

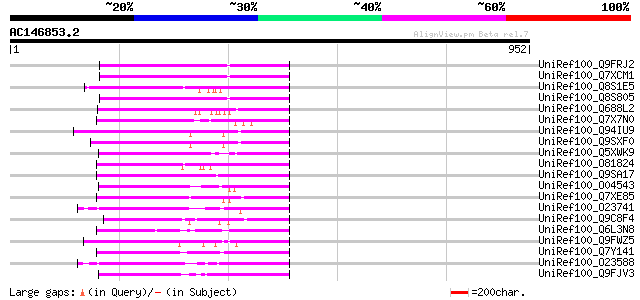
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146853.2 - phase: 0 /pseudo
(952 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FRJ2 Putative copia-like retrotransposon polyprotein... 275 4e-72
UniRef100_Q7XCM1 Putative gag-pol polyprotein [Oryza sativa] 275 4e-72
UniRef100_Q8S1E5 Putative gag/pol polyprotein [Oryza sativa] 274 8e-72
UniRef100_Q8S805 Putative copia-type polyprotein [Oryza sativa] 271 5e-71
UniRef100_Q688L2 Putative polyprotein [Oryza sativa] 255 5e-66
UniRef100_Q7X7N0 OSJNBa0035I04.5 protein [Oryza sativa] 242 3e-62
UniRef100_Q94IU9 Copia-like polyprotein [Arabidopsis thaliana] 211 1e-52
UniRef100_Q9SXF0 T3P18.3 [Arabidopsis thaliana] 207 9e-52
UniRef100_Q5XWK9 Gag-pol polyprotein-like [Solanum tuberosum] 207 2e-51
UniRef100_O81824 Hypothetical protein AT4g27210 [Arabidopsis tha... 205 5e-51
UniRef100_Q9SA17 F28K20.17 protein [Arabidopsis thaliana] 204 1e-50
UniRef100_O04543 F20P5.25 protein [Arabidopsis thaliana] 195 5e-48
UniRef100_Q7XE85 Putative pol polyprotein [Oryza sativa] 194 1e-47
UniRef100_O23741 SLG-Sc and SLA-Sc genes and Melmoth retrotransp... 194 1e-47
UniRef100_Q9C8F4 Ty1/copia-element polyprotein [Arabidopsis thal... 192 5e-47
UniRef100_Q6L3N8 Putative gag-pol polyprotein [Solanum demissum] 190 2e-46
UniRef100_Q9FWZ5 Putative retroelement polyprotein [Arabidopsis ... 188 6e-46
UniRef100_Q7Y141 Putative polyprotein [Oryza sativa] 188 8e-46
UniRef100_O23588 Retrotransposon like protein [Arabidopsis thali... 187 1e-45
UniRef100_Q9FJV3 Retroelement pol polyprotein-like [Arabidopsis ... 184 1e-44
>UniRef100_Q9FRJ2 Putative copia-like retrotransposon polyprotein [Oryza sativa]
Length = 1302
Score = 275 bits (704), Expect = 4e-72
Identities = 140/351 (39%), Positives = 213/351 (59%), Gaps = 7/351 (1%)
Query: 165 QKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYL 224
+ + +G V RLSCPY+S QNGK+ER +R+ N+ +RT+L+H++ P SFW AL+ AT+L
Sbjct: 536 RSLLSLHGAVLRLSCPYSSQQNGKAERILRTINDYVRTMLVHSAAPLSFWAEALQTATHL 595
Query: 225 MNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYP 284
+N P + +P ++L P+Y+HLRVFGCLC+P ++ +KL PRS CVF+GYP
Sbjct: 596 INRRPCRATGSLTPYQLLLGAPPTYDHLRVFGCLCYPNTIATAPHKLSPRSLACVFIGYP 655
Query: 285 SNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQHLMDQ 344
++HRGY+C ++S ++ RHV F E++FP+ P+P+ D+ ++ Q
Sbjct: 656 ADHRGYRCYDMVSRRVFTSRHVTFVEDVFPFRDAPSPRPSAPPPPDHGDDTIVLLPAPAQ 715
Query: 345 TQTGPPIPQPAHQPNHITSPCPNSPNITSPPQ--SNPTSPIQQNLPPIFQPTLQANTKPI 402
P PAH SP ++P+ +P + P SP + + P P A T
Sbjct: 716 HVVTPVGTAPAHDAASPPSPASSTPSSAAPAHDVAPPPSP-ETSSPASASPPRHAMT--- 771
Query: 403 TRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVP 462
TR++ GI KPN + Y + T SP P + +AL+DPNW+ AM E+++L+ N+TW LVP
Sbjct: 772 TRARAGISKPNPR-YAMTATSTLSPTPSSVRAALRDPNWRAAMQAEFDALLANRTWTLVP 830
Query: 463 RPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
RPP +I W+F+ K +DG+ +++KAR V G Q+ G+D GETFSP+
Sbjct: 831 RPPGARIITGKWVFKTKLHADGSLDKYKARWVVRGFNQRPGVDFGETFSPV 881
>UniRef100_Q7XCM1 Putative gag-pol polyprotein [Oryza sativa]
Length = 1417
Score = 275 bits (704), Expect = 4e-72
Identities = 140/351 (39%), Positives = 213/351 (59%), Gaps = 7/351 (1%)
Query: 165 QKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYL 224
+ + +G V RLSCPY+S QNGK+ER +R+ N+ +RT+L+H++ P SFW AL+ AT+L
Sbjct: 634 RSLLSLHGAVLRLSCPYSSQQNGKAERILRTINDYVRTMLVHSAAPLSFWAEALQTATHL 693
Query: 225 MNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYP 284
+N P + +P ++L P+Y+HLRVFGCLC+P ++ +KL PRS CVF+GYP
Sbjct: 694 INRRPCRATGSLTPYQLLLGAPPTYDHLRVFGCLCYPNTIATAPHKLSPRSLACVFIGYP 753
Query: 285 SNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQHLMDQ 344
++HRGY+C ++S ++ RHV F E++FP+ P+P+ D+ ++ Q
Sbjct: 754 ADHRGYRCYDMVSRRVFTSRHVTFVEDVFPFRDAPSPRPSAPPPPDHGDDTIVLLPAPAQ 813
Query: 345 TQTGPPIPQPAHQPNHITSPCPNSPNITSPPQ--SNPTSPIQQNLPPIFQPTLQANTKPI 402
P PAH SP ++P+ +P + P SP + + P P A T
Sbjct: 814 HVVTPVGTAPAHDAASPPSPASSTPSSAAPAHDVAPPPSP-ETSSPASASPPRHAMT--- 869
