

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146807.2 + phase: 0
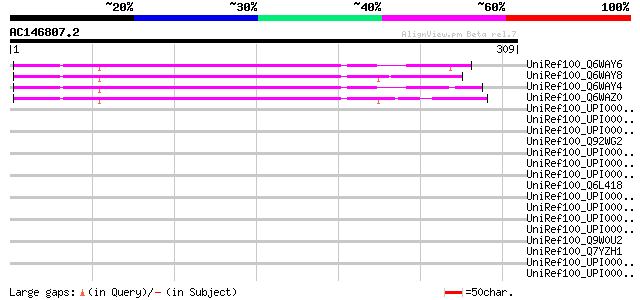
(309 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum] 166 7e-40
UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum] 164 3e-39
UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum] 163 5e-39
UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum] 163 6e-39
UniRef100_UPI0000360333 UPI0000360333 UniRef100 entry 39 0.18
UniRef100_UPI0000456379 UPI0000456379 UniRef100 entry 37 0.51
UniRef100_UPI0000246EF2 UPI0000246EF2 UniRef100 entry 37 0.87
UniRef100_Q92WG2 Putative cellulose synthase catalytic subunit p... 37 0.87
UniRef100_UPI0000365B62 UPI0000365B62 UniRef100 entry 36 1.1
UniRef100_UPI0000019A9C UPI0000019A9C UniRef100 entry 36 1.1
UniRef100_UPI00002F9E5E UPI00002F9E5E UniRef100 entry 36 1.1
UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demi... 36 1.1
UniRef100_UPI0000437B8E UPI0000437B8E UniRef100 entry 36 1.5
UniRef100_UPI00003625DE UPI00003625DE UniRef100 entry 36 1.5
UniRef100_UPI000021ACA2 UPI000021ACA2 UniRef100 entry 36 1.5
UniRef100_UPI00003C22F0 UPI00003C22F0 UniRef100 entry 35 1.9
UniRef100_Q9W0U2 CG7036-PA, isoform A [Drosophila melanogaster] 35 1.9
UniRef100_Q7YZH1 PHD zinc finger protein rhinoceros [Drosophila ... 35 1.9
UniRef100_UPI000004CC0D UPI000004CC0D UniRef100 entry 35 2.5
UniRef100_UPI0000319BA1 UPI0000319BA1 UniRef100 entry 35 2.5
>UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum]
Length = 562
Score = 166 bits (420), Expect = 7e-40
Identities = 102/284 (35%), Positives = 144/284 (49%), Gaps = 26/284 (9%)
Query: 3 AIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCVPLLYRWFISHLPSS--FHDNSEN 60
A+ IF NPVPTLLADTY+AIH R KG G I CC+PLL RWF+S LP S F D
Sbjct: 199 AVSIFIGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQST 257
Query: 61 WSYSQRIMALTPNEVVWITPTAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQFGFP 120
++QR+M+LT ++ W + V+ +I CG F NVPL+GTRG INYNP L++RQ GF
Sbjct: 258 HKWTQRVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFV 317
Query: 121 MKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKHLGVRSGIAHEAYTQWVIN 180
MK +P+ + ++ + + E AW + K LG + +A YT WV
Sbjct: 318 MKGRPLEAEVAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKE 377
Query: 181 RAEEIGMPYPAMRYVSASAPSIPLPLPPTTQGMYQEHLAMESREKQMWKARYNEAENLIM 240
R E + +PY M + PSI L + + + ME+R
Sbjct: 378 RVETLLLPYDRMEPLQEQPPSI---LAESVPAEHYKQALMENRR---------------- 418
Query: 241 TLDGKDEQKTHENLMLKKELVKVRREL---EEKDELLMRDSKRA 281
L GK++ E K + + + +L E+D +R KR+
