

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146842.1 + phase: 0
(216 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
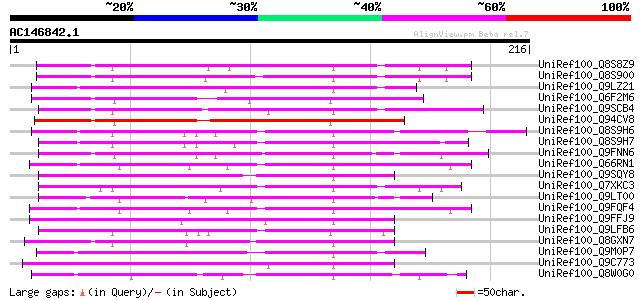
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8S8Z9 Syringolide-induced protein 1-3-1B [Glycine max] 114 2e-24
UniRef100_Q8S900 Syringolide-induced protein 1-3-1A [Glycine max] 110 2e-23
UniRef100_Q9LZ21 Putative I-box binding factor [Arabidopsis thal... 109 6e-23
UniRef100_Q6F2M6 Putative myb-like transcription factor [Oryza s... 105 1e-21
UniRef100_Q9SCB4 I-box binding factor [Lycopersicon esculentum] 104 1e-21
UniRef100_Q94CV8 Putative I-box binding factor [Oryza sativa] 97 2e-19
UniRef100_Q8S9H6 MYB-like transcription factor DVL1 [Antirrhinum... 96 8e-19
UniRef100_Q8S9H7 MYB-like transcription factor DIVARICATA [Antir... 95 1e-18
UniRef100_Q9FNN6 Similarity to I-box binding factor [Arabidopsis... 94 3e-18
UniRef100_Q66RN1 MYB transcription factor [Hevea brasiliensis] 89 6e-17
UniRef100_Q9SQY8 Hypothetical protein F13M14.13 [Arabidopsis tha... 89 1e-16
UniRef100_Q7XKC3 OSJNBa0064G10.8 protein [Oryza sativa] 89 1e-16
UniRef100_Q9LT00 Similarity to I-box binding factor [Arabidopsis... 88 2e-16
UniRef100_Q9FQF4 Hypothetical protein [Hevea brasiliensis] 88 2e-16
UniRef100_Q9FFJ9 Similarity to I-box binding factor [Arabidopsis... 87 3e-16
UniRef100_Q9LFB6 Hypothetical protein F7J8_180 [Arabidopsis thal... 86 5e-16
UniRef100_Q8GXN7 Putative MYB family transcription factor [Arabi... 86 5e-16
UniRef100_Q9M0P7 Hypothetical protein AT4g09450 [Arabidopsis tha... 84 3e-15
UniRef100_Q9C773 MYB-family transcription factor, putative; alte... 83 4e-15
UniRef100_Q8W0G0 P0529E05.19 protein [Oryza sativa] 83 4e-15
>UniRef100_Q8S8Z9 Syringolide-induced protein 1-3-1B [Glycine max]
Length = 236
Score = 114 bits (284), Expect = 2e-24
Identities = 77/197 (39%), Positives = 116/197 (58%), Gaps = 20/197 (10%)
Query: 12 EWTWDENKIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALIEEGLV 70
+WT +K+FE L E++ + RWE I V G+S+ EV+EHYE L+HD+ I+ G V
Sbjct: 16 QWTRYHDKLFERALLVVPEDLPD-RWEKIADQVPGKSAVEVREHYEALVHDVFEIDSGRV 74
Query: 71 DFSTNSDDFII------SKASTDENK------APPTKNKTKKVVRVKHWTEEEHRLFLEG 118
+ + DD + ++ ST +N + P + + + WTEEEHRLFL G
Sbjct: 75 EVPSYVDDSVATPPSGGAEISTWDNANQISFGSKPKQQGDNERKKGTPWTEEEHRLFLIG 134
Query: 119 IEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFY-ITSLKGN 176