Query: 403 TRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVP 462
TR++ GI KPN + Y + T SP P + +AL+DPNW+ AM E+++L+ N+TW LVP
Sbjct: 870 TRARAGISKPNPR-YAMTATSTLSPTPSSVRAALRDPNWRAAMQAEFDALLANRTWTLVP 928
Query: 463 RPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
RPP +I W+F+ K +DG+ +++KAR V G Q+ G+D GETFSP+
Sbjct: 929 RPPGARIITGKWVFKTKLHADGSLDKYKARWVVRGFNQRPGVDFGETFSPV 979
>UniRef100_Q8S1E5 Putative gag/pol polyprotein [Oryza sativa]
Length = 1090
Score = 274 bits (701), Expect = 8e-72
Identities = 170/419 (40%), Positives = 230/419 (54%), Gaps = 45/419 (10%)
Query: 138 EHTSTHNFKEKLKQFNVTMAVN-VNSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRST 196
E+ ST F LK F VN+ + G RLSCPYTS QNGK+ER +R+
Sbjct: 232 EYVSTQ-FGLPLKSFQADNGREFVNTAITTFLASRGTQLRLSCPYTSPQNGKAERMLRTI 290
Query: 197 NNIIRTLLIHASLPSSFWHHALEMATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFG 256
NN IRTLLI AS+P S+W AL ATYL+N P +I+ P ++L+ P ++HLRVFG
Sbjct: 291 NNSIRTLLIQASMPPSYWAEALATATYLLNRRPSSSIHQSLPFQLLHRTIPDFSHLRVFG 350
Query: 257 CLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHRGYKCLGLLSNKIIICRHVLFNENIFPYA 316
CLC+P ++ +KL PRST CVFLGYP++H+GY+CL L +++III RHV+F+E+ FP+A
Sbjct: 351 CLCYPNLSATTPHKLSPRSTACVFLGYPTSHKGYRCLDLSTHRIIISRHVVFDESQFPFA 410
Query: 317 KLHIPQPNTYTFLDNELSPYIIQHL-MDQTQ----------TGPPIPQPAHQPNHIT--- 362
P +++ FL LSP L ++Q + P +P P+ + + T
Sbjct: 411 ATP-PAASSFDFLLQGLSPADAPSLEVEQPRPLTVAPSTEVEQPYLPLPSRRLSAGTVTV 469
Query: 363 ---SPCPNSPNI------TSPP-----QSNPTSPIQQ--------NLPPIFQPTLQ---A 397
+P +P + +PP S SP + +PP + A
Sbjct: 470 ASEAPSAGAPLVGTSSADATPPGSATRASTIVSPFRHVYTRRPVTTVPPSSSTAVTNAVA 529
Query: 398 NTKP---ITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIK 454
+P +TRSQ G +P + T SP+P N SAL DPNW+ AM DEY L+
Sbjct: 530 APQPHSMVTRSQSGSLRPVDRLTYTATQAAASPVPANYHSALADPNWRAAMADEYKELVD 589
Query: 455 NKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
N TW LV RPP N+ WIF+HK SDG+ R+KAR V G QQ GID ETFSP+
Sbjct: 590 NGTWRLVSRPPRANIATGKWIFKHKFHSDGSLARYKARWVVRGYSQQHGIDYDETFSPV 648
>UniRef100_Q8S805 Putative copia-type polyprotein [Oryza sativa]
Length = 1803
Score = 271 bits (694), Expect = 5e-71
Identities = 139/351 (39%), Positives = 210/351 (59%), Gaps = 7/351 (1%)
Query: 165 QKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYL 224
+ + +G V RLSCPY+S QNGK+ER +R+ N+ +RT+L+H++ P SFW AL+ A +L
Sbjct: 634 RSLLSLHGAVLRLSCPYSSQQNGKAERILRTINDCVRTMLVHSAAPLSFWAEALQTAMHL 693
Query: 225 MNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYP 284
+N P + P ++L P+Y+HLRVFGCLC+P ++ +KL PRS CVF+GYP
Sbjct: 694 INRRPCRATGSLKPYQLLLGAPPTYDHLRVFGCLCYPNTIATAPHKLSPRSLACVFIGYP 753
Query: 285 SNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQHLMDQ 344
++HRGY+C ++S ++ RHV F E++FP+ P+P+ D+ ++ Q
Sbjct: 754 ADHRGYRCYDMVSRRVFTSRHVTFVEDVFPFRDAPSPRPSAPPPPDHGDDTIVLLPAPAQ 813
Query: 345 TQTGPPIPQPAHQPNHITSPCPNSPNITSPPQ--SNPTSPIQQNLPPIFQPTLQANTKPI 402
P PAH SP ++P+ +P + P SP + + P P A T
Sbjct: 814 HVVTPVGTAPAHDAASPPSPASSTPSSAAPAHDVAPPPSP-ETSSPASASPPRHAMT--- 869
Query: 403 TRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVP 462
TR++ GI KPN + Y + T SP P + AL+DPNW+ AM E+++L+ N+TW LVP
Sbjct: 870 TRARAGISKPNPR-YAMTATSTLSPTPSSVRVALRDPNWRAAMQAEFDALLANRTWTLVP 928
Query: 463 RPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
RPP +I W+F+ K +DG+ +++KAR V G Q+ G+D GETFSP+
Sbjct: 929 RPPGARIITGKWVFKTKLHADGSLDKYKARWVVRGFNQRPGVDFGETFSPV 979
>UniRef100_Q688L2 Putative polyprotein [Oryza sativa]
Length = 1679
Score = 255 bits (651), Expect = 5e-66
Identities = 164/436 (37%), Positives = 224/436 (50%), Gaps = 86/436 (19%)
Query: 161 NSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEM 220
NS + NG+ R+SCP+TS QNGK+ER +RS NNI+R++L A LP SFW AL
Sbjct: 827 NSAARTFFLTNGVHLRMSCPHTSPQNGKAERILRSLNNIVRSMLFQAKLPGSFWVEALHT 886
Query: 221 ATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVF 280
AT+L+N P KT++ +P LY PSY+HLRVFGC C+P ++ +KL PRST CVF
Sbjct: 887 ATHLINRHPTKTLDRHTPHFALYGTHPSYSHLRVFGCKCYPNLSATTPHKLAPRSTMCVF 946
Query: 281 LGYPSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLH--IPQPNTYTFLDNELSPYII 338