Sbjct: 419 -LRGKEQDTQLELYKAKADKLNLAHQLRGVREEDASRLRSKKRS 461
>UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum]
Length = 562
Score = 164 bits (415), Expect = 3e-39
Identities = 101/285 (35%), Positives = 146/285 (50%), Gaps = 16/285 (5%)
Query: 3 AIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCVPLLYRWFISHLPSS--FHDNSEN 60
A+ IF NPVPTLLADTY+AIH R KG G I CC+PLL RWF+S LP S F D
Sbjct: 199 AVSIFIGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQST 257
Query: 61 WSYSQRIMALTPNEVVWITPTAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQFGFP 120
++QR+M+LT ++ W + V+ +I CG F NVPL+GTRG INYNP L++RQ GF
Sbjct: 258 HKWTQRVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLIGTRGCINYNPVLSLRQLGFV 317
Query: 121 MKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKHLGVRSGIAHEAYTQWVIN 180
MK +P+ + + + + E AW + K LG + +A YT WV
Sbjct: 318 MKGRPLEAEVAESVCFEKRSDPARLEQIGRAWKSIGMKDGSVLGKKFAVAMPDYTDWVKE 377
Query: 181 RAEEIGMPYPAMRYVSASAPSIPLPLPPTTQGMYQEHLAMESR---------EKQMWKAR 231
R E + +PY M + P I L + + + ME+R + +++KA+
Sbjct: 378 RVETLLLPYDRMEPLQEQPPLI---LAESVPAEHYKQALMENRRLREKEQDTQMELYKAK 434
Query: 232 YNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRELEEKDELLMR 276
++ NL L G E+ K+ +V L+ + +R
Sbjct: 435 -ADSLNLAHQLRGVREEDASRLRSKKRSYEEVESMLDAEHRECLR 478
>UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum]
Length = 546
Score = 163 bits (413), Expect = 5e-39
Identities = 101/288 (35%), Positives = 144/288 (49%), Gaps = 26/288 (9%)
Query: 3 AIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCVPLLYRWFISHLPSS--FHDNSEN 60
AI IF NPVPTLLADTY+AIH R KG G I CC+PLL RWF+S LP+S F D
Sbjct: 183 AISIFIGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFMSLLPASGPFMDTQST 241
Query: 61 WSYSQRIMALTPNEVVWITPTAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQFGFP 120
++QR+M+LT ++ W + VK+++ CG+F NVPL+GT+G INYNP L++RQ GF
Sbjct: 242 LKWTQRVMSLTSYDIRWQSYRMDVKDVVMSCGEFRNVPLVGTKGCINYNPVLSLRQLGFI 301
Query: 121 MKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKHLGVRSGIAHEAYTQWVIN 180
M +P+ ++ + E AW + K LG + IA YT WV
Sbjct: 302 MSRRPLEAEIVESVYFEKRTDPVRLEQIGRAWKSIGVKDGSVLGKKFAIAMPDYTDWVKK 361
Query: 181 RAEEIGMPYPAMRYVSASAPSIPLPLPPTTQGMYQEHLAMESREKQMWKARYNEAENLIM 240
R E + +PY M + P I L + + + ME+R
Sbjct: 362 RVETLLLPYDRMEPLQEQPPLI---LADSVPAEHYKQALMENRR---------------- 402
Query: 241 TLDGKDEQKTHENLMLKKELVKVRRELEEKDELLMRDSKRARGRRNFY 288
L K++ E K + + + +L E+ D+ RAR ++ Y
Sbjct: 403 -LREKEQDTRMELYKAKADKLNLAHQLR---EMQGEDASRARSKKRSY 446
>UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum]