+ GKG W+ IS++ V TRT +QVASHAQK+FL Q S K +KRS+ + IT++ N
Sbjct: 135 LSKFGKGDWRSISRNVVVTRTPTQVASHAQKYFLRQ---NSVKKERKRSSIHDITTVDSN 191
Query: 177 SKPL-LNKDNIPSPSTS 192
S P+ ++++ +P P S
Sbjct: 192 SVPVPIDQNWVPPPGGS 208
>UniRef100_Q8S900 Syringolide-induced protein 1-3-1A [Glycine max]
Length = 233
Score = 110 bits (276), Expect = 2e-23
Identities = 77/200 (38%), Positives = 112/200 (55%), Gaps = 26/200 (13%)
Query: 12 EWTWDENKIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALIEEGLV 70
+WT +K+FE L E++ + RWE I V G+S+ EV+EHYE L+HD+ I+ G V
Sbjct: 13 QWTRYHDKLFERALLVVPEDLPD-RWEKIADQVPGKSAVEVREHYEALVHDVFEIDSGRV 71
Query: 71 DFSTNSDDFI---------------ISKASTDENKAPPTKNKTKKVVRVKHWTEEEHRLF 115
+ + DD + ++ S +N+ KK WTEEEHRLF
Sbjct: 72 EVPSYVDDSVAMPPSGGAGISTWDNANQISFGSKLKQQGENERKKGT---PWTEEEHRLF 128
Query: 116 LEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFY-ITSL 173
L G+ GKG W+ IS++ V TRT +QVASHAQK+FL Q S K +KRS+ + IT++
Sbjct: 129 LIGLSKFGKGDWRSISRNVVVTRTPTQVASHAQKYFLRQ---NSVKKERKRSSIHDITTV 185
Query: 174 KGNSKPL-LNKDNIPSPSTS 192
NS P+ +++ +P P S
Sbjct: 186 DSNSAPMPIDQTWVPPPGGS 205
>UniRef100_Q9LZ21 Putative I-box binding factor [Arabidopsis thaliana]
Length = 215
Score = 109 bits (272), Expect = 6e-23
Identities = 68/163 (41%), Positives = 95/163 (57%), Gaps = 7/163 (4%)
Query: 10 SKEWTWDENKIFETILFEYLEEVQEGRWENIGLVCGRSSTEVKEHYETLLHDLALIEEGL 69
S +WT E+K+FE L + E RWE I +S+ EV+EHYE L+HD+ I+ G
Sbjct: 3 SSQWTRSEDKMFEQALVLF-PEGSPNRWERIADQLHKSAGEVREHYEVLVHDVFEIDSGR 61
Query: 70 VDFSTNSDDFIISKASTDE--NKAPPTKNKTKKVVRVKHWTEEEHRLFLEGIEIHGKGKW 127
VD DD + A D + +K+ + R WTE EH+LFL G++ +GKG W
Sbjct: 62 VDVPDYMDDSAAAAAGWDSAGQISFGSKHGESERKRGTPWTENEHKLFLIGLKRYGKGDW 121
Query: 128 KLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFY 169
+ IS++ V TRT +QVASHAQK+FL Q S K +KRS+ +
Sbjct: 122 RSISRNVVVTRTPTQVASHAQKYFLRQ---NSVKKERKRSSIH 161
>UniRef100_Q6F2M6 Putative myb-like transcription factor [Oryza sativa]
Length = 182
Score = 105 bits (261), Expect = 1e-21
Identities = 68/166 (40%), Positives = 97/166 (57%), Gaps = 12/166 (7%)
Query: 10 SKEWTWDENKIFETILFEYLEEVQEGRWENIGL-VCGRSSTEVKEHYETLLHDLALIEEG 68
S+ W+ E+K+FE+ L + E RW + + GRS+ EV EHY+ L+ D+ LIE G
Sbjct: 25 SRPWSKAEDKVFESALVAFPEHTHN-RWALVASRLPGRSAHEVWEHYQVLVDDVDLIERG 83
Query: 69 LVDFSTNSDDFIISKASTDENKAPPTKNKT-KKVVRVKHWTEEEHRLFLEGIEIHGKGKW 127
+V DD D N A + + R WTEEEHRLFLEG+E +G+G W