LGYP H+GY+C LSN++II RHV+F+E+ FP+ +L + FL++ +P
Sbjct: 947 LGYPLYHKGYRCFDPLSNRVIISRHVVFDEHSFPFTELTNGVSNATDLDFLEDFTAPAQA 1006
Query: 339 Q-------HLMDQTQT----------------------------------GPPIPQPAHQ 357
+ TQT GPP P PA
Sbjct: 1007 PIGATRRPAVAPTTQTASSPMVHGLERPPPCSPTRPVSTPGGPSSPDSRLGPPSPTPALI 1066
Query: 358 PNHITSPCPNS----PNITSPPQSN--------PTSP-----IQQNLPPIFQ-------- 392
TSP P S P+ ++ P S+ PT P +LPP
Sbjct: 1067 GPASTSPGPPSAGPAPSASTCPASSTWETVARPPTPPGLPRLDGPHLPPAPHVPRRLRSV 1126
Query: 393 -------PTLQANTKPI-------TRSQHGIFKPNQKYYGLHTHVTK-SPLPRNPVSALK 437
P P+ TR++ G KP + L+ H S +P+ +AL
Sbjct: 1127 RATGAPTPLSGLEISPVVNDHVMTTRAKSGHHKPVHR---LNLHAAPLSLVPKTYRAALA 1183
Query: 438 DPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDG 497
DP W+ AM +EYN+L+ N+TWDLVPRP VNV+ WIF+HK +DG+ +R+KAR V G
Sbjct: 1184 DPLWRAAMEEEYNALLANRTWDLVPRPAGVNVVTGKWIFKHKFHADGSLDRYKARWVLRG 1243
Query: 498 AGQQVGIDCGETFSPM 513
Q+ G+D ETFSP+
Sbjct: 1244 FTQRPGVDFDETFSPV 1259
>UniRef100_Q7X7N0 OSJNBa0035I04.5 protein [Oryza sativa]
Length = 1246
Score = 242 bits (618), Expect = 3e-62
Identities = 149/378 (39%), Positives = 199/378 (52%), Gaps = 36/378 (9%)
Query: 160 VNSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALE 219
VN N G + RLSCPYTS QNGK+ER IR+ NN IRTLL+ AS+P S+W L
Sbjct: 451 VNHNTTSFLAGRGSLLRLSCPYTSPQNGKAERMIRTLNNSIRTLLLQASMPPSYWAEGLA 510
Query: 220 MATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCV 279
ATYL+N P ++N P ++L+ K P+Y+ LRVFGCLC+P ++ +KL P S CV
Sbjct: 511 TATYLLNRRPSSSVNNSIPFQLLHRKIPNYSMLRVFGCLCYPNLSATAAHKLAPYSAACV 570
Query: 280 FLGYPSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQ 339
FLGYPS+H+GY CL + + +III HV+F+E FP++ + + L + +P +I
Sbjct: 571 FLGYPSSHKGYCCLNISTRRIIISCHVIFDETQFPFSGDPVDASSLDFLLQDAPAPSVI- 629
Query: 340 HLMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTL---Q 396
P QP+ +P P N+ + S ++LP QP Q
Sbjct: 630 ---------APSLAGVEQPHLPHAPFP--VNVEQRLPTGAPSTKDEHLPYYVQPAAHCGQ 678
Query: 397 ANTKPITRSQH-GIFKPN----QKYYGLHTHVTKS--------PLPRNPVSALKDPNWK- 442
+ K T H IF Y +H T S PLP + P+ +
Sbjct: 679 DDGKFHTAGCHVSIFLKQGTIVTSYPAVHVFFTTSRHGGAIPVPLPTTTGADDSAPSHRC 738
Query: 443 -------MAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVG 495
AM +E+ +LI N TW LVPRPP NV+ WIF+HK SDG+ RHKAR V
Sbjct: 739 GCTHTSPAAMAEEFKALIDNGTWRLVPRPPGANVVTGKWIFKHKFHSDGSLARHKARWVV 798
Query: 496 DGAGQQVGIDCGETFSPM 513
G QQ GID ETFSP+
Sbjct: 799 HGYSQQHGIDYDETFSPV 816
>UniRef100_Q94IU9 Copia-like polyprotein [Arabidopsis thaliana]
Length = 1466
Score = 211 bits (536), Expect = 1e-52
Identities = 139/420 (33%), Positives = 205/420 (48%), Gaps = 27/420 (6%)
Query: 118 FCGLFQCHINPKFIQYLKN*EHTSTHNFKEKLKQFNVTMAVNVNSN-FQKMCEANGIVFR 176
F F + KFI + + K+K+F SN ++ +GI R
Sbjct: 546 FSWFFPLRMKSKFISVFIAYQKLVENQLGTKIKEFQSDGGGEFTSNKLKEHFREHGIHHR 605
Query: 177 LSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHKTINFE 236
+SCPYT QNG +ERK R + ++L H+ P FW A A YL N+LP +
Sbjct: 606 ISCPYTPQQNGVAERKHRHLVELGLSMLYHSHTPLKFWVEAFFTANYLSNLLPSSVLKEI 665
Query: 237 SPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHRGYKCLGLL 296
SP + L+ + Y LRVFG C+P K PRS +CVFLGY + ++GY+CL
Sbjct: 666 SPYETLFQQKVDYTPLRVFGTACYPCLRPLAKNKFDPRSLQCVFLGYHNQYKGYRCLYPP 725
Query: 297 SNKIIICRHVLFNENIFPY-AKLHIPQPNTYTFL-----DNELSPYII--QHLMDQTQTG 348
+ K+ I RHV+F+E FP+ K H P T L +L+P + L +
Sbjct: 726 TGKVYISRHVIFDEAQFPFKEKYHSLVPKYQTTLLQAWQHTDLTPPSVPSSQLQPLARQM 785
Query: 349 PPIPQPAHQP--NHITSPCPN-SPNITSPPQSNPTSPIQQNLPPIF----------QPTL 395
P+ +QP N+ T N + +S ++ + P+ Q +L
Sbjct: 786 TPMATSENQPMMNYETEEAVNVNMETSSDEETESNDEFDHEVAPVLNDQNEDNALGQGSL 845
Query: 396 QANTKPITRSQHGIFKPNQKYYGLHTHVTKSPL--PRNPVSALKDPNWKMAMNDEYNSLI 453
+ ITRS+ GI KPN +Y + V+KS P+ +A+K P+W A+ DE + +
Sbjct: 846 ENLHPMITRSKDGIQKPNPRYALI---VSKSSFDEPKTITTAMKHPSWNAAVMDEIDRIH 902
Query: 454 KNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
TW LVP D+N++ S W+F+ K K DGT ++ KARLV G Q+ G+D ETFSP+
Sbjct: 903 MLNTWSLVPATEDMNILTSKWVFKTKLKPDGTIDKLKARLVAKGFDQEEGVDYLETFSPV 962
>UniRef100_Q9SXF0 T3P18.3 [Arabidopsis thaliana]