Length = 562
Score = 163 bits (412), Expect = 6e-39
Identities = 104/300 (34%), Positives = 149/300 (49%), Gaps = 23/300 (7%)
Query: 3 AIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCVPLLYRWFISHLPSS--FHDNSEN 60
A+ IF NPVPTLLADTY+AIH R KG G I CC+PLL RWF+S LP S F D
Sbjct: 199 AVSIFIGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQST 257
Query: 61 WSYSQRIMALTPNEVVWITPTAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQFGFP 120
++QR+M+LT ++ W + V+ +I CG F NVPL+GTRG INYNP L++RQ GF
Sbjct: 258 HKWTQRVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFV 317
Query: 121 MKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKHLGVRSGIAHEAYTQWVIN 180
MK +P+ + ++ + + E AW + K LG + +A YT WV
Sbjct: 318 MKGRPLEAEIAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKE 377
Query: 181 RAEEIGMPYPAMRYVSASAPSIPLPLPPTTQGMYQEHLAMESR---------EKQMWKAR 231
R E + +PY M + P I L + + + ME+R + +++KA+
Sbjct: 378 RVETLLLPYDRMEPLQEQPPLI---LAESVPAEHYKQALMENRRLREKEQDTQMELYKAK 434
Query: 232 YNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRELEEKDELLMRDSKRARGRRNFYARY 291
NL L G E+ L +R EE + +L + + + A Y
Sbjct: 435 AGRL-NLAHQLRGVREEDA-------SRLRSKKRSYEEMESMLDAEHRECLRLQRVEASY 486
>UniRef100_UPI0000360333 UPI0000360333 UniRef100 entry
Length = 896
Score = 38.9 bits (89), Expect = 0.18
Identities = 29/86 (33%), Positives = 39/86 (44%), Gaps = 4/86 (4%)
Query: 198 SAPSIPLPLPPT---TQGMYQEHLAMESREKQMWKAR-YNEAENLIMTLDGKDEQKTHEN 253
+APS P + P T Y+ + +E K R Y E + D E N
Sbjct: 2 AAPSFPAKMSPIKALTMKDYENQITALKKENFNLKLRIYFMEERMQQKCDDSTEDIFKTN 61
Query: 254 LMLKKELVKVRRELEEKDELLMRDSK 279
+ LK EL ++REL EK ELL+ SK
Sbjct: 62 IELKVELESMKRELAEKQELLVSASK 87
>UniRef100_UPI0000456379 UPI0000456379 UniRef100 entry
Length = 602
Score = 37.4 bits (85), Expect = 0.51
Identities = 27/84 (32%), Positives = 41/84 (48%), Gaps = 11/84 (13%)
Query: 224 EKQMWKARYNEAENLIMTLDGKDEQKTHENLM---LKKELVKVRRELEEKDELLMRDSKR 280
E++ WK+R N AEN +TL EQ+ L LKKE +++ ELE + R
Sbjct: 305 EQKQWKSRTNGAENKTLTLAEYHEQEEIFKLRLGHLKKEEAEIQAELERLE--------R 356
Query: 281 ARGRRNFYARYCGSDSESESEDHP 304
R R + ++ S+ +DHP
Sbjct: 357 VRNLRIGELKRIHNEDNSQFKDHP 380
>UniRef100_UPI0000246EF2 UPI0000246EF2 UniRef100 entry
Length = 701
Score = 36.6 bits (83), Expect = 0.87
Identities = 25/76 (32%), Positives = 40/76 (51%), Gaps = 6/76 (7%)
Query: 222 SREKQMWKARYNEAENLIMTLDGKDEQKTHENLM---LKKELVKVRRELEEKD---ELLM 275
+ E++ WK++ N AEN +TL EQ+ L LKKE +++ ELE + +L +
Sbjct: 308 TNEQKQWKSKTNGAENETLTLKEYHEQEEIFKLRLGHLKKEEAEIQAELERLERVRKLHI 367
Query: 276 RDSKRARGRRNFYARY 291
R+ KR N +Y