Sbjct: 84 MVASPGCWDD--------DNNSAGHGRGSGGDERRRGVPWTEEEHRLFLEGLEKYGRGDW 135
Query: 128 KLISQ-HVRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITS 172
+ IS+ V+TRT +QVASHAQK F+ Q + +S+ K++S IT+
Sbjct: 136 RNISRWSVKTRTPTQVASHAQKFFIRQANASSRGDSKRKSIHDITA 181
>UniRef100_Q9SCB4 I-box binding factor [Lycopersicon esculentum]
Length = 191
Score = 104 bits (260), Expect = 1e-21
Identities = 66/189 (34%), Positives = 112/189 (58%), Gaps = 10/189 (5%)
Query: 13 WTWDENKIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALIEEGLVD 71
WT D++K+FE L Y E + RW+ I V G+++ ++ HY+ L+HD+ I+ G +D
Sbjct: 7 WTRDDDKLFEHGLVLYPENSAD-RWQLIADHVPGKTADDIMAHYDDLVHDVYEIDSGRID 65
Query: 72 FSTNSDDFIISKASTDENKAPPTKNKTKKVVRVKH--WTEEEHRLFLEGIEIHGKGKWKL 129
+ +DD + + D + K+ ++ R K WTE+EHRLFL G++ +GKG W+
Sbjct: 66 LPSYTDDPV--ELEGDCQITSGSNKKSNEIERKKGTPWTEDEHRLFLIGLDKYGKGDWRS 123
Query: 130 ISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITSLKGNSKPLLNKDNIPS 188
IS++ V +RT +QVASHAQK+F+ Q + K +KRS+ + + ++ P+ + + +
Sbjct: 124 ISRNVVVSRTPTQVASHAQKYFIRQ---QAMKKERKRSSIHDITTAVDTNPVPPQSSFQN 180
Query: 189 PSTSWDGNF 197
S S + NF
Sbjct: 181 QSGSQNFNF 189
>UniRef100_Q94CV8 Putative I-box binding factor [Oryza sativa]
Length = 173
Score = 97.4 bits (241), Expect = 2e-19
Identities = 58/156 (37%), Positives = 94/156 (60%), Gaps = 8/156 (5%)
Query: 11 KEWTWDENKIFETILFEYLEEVQEGRWENIGL-VCGRSSTEVKEHYETLLHDLALIEEGL 69
+ W+ E+K+FE+ L E+V + RW + + GR+ E EHY+ L+ D+ LI G
Sbjct: 16 RPWSKVEDKVFESALVLCPEDVPD-RWALVAAQLPGRTPQEALEHYQVLVADIDLIMRGA 74
Query: 70 VDFSTNSDDFIISKASTDENKAPPTKNKTKKVVRVKHWTEEEHRLFLEGIEIHGKGKWKL 129
VD + DD ++ + K + ++ R W+E+EHRLFLEG++ +G+G W+
Sbjct: 75 VDAPGSWDD-----NDGNDRRGGGGKPRGEERRRGVPWSEDEHRLFLEGLDRYGRGDWRN 129
Query: 130 ISQ-HVRTRTASQVASHAQKHFLHQLDGTSKKTYKK 164
IS+ VRTRT +QVASHAQK+F+ Q + ++ + +K
Sbjct: 130 ISRFSVRTRTPTQVASHAQKYFIRQANAGARDSKRK 165
>UniRef100_Q8S9H6 MYB-like transcription factor DVL1 [Antirrhinum majus]
Length = 291
Score = 95.5 bits (236), Expect = 8e-19
Identities = 76/225 (33%), Positives = 115/225 (50%), Gaps = 32/225 (14%)
Query: 10 SKEWTWDENKIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALIEEG 68
S +WT +NK FE L + +E RWE + +V G++ +V HY+ L D+ IE G
Sbjct: 22 SPKWTAADNKAFENALAVF-DEYTPHRWERVAEIVPGKTVWDVIRHYKELEDDVTSIEAG 80
Query: 69 LVD---FSTNS------------DDFIISKA--STDENKAPPTKNKTKKVVRVKHWTEEE 111
LV ++T+ D F+ S + + P+ + KK V WTEEE