Length = 1309
Score = 207 bits (528), Expect = 9e-52
Identities = 134/390 (34%), Positives = 196/390 (49%), Gaps = 27/390 (6%)
Query: 148 KLKQFNVTMAVNVNSN-FQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIH 206
K+K+F SN ++ +GI R+SCPYT QNG +ERK R + ++L H
Sbjct: 419 KIKEFQSDGGGEFTSNKLKEHFREHGIHHRISCPYTPQQNGVAERKHRHLVELGLSMLYH 478
Query: 207 ASLPSSFWHHALEMATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSS 266
+ P FW A A YL N+LP + SP + L+ + Y LRVFG C+P
Sbjct: 479 SHTPLKFWVEAFFTANYLSNLLPSSVLKEISPYETLFQQKVDYTPLRVFGTACYPCLRPL 538
Query: 267 KIYKLQPRSTKCVFLGYPSNHRGYKCLGLLSNKIIICRHVLFNENIFPY-AKLHIPQPNT 325
K PRS +CVFLGY + ++GY+CL + K+ I RHV+F+E FP+ K H P
Sbjct: 539 AKNKFDPRSLQCVFLGYHNQYKGYRCLYPPTGKVYISRHVIFDEAQFPFKEKYHSLVPKY 598
Query: 326 YTFL-----DNELSPYII--QHLMDQTQTGPPIPQPAHQP--NHITSPCPN-SPNITSPP 375
T L +L+P + L + P+ +QP N+ T N + +S
Sbjct: 599 QTTLLQAWQHTDLTPPSVPSSQLQPLARQVTPMATSENQPMMNYETEEAVNVNMETSSDE 658
Query: 376 QSNPTSPIQQNLPPIF----------QPTLQANTKPITRSQHGIFKPNQKYYGLHTHVTK 425
++ + P+ Q +L+ ITRS+ GI KPN +Y + V+K
Sbjct: 659 ETESNDEFDHEVAPVLNDQNEDNALGQGSLENLHPMITRSKDGIQKPNPRYALI---VSK 715
Query: 426 SPL--PRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSD 483
S P+ +A+K P W A+ DE + + TW LVP D+N++ S W+F+ K K D
Sbjct: 716 SSFDEPKTITTAMKHPGWNAAVMDEIDRIHMLNTWSLVPATEDMNILTSKWVFKTKLKPD 775
Query: 484 GTFERHKARLVGDGAGQQVGIDCGETFSPM 513
GT ++ KARLV G Q+ G+D ETFSP+
Sbjct: 776 GTIDKLKARLVAKGFDQEEGVDYLETFSPV 805
>UniRef100_Q5XWK9 Gag-pol polyprotein-like [Solanum tuberosum]
Length = 1212
Score = 207 bits (526), Expect = 2e-51
Identities = 131/353 (37%), Positives = 183/353 (51%), Gaps = 27/353 (7%)
Query: 164 FQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATY 223
F+K GIV + SCPYT QNG +ERK R ++ RTLLI +S+PS +W AL A Y
Sbjct: 584 FKKFLLDKGIVSQHSCPYTPQQNGVAERKNRHLLDVTRTLLIESSVPSKYWVEALSTAVY 643
Query: 224 LMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGY 283
L+N LP K +N ESP LYH++P+Y+ FGC+CF P S+ KL +STKC F+GY
Sbjct: 644 LINRLPSKVLNLESPYFRLYHQNPNYSDFHTFGCVCFVHLPPSQCNKLSVQSTKCAFMGY 703
Query: 284 PSNHRGYKCLGLLSNKIIICRHVLFNEN--IFP-YAKLHIPQPNTYTFLDNELSPYIIQH 340
++ +G+ C S+K I R+V+F EN FP L P TF D S +
Sbjct: 704 STSQKGFICYDPCSHKFRISRNVVFFENQYFFPTIVDLSSVSPLLPTFEDLSSSFKRFKP 763
Query: 341 LMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANTK 400
+ P +P P P T+P S N S+ + P++
Sbjct: 764 GFVYERRRPTLPYPNTDPPPETAPQLESEN------SSRSGPLE---------------- 801
Query: 401 PITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDL 460
P RS PN +YG + ++ +P A K W+ AM +E +L +N TWD+
Sbjct: 802 PTRRSTRVSRTPN--WYGFSSTLSNISVPSCYSQASKHECWQKAMEEELLALKENDTWDI 859
Query: 461 VPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
V P +V I W++ K SDGT +R+KARLV G Q+ G+D ETF+P+
Sbjct: 860 VSCPSNVRPIGCKWVYSIKLHSDGTLDRYKARLVVLGNRQEYGVDYEETFAPV 912
>UniRef100_O81824 Hypothetical protein AT4g27210 [Arabidopsis thaliana]
Length = 1318
Score = 205 bits (522), Expect = 5e-51
Identities = 135/399 (33%), Positives = 197/399 (48%), Gaps = 47/399 (11%)
Query: 160 VNSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALE 219
V+ F + +++GI +LSCP+T QNG +ERK R + ++L + +P FW A
Sbjct: 402 VSHKFLQHLQSHGIQQQLSCPHTPQQNGLAERKHRHLVELGLSMLFQSHVPHKFWVEAFF 461
Query: 220 MATYLMNILPHKTINFE-SPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKC 278
A +L+N+LP + SP + LY K P Y LR FG CFP K P S KC
Sbjct: 462 TANFLINLLPTSALKESISPYEKLYDKKPDYTSLRSFGSACFPTLRDYAENKFNPCSLKC 521
Query: 279 VFLGYPSNHRGYKCLGLLSNKIIICRHVLFNENIFP----YAKLHIPQPNT---YTFLDN 331
VFLGY ++GY+CL + ++ I RHV+F+E+++P Y LH PQP T +L +
Sbjct: 522 VFLGYNEKYKGYRCLYPPTGRLYISRHVIFDESVYPFSHTYKHLH-PQPRTPLLAAWLRS 580
Query: 332 ELSPYIIQHLMDQTQTG-------PPIPQP--------------AHQPNHITSPCPN--- 367
SP +++ PP+PQ +H N T P+
Sbjct: 581 SDSPAPSTSTSPSSRSPLFTSADFPPLPQRKTPLLPTLVPISSVSHASNITTQQSPDFDS 640
Query: 368 ------------SPNITSPPQSNPTSPIQQNLPPIFQPTLQANTKP-ITRSQHGIFKPNQ 414
+ +S S+ IQQ + Q N P +TR++ GI KPN
Sbjct: 641 ERTTDFDSASIGDSSHSSQAGSDSEETIQQASVNVHQTHASTNVHPMVTRAKVGISKPNP 700