Sbjct: 368 REVKRIHNEDNSQFKY 383
>UniRef100_Q92WG2 Putative cellulose synthase catalytic subunit protein [Rhizobium
meliloti]
Length = 664
Score = 36.6 bits (83), Expect = 0.87
Identities = 26/92 (28%), Positives = 41/92 (44%), Gaps = 7/92 (7%)
Query: 34 YILCCVPLLYRWFISHLPSSFHDNSENWSYSQRIMALTPNEVVWITPTAQVKEIITGCGD 93
+I+ +P++Y WF LP F D ++ S ++A + W+TPT + + T G
Sbjct: 380 FIVLLIPIVYLWF-GALPLYFTDVADYVSNQVPLLAAYFLLMFWLTPTRYLPLVSTAVGT 438
Query: 94 FLNVPLLGTRGGINYNPELAMRQFGFPMKAKP 125
F +L T +R FG P K P
Sbjct: 439 FSTFRMLPT------VLSSLVRPFGKPFKVTP 464
>UniRef100_UPI0000365B62 UPI0000365B62 UniRef100 entry
Length = 664
Score = 36.2 bits (82), Expect = 1.1
Identities = 31/123 (25%), Positives = 56/123 (45%), Gaps = 44/123 (35%)
Query: 161 KHLGVRSGI-AHEAYTQWVINRAEEIGMPYPAMRYVSASAPSIPLPLPPTTQGMYQEHLA 219
+HL +R+ + AHEA+T+W + S+SAP P P+P + + E +A
Sbjct: 486 EHLCIRAYLEAHEAFTEWFSH---------------SSSAPQKPAPVP---EAKFTEKVA 527
Query: 220 MESREKQ------MWKARYN--------EAENLIMTLDG-----------KDEQKTHENL 254
E REK+ W +R + N+++ +DG +D +++H+
Sbjct: 528 NEMREKEYEVSLAAWSSRLDVLTEDVKERIYNVLLFVDGGWMVDNRQDSEQDSERSHQMA 587
Query: 255 MLK 257
ML+
Sbjct: 588 MLR 590
>UniRef100_UPI0000019A9C UPI0000019A9C UniRef100 entry
Length = 811
Score = 36.2 bits (82), Expect = 1.1
Identities = 31/123 (25%), Positives = 56/123 (45%), Gaps = 44/123 (35%)
Query: 161 KHLGVRSGI-AHEAYTQWVINRAEEIGMPYPAMRYVSASAPSIPLPLPPTTQGMYQEHLA 219
+HL +R+ + AHEA+T+W + S+SAP P P+P + + E +A
Sbjct: 633 EHLCIRAYLEAHEAFTEWFSH---------------SSSAPQKPAPVP---EAKFTEKVA 674
Query: 220 MESREKQ------MWKARYN--------EAENLIMTLDG-----------KDEQKTHENL 254
E REK+ W +R + N+++ +DG +D +++H+
Sbjct: 675 NEMREKEYEVSLAAWSSRLDVLTEDVKERIYNVLLFVDGGWMVDNRQDSEQDSERSHQMA 734
Query: 255 MLK 257
ML+
Sbjct: 735 MLR 737
>UniRef100_UPI00002F9E5E UPI00002F9E5E UniRef100 entry
Length = 687
Score = 36.2 bits (82), Expect = 1.1
Identities = 24/104 (23%), Positives = 45/104 (43%), Gaps = 13/104 (12%)
Query: 211 QGMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENL-------------MLK 257
+G E + ++K+ K + E + +GK E K EN+ M K
Sbjct: 93 EGEKTEKKGSKKKKKEAKKKAEKKDEEKVKEAEGKKETKEEENIDKKVAKRKEKEDKMKK 152
Query: 258 KELVKVRRELEEKDELLMRDSKRARGRRNFYARYCGSDSESESE 301
KE K +R+ EE++ + ++ ++A+ + AR + E E
Sbjct: 153 KEEEKAKRKAEEEERIKKKEEEKAKKKEEEKAREAEKAKKKEEE 196
>UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demissum]
Length = 607
Score = 36.2 bits (82), Expect = 1.1
Identities = 49/194 (25%), Positives = 66/194 (33%), Gaps = 33/194 (17%)
Query: 13 VPTLLADTYHAIHDRTLKGRGYILCCVPLLYRWFISHL---------------------- 50