Sbjct: 81 LVPVPGYNTSLPFTLEWGSGHGFDGFMQSYVVGGRKSSCSRPSDQERKKGVP---WTEEE 137
Query: 112 HRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYI 170
H+LFL G++ +GKG W+ IS++ V TRT +QVASHAQK+F+ QL G K ++ S I
Sbjct: 138 HKLFLMGLKKYGKGDWRNISRNFVITRTPTQVASHAQKYFIRQLSGGKDK--RRASIHDI 195
Query: 171 TSLKGNSKPLLNKDNIPSPSTSWDGNFHPLLYKDNYVPALPSPHL 215
T++ N ++N S+ PL ++ N A H+
Sbjct: 196 TTVNLNDGQTFPRENKIKQSS-------PLAHQSNSAAATSKLHI 233
>UniRef100_Q8S9H7 MYB-like transcription factor DIVARICATA [Antirrhinum majus]
Length = 307
Score = 95.1 bits (235), Expect = 1e-18
Identities = 72/198 (36%), Positives = 102/198 (51%), Gaps = 25/198 (12%)
Query: 13 WTWDENKIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALIEEGLVD 71
WT ENK FE L + +E RWE + V G++ +V Y+ L D++ IE G V
Sbjct: 26 WTAAENKAFENALAVF-DENTPNRWERVAERVPGKTVGDVMRQYKELEDDVSSIEAGFVP 84
Query: 72 ---FSTNS------------DDFIISKASTDENKAP--PTKNKTKKVVRVKHWTEEEHRL 114
+ST+S D F S + + P++ + KK V WTEEEH+L
Sbjct: 85 VPGYSTSSPFTLEWGSGHGFDGFKQSYGTGGRKSSSGRPSEQERKKGVP---WTEEEHKL 141
Query: 115 FLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITSL 173
FL G++ +GKG W+ IS++ V TRT +QVASHAQK+F+ QL G K + +L
Sbjct: 142 FLMGLKKYGKGDWRNISRNFVITRTPTQVASHAQKYFIRQLSGGKDKRRASIHDITTVNL 201
Query: 174 KGNSKPLLNKDNIPSPST 191
N P + DN PS+
Sbjct: 202 SDNQTP--SPDNKKPPSS 217
>UniRef100_Q9FNN6 Similarity to I-box binding factor [Arabidopsis thaliana]
Length = 298
Score = 93.6 bits (231), Expect = 3e-18
Identities = 69/210 (32%), Positives = 115/210 (53%), Gaps = 28/210 (13%)
Query: 13 WTWDENKIFETILFEYLEEVQEGRWENIGL-VCGRSSTEVKEHYETLLHDLALIEEGLVD 71
W+ +++ FE L +E +E RWE I V G+S ++KEHYE L+ D+ IE G V
Sbjct: 12 WSREDDIAFERALANNTDESEE-RWEKIAADVPGKSVEQIKEHYELLVEDVTRIESGCVP 70
Query: 72 FSTNS----------DDFIISKA-----STDENKAPPTKNKTKKVVRVKHWTEEEHRLFL 116
D+ SK + + N+A +K+ ++ + WTE+EHRLFL
Sbjct: 71 LPAYGSPEGSNGHAGDEGASSKKGGNSHAGESNQAGKSKSDQERRKGIA-WTEDEHRLFL 129
Query: 117 EGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITSLKG 175
G++ +GKG W+ IS++ V TRT +QVASHAQK+F+ +L+ +K ++RS+ + + G
Sbjct: 130 LGLDKYGKGDWRSISRNFVVTRTPTQVASHAQKYFI-RLNSMNKD--RRRSSIHDITSVG 186
Query: 176 NSK------PLLNKDNIPSPSTSWDGNFHP 199
N+ P+ ++N + + + + N P
Sbjct: 187 NADVSTPQGPITGQNNSNNNNNNNNNNSSP 216
>UniRef100_Q66RN1 MYB transcription factor [Hevea brasiliensis]
Length = 310
Score = 89.4 bits (220), Expect = 6e-17
Identities = 66/205 (32%), Positives = 105/205 (51%), Gaps = 25/205 (12%)