Query: 415 KYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMW 474
+Y L +H P P+ +ALK P W AM +E + + +TW LVP D++V+ S W
Sbjct: 701 RYVFL-SHKVSYPEPKTVTAALKHPGWTGAMTEEIGNCSETQTWSLVPYKSDMHVLGSKW 759
Query: 475 IFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
+FR K +DGT + KAR+V G Q+ GID ET+SP+
Sbjct: 760 VFRTKLHADGTLNKLKARIVAKGFLQEEGIDYLETYSPV 798
>UniRef100_Q9SA17 F28K20.17 protein [Arabidopsis thaliana]
Length = 1415
Score = 204 bits (519), Expect = 1e-50
Identities = 122/363 (33%), Positives = 189/363 (51%), Gaps = 14/363 (3%)
Query: 160 VNSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALE 219
V++ + +GI R+SCPYT QNG +ERK R + ++L H+ P FW +
Sbjct: 587 VSNKLKTHLSEHGIHHRISCPYTPQQNGLAERKHRHLVELGLSMLFHSHTPQKFWVESFF 646
Query: 220 MATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCV 279
A Y++N LP + SP + L+ + P Y+ LRVFG C+P K PRS +CV
Sbjct: 647 TANYIINRLPSSVLKNLSPYEALFGEKPDYSSLRVFGSACYPCLRPLAQNKFDPRSLQCV 706
Query: 280 FLGYPSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLH---IPQ---PNTYTFLDNEL 333
FLGY S ++GY+C + K+ I R+V+FNE+ P+ + + +PQ P + N++
Sbjct: 707 FLGYNSQYKGYRCFYPPTGKVYISRNVIFNESELPFKEKYQSLVPQYSTPLLQAWQHNKI 766
Query: 334 SPYIIQHLMDQTQTGP---PIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPI 390
S + Q + P + +T P P S N S + N P+ + +
Sbjct: 767 SEISVPAAPVQLFSKPIDLNTYAGSQVTEQLTDPEPTSNNEGSDEEVN---PVAEEIAAN 823
Query: 391 FQPTLQANTKPITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYN 450
+ + ++ TRS+ GI KPN + Y L T + P+ SA+K P W A+++E N
Sbjct: 824 QEQVINSHAM-TTRSKAGIQKPNTR-YALITSRMNTAEPKTLASAMKHPGWNEAVHEEIN 881
Query: 451 SLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETF 510
+ TW LVP D+N++ S W+F+ K DG+ ++ KARLV G Q+ G+D ETF
Sbjct: 882 RVHMLHTWSLVPPTDDMNILSSKWVFKTKLHPDGSIDKLKARLVAKGFDQEEGVDYLETF 941
Query: 511 SPM 513
SP+
Sbjct: 942 SPV 944
>UniRef100_O04543 F20P5.25 protein [Arabidopsis thaliana]
Length = 1315
Score = 195 bits (496), Expect = 5e-48
Identities = 121/368 (32%), Positives = 177/368 (47%), Gaps = 37/368 (10%)
Query: 163 NFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMAT 222
NF + + GIV SCP T QN ERK + N+ R+L + +P S+W + A
Sbjct: 538 NFTQFYHSKGIVPYHSCPETPQQNSVVERKHQHILNVARSLFFQSHIPISYWGDCILTAV 597
Query: 223 YLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLG 282
YL+N LP + + P +VL P+Y+H++VFGCLC+ +K PR+ C F+G
Sbjct: 598 YLINRLPAPILEDKCPFEVLTKTVPTYDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIG 657
Query: 283 YPSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQHLM 342
YPS +GYK L L ++ II+ RHV+F+E +FP+ + Q F D
Sbjct: 658 YPSGFKGYKLLDLETHSIIVSRHVVFHEELFPFLGSDLSQEEQNFFPDLN---------- 707
Query: 343 DQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANTKP- 401
P P Q + +P +S ++ P +NPT+ + + P Q + + KP
Sbjct: 708 -------PTPPMQRQSSDHVNPSDSSSSVEILPSANPTNNVPE---PSVQTSHRKAKKPA 757
Query: 402 ----------ITRSQHGIFK------PNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAM 445
++ + H I K N Y + K+ P N A K W+ AM
Sbjct: 758 YLQDYYCHSVVSSTPHEIRKFLSYDRINDPYLTFLACLDKTKEPSNYTEAEKLQVWRDAM 817
Query: 446 NDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGID 505
E++ L TW++ P D I WIF+ K SDG+ ER+KARLV G Q+ GID
Sbjct: 818 GAEFDFLEGTHTWEVCSLPADKRCIGCRWIFKIKYNSDGSVERYKARLVAQGYTQKEGID 877
Query: 506 CGETFSPM 513
ETFSP+
Sbjct: 878 YNETFSPV 885
>UniRef100_Q7XE85 Putative pol polyprotein [Oryza sativa]
Length = 1688
Score = 194 bits (493), Expect = 1e-47
Identities = 131/394 (33%), Positives = 196/394 (49%), Gaps = 46/394 (11%)
Query: 160 VNSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALE 219
+++ F++ + G + +LSCP +QNG +ERK R RTLLI + +P+ FW A+
Sbjct: 456 MSNAFREFLVSQGTLPQLSCPGAHAQNGVAERKHRHIIETARTLLIASFVPAHFWAEAIS 515
Query: 220 MATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCV 279
A YL+N+ P ++ SP +VL+ P Y+HLRVFGC C+ L + KL +S +CV
Sbjct: 516 TAVYLINMQPSSSLQGRSPGEVLFGSPPRYDHLRVFGCTCYVLLAPRERTKLTAQSVECV 575
Query: 280 FLGYPSNHRGYKCLGLLSNKIIICRHVLFNEN-IFPYAKLHIPQ--PNTYTFLDNELSPY 336
FLGY H+GY+C + +I I R V F+EN F Y+ + P N+ +FL L P
Sbjct: 576 FLGYSLEHKGYRCYDPSARRIRISRDVTFDENKPFFYSSTNQPSSPENSISFL--YLPPI 633