VP +LAD Y A+ G Y C LL W I H+
Sbjct: 214 VPMILADIYRAL-TICKNGGDYFEGCNMLLQLWMIEHIRHHPYVVDFKVECNDYIGGHEE 272
Query: 51 ---PSSFHDNSENWSYSQRIMALTPNEVVWITPTAQVKEIITGCGDFLNVPLLGTRGGIN 107
SF E W + + LT +++VW E+I + L+G RG
Sbjct: 273 RIKDHSFPKGIEAWK--KYLNNLTADKIVWNYHWFPSAEVIYMSTFRSFIVLMGLRGVQP 330
Query: 108 YNPELAMRQFGFPMKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKHLGVR- 166
Y P MRQ G P + ++ P + E F W C S H V
Sbjct: 331 YMPLRVMRQLGRRQFLPPNEDIREFMYEFHPEIPLRKSEIF-KIWGG-CMLSSPHDRVED 388
Query: 167 --SGIAHEAYTQWV 178
G +AY +WV
Sbjct: 389 RTKGEVDQAYLEWV 402
>UniRef100_UPI0000437B8E UPI0000437B8E UniRef100 entry
Length = 786
Score = 35.8 bits (81), Expect = 1.5
Identities = 26/86 (30%), Positives = 40/86 (46%), Gaps = 2/86 (2%)
Query: 196 SASAPSIPL-PLPPTTQGM-YQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHEN 253
S SAP+ P P PP E S E++ + + +E E L E+K E
Sbjct: 190 SKSAPTTPASPAPPIAATCPLSEEKTKTSEEQEEKQRQRDEKERLKQEAKAAKEKKKEEA 249
Query: 254 LMLKKELVKVRRELEEKDELLMRDSK 279
+K+E + ++E +EKDE R+ K
Sbjct: 250 RKMKEEKEREKKEKKEKDEKERREKK 275
>UniRef100_UPI00003625DE UPI00003625DE UniRef100 entry
Length = 1970
Score = 35.8 bits (81), Expect = 1.5
Identities = 20/62 (32%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query: 211 QGMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRELEEK 270
Q + +E+ ++S E WK +Y+ + L+ D E++ E LK+E+ K+RRE E
Sbjct: 1379 QNLMEENQRLKS-EDGYWKDQYDGKQALLRQYDTDKERQEMEKNSLKREIEKLRREQREI 1437
Query: 271 DE 272
+E
Sbjct: 1438 EE 1439
>UniRef100_UPI000021ACA2 UPI000021ACA2 UniRef100 entry
Length = 871
Score = 35.8 bits (81), Expect = 1.5
Identities = 25/85 (29%), Positives = 44/85 (51%), Gaps = 3/85 (3%)
Query: 225 KQMWKARYNEAENLIMTLDG--KDEQKTHENLMLKKELVKVRRELEEKDELLMRDSKRAR 282
+++ KAR E + L + G K+++K + + +E + R E E K E RDSK ++
Sbjct: 529 EELEKAREREVKELHDKIKGQQKEQEKARKEEIKNEERERKRLEKEAKMEQ-KRDSKESQ 587
Query: 283 GRRNFYARYCGSDSESESEDHPTTS 307
GR +F ++ +D + E TS
Sbjct: 588 GRSHFLSKKTTNDQDDPQEPTDMTS 612
>UniRef100_UPI00003C22F0 UPI00003C22F0 UniRef100 entry
Length = 176
Score = 35.4 bits (80), Expect = 1.9
Identities = 28/100 (28%), Positives = 48/100 (48%), Gaps = 4/100 (4%)
Query: 190 PAMRYVSASAPSIPLPLPPTTQGMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQK 249
PA + S +PL + G+ + A RE++M KAR + LI T+DG+D+
Sbjct: 77 PASKLASFQGAYLPLLRTHLSNGLKKRDKA---RERKMDKAREQSRKKLIETVDGRDKII 133
Query: 250 THENLMLKKELVKVRRELEEKDELLMRDSKRARGRRNFYA 289
T++ + K+ + +R+ K L +R K +R A
Sbjct: 134 TNK-MGSKRGAGRRKRQRALKKGLALRKEKSKEAKRKVQA 172
>UniRef100_Q9W0U2 CG7036-PA, isoform A [Drosophila melanogaster]