Query: 9 KSKEWTWDENKIFETILFEYLEEVQEGRWENIGLVC-GRSSTEVKEHYETLLHDLALIEE 67
++ +WT +ENK FE L Y ++ + RW+ + V G++ +V + Y L D++ IE
Sbjct: 25 RATKWTPEENKQFENALALY-DKDEPDRWQRVAAVIPGKTVGDVIKQYRELEEDVSDIEA 83
Query: 68 GLVDFS--TNSDDFIISKASTDEN----------------KAPPTKNKTKKVVRVKHWTE 109
GL+ ++SD F + + ++ A ++ + KK V WTE
Sbjct: 84 GLIPIPGYSSSDAFTLEWFNNNQGYDGFRHYYTPGGKRTTAARSSEQERKKGVP---WTE 140
Query: 110 EEHRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNF 168
EEHR FL G++ +GKG W+ IS++ V TRT +QVASHAQK+F+ Q G K +
Sbjct: 141 EEHRQFLMGLQKYGKGDWRNISRNFVTTRTPTQVASHAQKYFIRQSTGGKDKRRSSIHDI 200
Query: 169 YITSLKGNSKPLLN-KDNIPSPSTS 192
+L P + K + P ST+
Sbjct: 201 TTVNLPDTKSPSPDEKKSSPDHSTT 225
>UniRef100_Q9SQY8 Hypothetical protein F13M14.13 [Arabidopsis thaliana]
Length = 287
Score = 88.6 bits (218), Expect = 1e-16
Identities = 54/149 (36%), Positives = 77/149 (51%), Gaps = 6/149 (4%)
Query: 13 WTWDENKIFETILFEYLEEVQEGRWENIGLVCGRSSTEVKEHYETLLHDLALIEEGLVDF 72
W D++K FE L + E ENI + EV +Y+ L+ D+ LIE G
Sbjct: 8 WKRDDDKRFELALVRFPAEGSPDFLENIAQFLQKPLKEVYSYYQALVDDVTLIESGKYPL 67
Query: 73 STNSDDFIISKASTDENKAPPTKNKTKKVVRVKHWTEEEHRLFLEGIEIHGKGKWKLISQ 132
+D +S ++K T K + W+ EEHRLFL+G+ +GKG WK IS+
Sbjct: 68 PKYPEDDYVSLPEATKSKTQGTGKK-----KGIPWSPEEHRLFLDGLNKYGKGDWKSISR 122
Query: 133 H-VRTRTASQVASHAQKHFLHQLDGTSKK 160
V +R+ QVASHAQK+FL Q + K+
Sbjct: 123 ECVTSRSPMQVASHAQKYFLRQKNKKGKR 151
>UniRef100_Q7XKC3 OSJNBa0064G10.8 protein [Oryza sativa]
Length = 318
Score = 88.6 bits (218), Expect = 1e-16
Identities = 66/205 (32%), Positives = 104/205 (50%), Gaps = 35/205 (17%)
Query: 13 WTWDENKIFETILFEYLEEVQEGR--WENIG-LVCGRSSTEVKEHYETLLHDLALIEEGL 69
WT + K FE L ++ +EG WE + V G+++ EV+ HYE L+ D+ IE G
Sbjct: 33 WTREREKAFENALATVGDDEEEGDGLWEKLAEAVEGKTADEVRRHYELLVEDVDGIEAGR 92
Query: 70 VDFSTNSDDFIISKAST-------------------DENKAPPTKNKTKKVVRVKHWTEE 110
V + D + + S ++ A ++ + +K + WTE+
Sbjct: 93 VPLLVYAGDGGVEEGSAGGGKKGGGGGGGGGGGGHGEKGSAKSSEQERRKGIA---WTED 149
Query: 111 EHRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFY 169
EHRLFL G+E +GKG W+ IS++ V +RT +QVASHAQK+F+ S ++RS+ +
Sbjct: 150 EHRLFLLGLEKYGKGDWRSISRNFVISRTPTQVASHAQKYFIRL---NSMNRERRRSSIH 206
Query: 170 -ITSLKGNSK-----PLLNKDNIPS 188
ITS+ P+ + N PS
Sbjct: 207 DITSVNNGDTSAAQGPITGQPNGPS 231
>UniRef100_Q9LT00 Similarity to I-box binding factor [Arabidopsis thaliana]
Length = 337