Query: 337 IIQHLMDQTQTGP---PIPQPAHQPNHITSPCPN-SPNITSPPQSN-PTSPIQQNLPPIF 391
+ + P PIP P ++ P P+ SP+ SPP S+ P S ++P
Sbjct: 634 PSPESLPSSPITPSPSPIPPSVPSPTYVPPPPPSPSPSPVSPPPSHIPASSSPPHVPSTI 693
Query: 392 ----------------------QPTLQANTKPI----------TRSQHGIFKPNQKYYGL 419
QPTL+ T + R++ + PN+ + +
Sbjct: 694 TLDTFPFHYSRRPKIPNESQPSQPTLEDPTCSVDDSSPAPRYNLRARDALRAPNRDDFVV 753
Query: 420 HTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHK 479
P A+ P+WK+AM++E +L + TWD+VP P I W+++ K
Sbjct: 754 GVVFE----PSTYQEAIVLPHWKLAMSEELAALERTNTWDVVPLPSHAVPITCKWVYKVK 809
Query: 480 EKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
KSDG ER+KARLV G Q G D ETF+P+
Sbjct: 810 TKSDGQVERYKARLVARGFQQAHGRDYDETFAPV 843
>UniRef100_O23741 SLG-Sc and SLA-Sc genes and Melmoth retrotransposon sequence
[Brassica oleracea]
Length = 1131
Score = 194 bits (492), Expect = 1e-47
Identities = 129/400 (32%), Positives = 188/400 (46%), Gaps = 53/400 (13%)
Query: 125 HINPKFIQYLKN*EHTSTHNFKEKLKQFNVTMAVNVNSNFQKMCEANGIVFRLSCPYTSS 184
HI P F+ ++ + K+K A ++ F ++ + GIV SCP T
Sbjct: 620 HIFPTFVNQIET-------QYNTKIKSVRRDNAPELS--FTELFKEKGIVSYHSCPETLE 670
Query: 185 QNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHKTINFESPLKVLYH 244
QN ERK + N+ R L+ + +P +W + A +L+N P + +SP +VL
Sbjct: 671 QNSVLERKHQHLLNVARALMFQSQVPLQYWGDCVLTAAFLINRTPSPLLANKSPYEVLMG 730
Query: 245 KDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHRGYKCLGLLSNKIIICR 304
K P Y+ LR FGCLC+ + +K PRS CVFLGYPS ++GYK L L SNKI I R
Sbjct: 731 KAPQYDQLRTFGCLCYGSTSPKQRHKFMPRSRACVFLGYPSGYKGYKLLDLESNKIYISR 790
Query: 305 HVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQHLMDQTQTGPPIPQPAHQPNHITSP 364
+V F+E+IFP AK ++ F P +T P
Sbjct: 791 NVTFHEDIFPMAKHQKMDESSLHFF----------------------------PPKVTVP 822
Query: 365 CPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANTKPI-TRSQHGIFKP---NQKYYGLH 420
SPNI+S P S + I + Q T+ A+ K S H P Y +
Sbjct: 823 SAPSPNISSSPFSTLSPQISKR-----QRTVPAHLKDFHCYSVHDSAYPISSTLSYSQIS 877
Query: 421 TH-------VTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSM 473
+H +T P+P++ + W + + E +++ +N TWD+VP P I
Sbjct: 878 SHHLAYINSITNIPIPQSYAEVRQSKEWTESADKELDAMEENDTWDVVPLPKGKKAIGCR 937
Query: 474 WIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
W+ K +DGT ER K+RLVG G Q+ G+D ETFSP+
Sbjct: 938 WVHTLKFNADGTLERRKSRLVGKGYTQKEGLDYIETFSPV 977
>UniRef100_Q9C8F4 Ty1/copia-element polyprotein [Arabidopsis thaliana]
Length = 1152
Score = 192 bits (487), Expect = 5e-47
Identities = 126/371 (33%), Positives = 187/371 (49%), Gaps = 36/371 (9%)
Query: 172 GIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHK 231
GIV ++SC YT QNG+ ERK R N+ R+LL A LP SFW ++ A YL+N P
Sbjct: 656 GIVHQISCVYTHQQNGRVERKHRHILNVARSLLFQAELPISFWEESVLTAAYLINRTPTP 715
Query: 232 TINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHRGYK 291
++ ++P K+LY + PSY LRVFG LCF + ++ K Q R KC+F+GYP +G++
Sbjct: 716 ILDGKTPYKILYSQPPSYASLRVFGSLCFARKHTGRLDKFQERGRKCIFVGYPHGQKGWR 775
Query: 292 CLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTF------LDNELSPYIIQHLMDQT 345
+ S + R V+F E+IFP+A + N TF + + + PY + L D
Sbjct: 776 IYDIESQIFFVSRDVVFQEDIFPFAD----KKNKDTFSSPAAVIPSPILPYDDEFL-DIY 830
Query: 346 QTGP-PIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPI-------QQNLPPIFQPTLQA 397
Q G P P + P+SP IT+ P + P+ Q+N+ T A
Sbjct: 831 QIGDVPATNPLPAIIDVNDSPPSSPIITATPAAASPPPLRRGLRQRQENVRLKDYQTYSA 890
Query: 398 NTK--------------PITRSQHG-IFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWK 442
+ P+ G IF P+ +++ + P N A+++ W+
Sbjct: 891 QCESTQTLSDNIGTCIYPMANYVSGEIFSPSNQHFLAAISMVDPPQTYN--QAIREKEWR 948
Query: 443 MAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQV 502
A+ E ++L TWD+ P V I S W+FR K S+GT ER+KARLV G Q+
Sbjct: 949 NAVFFEVDALEDQGTWDITKLPQGVKAIGSKWVFRIKYNSNGTVERYKARLVALGNHQKE 1008
Query: 503 GIDCGETFSPM 513
GID +TF+P+
Sbjct: 1009 GIDFTKTFAPV 1019
>UniRef100_Q6L3N8 Putative gag-pol polyprotein [Solanum demissum]
Length = 1333
Score = 190 bits (482), Expect = 2e-46
