: 1121
effective length of database: 460,108,200
effective search space: 515781292200
effective search space used: 515781292200
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 81 (35.8 bits)
Medicago: description of AC146856.10