Score = 87.8 bits (216), Expect = 2e-16
Identities = 66/182 (36%), Positives = 95/182 (51%), Gaps = 23/182 (12%)
Query: 13 WTWDENKIFETILFEYLEEVQEGRWENIGLVC-GRSSTEVKEHYETLLHDLALIEEGLVD 71
W+ D++ FE L Y ++ E RW+ I V G++ +V EHY L D+ LIE G V
Sbjct: 13 WSKDDDIAFEKALAIYNDKT-EIRWKKIATVVPGKTLEQVIEHYNILARDVMLIESGCVR 71
Query: 72 FSTNSDDFI-------------ISKASTDENKAPPTKNKTK---KVVRVKHWTEEEHRLF 115
+ DDF+ I + D K K+K K R W EHR F
Sbjct: 72 LP-DYDDFLEEPNHNAFGKERSILEGGNDRKYESKHKGKSKLKQKRRRGVPWKPFEHRQF 130
Query: 116 LEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITSLK 174
L G++ +GKG W+ IS+H V TRT++QVASHAQK+F H ++ KK +KR + + ++
Sbjct: 131 LHGLKKYGKGDWRSISRHCVVTRTSTQVASHAQKYFAH-INSEDKK--RKRPSIHDITIA 187
Query: 175 GN 176
N
Sbjct: 188 EN 189
>UniRef100_Q9FQF4 Hypothetical protein [Hevea brasiliensis]
Length = 310
Score = 87.8 bits (216), Expect = 2e-16
Identities = 65/205 (31%), Positives = 105/205 (50%), Gaps = 25/205 (12%)
Query: 9 KSKEWTWDENKIFETILFEYLEEVQEGRWENIGLVC-GRSSTEVKEHYETLLHDLALIEE 67
++ +WT +ENK FE L Y ++ + RW+ + V G++ +V + Y L D++ IE
Sbjct: 25 RATKWTPEENKQFENALALY-DKDEPDRWQKVAAVIPGKTVGDVIKQYRELEEDVSDIEA 83
Query: 68 GLVDFS--TNSDDFIISKASTDEN----------------KAPPTKNKTKKVVRVKHWTE 109
GL+ ++SD F + + ++ A ++ + KK V WTE
Sbjct: 84 GLIPIPGYSSSDAFTLEWFNNNQGYDGFRHYYTPGGKRTTAARSSEQERKKGVP---WTE 140
Query: 110 EEHRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNF 168
EEHR FL G++ +GKG W+ IS++ V TRT +QVASHAQK+F+ Q G + +
Sbjct: 141 EEHRQFLMGLQKYGKGDWRNISRNFVTTRTPTQVASHAQKYFIRQSTGGKDERRSSIHDI 200
Query: 169 YITSLKGNSKPLLN-KDNIPSPSTS 192
+L P + K + P ST+
Sbjct: 201 TTVNLPDTKSPSPDEKKSSPDHSTT 225
>UniRef100_Q9FFJ9 Similarity to I-box binding factor [Arabidopsis thaliana]
Length = 277
Score = 87.0 bits (214), Expect = 3e-16
Identities = 57/161 (35%), Positives = 85/161 (52%), Gaps = 9/161 (5%)
Query: 9 KSKEWTWDENKIFETILFEYLEEVQEGRWENIGLVCGRSSTEVKEHYETLLHDLALIEEG 68
+S WT +ENK FE L Y ++ + ++ ++ G++ ++V Y L DL IE G
Sbjct: 27 QSSSWTKEENKKFERALAVYADDTPDRWFKVAAMIPGKTISDVMRQYSKLEEDLFDIEAG 86
Query: 69 LV------DFSTNSDDFIISKASTDENKAPPTKNKTKKVVRVK--HWTEEEHRLFLEGIE 120
LV + D ++S D + P + R K WTEEEHR FL G+
Sbjct: 87 LVPIPGYRSVTPCGFDQVVSPRDFDAYRKLPNGARGFDQDRRKGVPWTEEEHRRFLLGLL 146
Query: 121 IHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKK 160
+GKG W+ IS++ V ++T +QVASHAQK++ QL G K
Sbjct: 147 KYGKGDWRNISRNFVGSKTPTQVASHAQKYYQRQLSGAKDK 187