Identities = 120/359 (33%), Positives = 183/359 (50%), Gaps = 38/359 (10%)
Query: 160 VNSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALE 219
++++F CE NGI L+ PYT QNG +ERK R+ + R+ L LP FW A+
Sbjct: 578 LSNDFNLFCEENGIRRELTAPYTPEQNGVAERKNRTVVEMARSSLKAKGLPDYFWGEAVA 637
Query: 220 MATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCV 279
Y +NI P K + +PL+ K P +HLR+FGC+ + L KL +STKC+
Sbjct: 638 TVVYFLNISPTKDVWNTTPLEAWNGKKPRVSHLRIFGCIAYALVNFHS--KLDEKSTKCI 695
Query: 280 FLGYPSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQ 339
F+GY + Y+ +S K+II R+V+FNE++ ++ F
Sbjct: 696 FVGYSLQSKAYRLYNPISGKVIISRNVVFNEDV------------SWNFNSG-------- 735
Query: 340 HLMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSP-PQSNPTSPIQQNLPPIFQPTLQAN 398
++M Q P + A + + P S +++SP S +P + ++ PI
Sbjct: 736 NMMSNIQLLPTDEESAVDFGNSPNSSPVSSSVSSPIAPSTTVAPDESSVEPI-------- 787
Query: 399 TKPITRSQHGIFKPNQKYYG-LHTHVTKSPLPRNPV---SALKDPNWKMAMNDEYNSLIK 454
P+ RS KPN KY ++T + L +P+ A++ WK AM +E ++ +
Sbjct: 788 --PLRRSTRE-KKPNPKYSNTVNTSCQFALLVSDPICYEEAVEQSEWKNAMIEEIQAIER 844
Query: 455 NKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
N TW+LV P NVI W+FR K +DG+ ++HKARLV G QQ G+D ETFSP+
Sbjct: 845 NSTWELVDAPEGKNVIGLKWVFRTKYNADGSIQKHKARLVAKGYSQQQGVDFDETFSPV 903
>UniRef100_Q9FWZ5 Putative retroelement polyprotein [Arabidopsis thaliana]
Length = 1404
Score = 188 bits (478), Expect = 6e-46
Identities = 134/422 (31%), Positives = 192/422 (44%), Gaps = 48/422 (11%)
Query: 136 N*EHTSTHNFKEKLKQFNVTMAVNVNSN-FQKMCEANGIVFRLSCPYTSSQNGKSERKIR 194
N E T+ F K+K F S F+ GI+ + SCPYT QNG +ERK R
Sbjct: 559 NFETYVTNQFNAKIKVFRTDNGGEYTSQKFRDHLAKRGIIHQTSCPYTPQQNGVAERKNR 618
Query: 195 STNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHKTINFESPLKVLYHKDPSYNHLRV 254
+ R+++ H S+P FW A+ A YL+N P K ++ SP +VL + P +HLRV
Sbjct: 619 HLMEVARSMMFHTSVPKRFWGDAVLTACYLINRTPTKVLSDLSPFEVLNNTKPFIDHLRV 678
Query: 255 FGCLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHRGYKCLGLLSNKIIICRHVLFN----- 309
FGC+CF L P + KL +STKC+FLGY + +GYKC N+ I R V F
Sbjct: 679 FGCVCFVLIPGEQRSKLDAKSTKCMFLGYSTTQKGYKCFDPTKNRTFISRDVKFLENQDY 738
Query: 310 ------ENIFPYAKLHIPQPNTYTFLDNEL---SPYIIQHLMDQTQTGPPIPQP------ 354
EN+ + T FL + L S QH + TQ + Q
Sbjct: 739 NNKKDWENLKDLTHSTSDRVETLKFLLDHLGNDSTSTTQHQPEMTQDQEDLNQENEEVSL 798
Query: 355 AHQPN--HITSPCPNSPNITSPPQ-----SNPTSPIQQNLPPIFQPTLQANTKPITRSQH 407
HQ N H+ PN+ + Q S+ Q LPP P L+ +T+ R +
Sbjct: 799 QHQENLTHVQEDPPNTQEHSEHVQEIQDDSSEDEEPTQVLPP--PPPLRRSTR--IRRKK 854
Query: 408 GIFKPNQ----------------KYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNS 451
F N + + +++ +P+ A++ W+ A+ DE N+
Sbjct: 855 EFFNSNAVAHPFQATCSLALVPLDHQAFLSKISEHWIPQTYEEAMEVKEWRDAIADEINA 914
Query: 452 LIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFS 511
+ +N TWD P + S W+F K KS+G ER+K RLV G Q G D ETF+
Sbjct: 915 MKRNHTWDEDDLPKGKKTVSSRWVFTIKYKSNGDIERYKTRLVARGFTQTYGSDYMETFA 974
Query: 512 PM 513
P+
Sbjct: 975 PV 976
>UniRef100_Q7Y141 Putative polyprotein [Oryza sativa]
Length = 1335
Score = 188 bits (477), Expect = 8e-46
Identities = 114/357 (31%), Positives = 178/357 (48%), Gaps = 21/357 (5%)
Query: 160 VNSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALE 219
++ F+K CE GI +L+ Y++ QNG +ERK R+ N++ ++L +P SFW A+
Sbjct: 592 ISKEFEKYCENAGIRRQLTAGYSAQQNGVAERKNRTINDMANSMLQDKGMPKSFWAEAVN 651
Query: 220 MATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCV 279
A Y++N P K + +P + Y K P H+RVFGC+C+ P+ K K +S +C+
Sbjct: 652 TAVYILNRSPTKAVTNRTPFEAWYGKKPVIGHMRVFGCICYAQVPAQKRVKFDNKSDRCI 711
Query: 280 FLGYPSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQ 339
F+GY +GY+ L KIII R +F+E+ T+ + E S +
Sbjct: 712 FVGYADGIKGYRLYNLEKKKIIISRDAIFDESA------------TWNWKSPEASSTPLL 759
Query: 340 HLMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANT 399
T P + +H SP P+SP ++S S+ +SP + Q + +
Sbjct: 760 PTTTITLGQPHMHGTHEVEDHTPSPQPSSP-MSSSSASSDSSPSSEE-----QISTPESA 813
Query: 400 KPITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVS---ALKDPNWKMAMNDEYNSLIKNK 456
RS + + + G H + P S A K NW AM DE + + KN