Length = 3201
Score = 35.4 bits (80), Expect = 1.9
Identities = 26/86 (30%), Positives = 38/86 (43%), Gaps = 7/86 (8%)
Query: 196 SASAPSIPLPLPPTTQGMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLM 255
+A P+ P P P + M + R + W++R DE+ TH
Sbjct: 1649 AAPLPASPTPTPTSNDEMSDAGSDLSERRRMRWRSRRRRRRR----SHEPDEEHTHHTQH 1704
Query: 256 LKKELVKVRRELEE--KDELLMRDSK 279
L E+ ++ RELEE K+ELL SK
Sbjct: 1705 LLNEM-EMARELEEERKNELLANASK 1729
>UniRef100_Q7YZH1 PHD zinc finger protein rhinoceros [Drosophila melanogaster]
Length = 3241
Score = 35.4 bits (80), Expect = 1.9
Identities = 26/86 (30%), Positives = 38/86 (43%), Gaps = 7/86 (8%)
Query: 196 SASAPSIPLPLPPTTQGMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLM 255
+A P+ P P P + M + R + W++R DE+ TH
Sbjct: 1689 AAPLPASPTPTPTSNDEMSDAGSDLSERRRMRWRSRRRRRRR----SHEPDEEHTHHTQH 1744
Query: 256 LKKELVKVRRELEE--KDELLMRDSK 279
L E+ ++ RELEE K+ELL SK
Sbjct: 1745 LLNEM-EMARELEEERKNELLANASK 1769
>UniRef100_UPI000004CC0D UPI000004CC0D UniRef100 entry
Length = 364
Score = 35.0 bits (79), Expect = 2.5
Identities = 20/76 (26%), Positives = 35/76 (45%)
Query: 207 PPTTQGMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRE 266
PP +E+ ESR ++ K+ +N+ ++ QKT +NL K ++ R
Sbjct: 167 PPVDLSSDEEYYVEESRSARLRKSGKEHIDNIKKAFSKENMQKTRQNLDKKVNRIRTRIV 226
Query: 267 LEEKDELLMRDSKRAR 282
E+ E L + +R R
Sbjct: 227 TPERRERLRQSGERLR 242
>UniRef100_UPI0000319BA1 UPI0000319BA1 UniRef100 entry
Length = 127
Score = 35.0 bits (79), Expect = 2.5
Identities = 24/93 (25%), Positives = 46/93 (48%), Gaps = 10/93 (10%)
Query: 93 DFLNVPLLGTRGGINYNPELAMRQFGFPMKAKPI-----NLATSPEFFYYSNAPTGQREA 147
D + +LGTRGG+ + + + +G +K K + +LA+ E ++++ G +A
Sbjct: 39 DNFSCEVLGTRGGVTWPDAIITKPYGNGVKRKKLGIEKPSLASVAELIHFTSLIEGNEKA 98
Query: 148 FIGAWSKVCRKSVKHLGVRSGIAHEAYTQWVIN 180
I A +SV+ +G+ + A T+ IN
Sbjct: 99 IIPA-----HQSVELIGIIEALYKSAITREKIN 126
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.134 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 562,805,603
Number of Sequences: 2790947
Number of extensions: 24824036
Number of successful extensions: 72742
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 43
Number of HSP's that attempted gapping in prelim test: 72674
Number of HSP's gapped (non-prelim): 96
length of query: 309
length of database: 848,049,833
effective HSP length: 127
effective length of query: 182
effective length of database: 493,599,564
effective search space: 89835120648
effective search space used: 89835120648
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146807.2