>UniRef100_Q9LFB6 Hypothetical protein F7J8_180 [Arabidopsis thaliana]
Length = 267
Score = 86.3 bits (212), Expect = 5e-16
Identities = 64/177 (36%), Positives = 94/177 (52%), Gaps = 32/177 (18%)
Query: 13 WTWDENKIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALIEEGLVD 71
WT +ENK FE L ++ W I L+ G++ +V + Y+ L D++ IE GL+
Sbjct: 30 WTAEENKRFEKALAYLDDKDNLESWSKIADLIPGKTVADVIKRYKELEDDVSDIEAGLIP 89
Query: 72 F---------STNSD------------DFII-----SKASTDENKAPPTKNKTKKVVRVK 105
+ NSD D+++ S A TD ++P + + KK V
Sbjct: 90 IPGYGGDASSAANSDYFFGLENSSYGYDYVVGGKRSSPAMTDCFRSPMPEKERKKGVP-- 147
Query: 106 HWTEEEHRLFLEGIEIHGKGKWKLISQ-HVRTRTASQVASHAQKHFLHQL-DGTSKK 160
WTE+EH FL G++ +GKG W+ I++ V TRT +QVASHAQK+FL QL DG K+
Sbjct: 148 -WTEDEHLRFLMGLKKYGKGDWRNIAKSFVTTRTPTQVASHAQKYFLRQLTDGKDKR 203
>UniRef100_Q8GXN7 Putative MYB family transcription factor [Arabidopsis thaliana]
Length = 298
Score = 86.3 bits (212), Expect = 5e-16
Identities = 57/178 (32%), Positives = 90/178 (50%), Gaps = 27/178 (15%)
Query: 7 DGKSKEWTWDENKIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALI 65
+ + +WT +ENK FE L Y ++ + RW + ++ G++ +V + Y L D++ I
Sbjct: 23 ENRGTKWTAEENKKFENALAFYDKDTPD-RWSRVAAMLPGKTVGDVIKQYRELEEDVSDI 81
Query: 66 EEGLVDF----------------------STNSDDFIISKASTDENKAPPTKNKTKKVVR 103
E GL+ N + + S A A T +K +
Sbjct: 82 EAGLIPIPGYASDSFTLDWGGYDGASGNNGFNMNGYYFSAAGGKRGSAARTAEHERK--K 139
Query: 104 VKHWTEEEHRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKK 160
WTEEEHR FL G++ +GKG W+ I+++ V TRT +QVASHAQK+F+ Q++G K
Sbjct: 140 GVPWTEEEHRQFLMGLKKYGKGDWRNIARNFVTTRTPTQVASHAQKYFIRQVNGGKDK 197
>UniRef100_Q9M0P7 Hypothetical protein AT4g09450 [Arabidopsis thaliana]
Length = 200
Score = 84.0 bits (206), Expect = 3e-15
Identities = 57/163 (34%), Positives = 83/163 (49%), Gaps = 12/163 (7%)
Query: 12 EWTWDENKIFETILFEYLEEVQEGRWENIGLVCGRSSTEVKEHYETLLHDLALIEEGLVD 71
+WT ++K FE L + + E ENI + EV+ +Y L+HD+ IE G
Sbjct: 6 QWTRVDDKRFELALLQ-IPEGSPNFIENIAYYLQKPVKEVEYYYCALVHDIERIESGKYV 64
Query: 72 FSTNSDDFIISKASTDENKAPPTKNKTKKVVRVKHWTEEEHRLFLEGIEIHGKGKWKLIS 131
+D + E+K K W+EEE RLFLEG+ GKG WK IS
Sbjct: 65 LPKYPEDDYVKLTEAGESKGNGKKTGIP-------WSEEEQRLFLEGLNKFGKGDWKNIS 117
Query: 132 QH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITSL 173
++ V++RT++QVASHAQK+F Q + T KR + + +L
Sbjct: 118 RYCVKSRTSTQVASHAQKYFARQ---KQESTNTKRPSIHDMTL 157
>UniRef100_Q9C773 MYB-family transcription factor, putative; alternative splicing