Sbjct: 814 PRRVRSMVELLESTSQQRGSEQHEFCNYSVVEPQSFQEAEKHDNWIKAMEDEIHMIEKNN 873
Query: 457 TWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
TW+LV RP D VI W+++ K DG+ +++KARLV G Q+ GID ET++P+
Sbjct: 874 TWELVDRPRDREVIGVKWVYKTKLNPDGSVQKYKARLVAKGFKQKPGIDYYETYAPV 930
>UniRef100_O23588 Retrotransposon like protein [Arabidopsis thaliana]
Length = 1433
Score = 187 bits (475), Expect = 1e-45
Identities = 122/391 (31%), Positives = 188/391 (47%), Gaps = 37/391 (9%)
Query: 125 HINPKFIQYLKN*EHTSTHNFKEKLKQFNVTMAVNVNSNFQKMCEANGIVFRLSCPYTSS 184
H+ P FI + HT ++ KLK A + F + A+GIV SCP T
Sbjct: 646 HVFPAFINMV----HTQ---YQTKLKSVRSDNAHELK--FTDLFAAHGIVAYHSCPETPE 696
Query: 185 QNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHKTINFESPLKVLYH 244
QN ERK + N+ R LL +++P FW + A +L+N LP +N +SP + L +
Sbjct: 697 QNSVVERKHQHILNVARALLFQSNIPLEFWGDCVLTAVFLINRLPTPVLNNKSPYEKLKN 756
Query: 245 KDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHRGYKCLGLLSNKIIICR 304
P+Y L+ FGCLC+ + +K +PR+ CVFLGYP ++GYK L + ++ + I R
Sbjct: 757 IPPAYESLKTFGCLCYSSTSPKQRHKFEPRARACVFLGYPLGYKGYKLLDIETHAVSISR 816
Query: 305 HVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQHLMDQTQTGPPIPQPAHQPNHITSP 364
HV+F+E+IFP+ I D P + PA + P
Sbjct: 817 HVIFHEDIFPFISSTIKD--------------------DIKDFFPLLQFPARTDD---LP 853
Query: 365 CPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANTKPITRSQ--HGIFKPNQKYYGLHTH 422
+ I + P + +S + L P F P + KP Q H + ++ +
Sbjct: 854 LEQTSIIDTHPHQDVSS--SKALVP-FDPLSKRQKKPPKHLQDFHCYNNTTEPFHAFINN 910
Query: 423 VTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKS 482
+T + +P+ A W AM +E ++++ TW +V PP+ I W+F K +
Sbjct: 911 ITNAVIPQRYSEAKDFKAWCDAMKEEIGAMVRTNTWSVVSLPPNKKAIGCKWVFTIKHNA 970
Query: 483 DGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
DG+ ER+KARLV G Q+ G+D ETFSP+
Sbjct: 971 DGSIERYKARLVAKGYTQEEGLDYEETFSPV 1001
>UniRef100_Q9FJV3 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1475
Score = 184 bits (467), Expect = 1e-44
Identities = 113/350 (32%), Positives = 172/350 (48%), Gaps = 23/350 (6%)
Query: 164 FQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATY 223
F+ + +A GI+ SCP T QN ERK + N+ R L+ A++P FW + A +
Sbjct: 718 FEALYQAKGIISYHSCPETPQQNSVVERKHQHILNVARALMFEANMPLEFWGDCILSAVF 777
Query: 224 LMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGY 283
L+N LP ++ +SP ++L+ K P Y L+VFGCLC+ + +K PR+ CVFLGY
Sbjct: 778 LINRLPTPLLSNKSPFELLHLKVPDYTSLKVFGCLCYESTSPQQRHKFAPRARACVFLGY 837
Query: 284 PSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQHLMD 343
PS ++GYK L L +N I I RHV+F E +FP F D + P + L+D
Sbjct: 838 PSGYKGYKLLDLETNTIHISRHVVFYETVFP-------------FTDKTIIPRDVFDLVD 884
Query: 344 QTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANTKPIT 403
P+ + P S ++P ++S +S P +Q P + + +
Sbjct: 885 ------PVHENIENP---PSTSESAPKVSSKRESRPPGYLQDYFCNAV-PDVTKDVRYPL 934
Query: 404 RSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPR 463
+ + ++++ V K P P A K W AM E ++L TW +
Sbjct: 935 NAYINYTQLSEEFTAYICAVNKYPEPCTYAQAKKIKEWLDAMEIEIDALESTNTWSVCSL 994
Query: 464 PPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
P I W+F+ K +DG+ ER KARLV G Q+ G+D +TFSP+
Sbjct: 995 PQGKKPIGCKWVFKVKLNADGSLERFKARLVAKGYTQREGLDYYDTFSPV 1044
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.337 0.147 0.487
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,565,456,948
Number of Sequences: 2790947
Number of extensions: 67103772
Number of successful extensions: 288517
Number of sequences better than 10.0: 1060
Number of HSP's better than 10.0 without gapping: 733
Number of HSP's successfully gapped in prelim test: 356
Number of HSP's that attempted gapping in prelim test: 283255
Number of HSP's gapped (non-prelim): 3333
length of query: 952
length of database: 848,049,833
effective HSP length: 137
effective length of query: 815
effective length of database: 465,690,094
effective search space: 379537426610
effective search space used: 379537426610
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 80 (35.4 bits)
Medicago: description of AC146853.2