isoform 2 of 2;71559-70643 [Arabidopsis thaliana]
Length = 263
Score = 83.2 bits (204), Expect = 4e-15
Identities = 55/158 (34%), Positives = 82/158 (51%), Gaps = 3/158 (1%)
Query: 6 NDGKSKEWTWDENKIFETILFEYLEEVQEGRWENIGLVCGRSSTEVKEHYETLLHDLALI 65
+ S WT +ENK+FE L Y E+ + ++ ++ G++ +V + Y L D+ I
Sbjct: 26 HSSSSGSWTKEENKMFERALAIYAEDSPDRWFKVASMIPGKTVFDVMKQYSKLEEDVFDI 85
Query: 66 EEGLVDFSTNSDDFIISKASTDENKAPPTKNKTKKVVRVKH--WTEEEHRLFLEGIEIHG 123
E G V TD + P+ + R K WTEEEHR FL G+ +G
Sbjct: 86 EAGRVPIPGYPAASSPLGFDTDMCRKRPSGARGSDQDRKKGVPWTEEEHRRFLLGLLKYG 145
Query: 124 KGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKK 160
KG W+ IS++ V ++T +QVASHAQK++ QL G K
Sbjct: 146 KGDWRNISRNFVVSKTPTQVASHAQKYYQRQLSGAKDK 183
>UniRef100_Q8W0G0 P0529E05.19 protein [Oryza sativa]
Length = 508
Score = 83.2 bits (204), Expect = 4e-15
Identities = 66/204 (32%), Positives = 103/204 (50%), Gaps = 38/204 (18%)
Query: 10 SKEWTWDENKIFETILFEYLEEVQEGRWENIGLVCGRSST-EVKEHYETLLHDLALIEEG 68
++ W+ +ENK+FE L + ++ RWE + + R + +V HY L +D+ IE G
Sbjct: 29 AEAWSAEENKVFERALAQ-VDLDSPNRWEMVAAMLPRKTVIDVMNHYRDLENDVGSIEAG 87
Query: 69 LVDFSTNSDDFIISKASTD--------------------ENKAPPTKNKTKKVVRVKHWT 108
LV F S +S AS+ +AP + K + WT
Sbjct: 88 LVPFPHYSSS--LSPASSGFTLQDWDGSDGGFRRGCYLKRGRAPDQERK-----KGVPWT 140
Query: 109 EEEHRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSN 167
EEEH+ FL G++ +G+G W+ IS++ V +RT +QVASHAQK+F+ G K+RS+
Sbjct: 141 EEEHKSFLMGLKKYGRGDWRNISRYFVTSRTPTQVASHAQKYFIRLNSGGKD---KRRSS 197
Query: 168 FY-ITSLKGNSKPLLNKDNIPSPS 190
+ IT++ + N PSPS
Sbjct: 198 IHDITTVNLPEEDTSN----PSPS 217
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.312 0.131 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 412,975,980
Number of Sequences: 2790947
Number of extensions: 17502963
Number of successful extensions: 38101
Number of sequences better than 10.0: 535
Number of HSP's better than 10.0 without gapping: 298
Number of HSP's successfully gapped in prelim test: 237
Number of HSP's that attempted gapping in prelim test: 37418
Number of HSP's gapped (non-prelim): 715
length of query: 216
length of database: 848,049,833
effective HSP length: 122
effective length of query: 94
effective length of database: 507,554,299
effective search space: 47710104106
effective search space used: 47710104106
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC146842.1


