

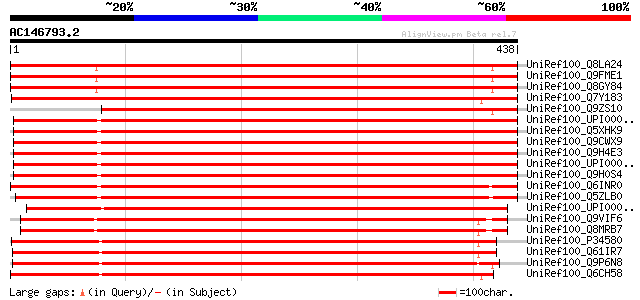
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146793.2 - phase: 0
(438 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LA24 Replication protein A1-like [Arabidopsis thaliana] 648 0.0
UniRef100_Q9FME1 ATP-dependent RNA helicase-like protein [Arabid... 647 0.0
UniRef100_Q8GY84 Putative replication protein A1 [Arabidopsis th... 647 0.0
UniRef100_Q7Y183 Putative RNA helicase [Oryza sativa] 568 e-160
UniRef100_Q9ZS10 RNA helicase [Arabidopsis thaliana] 553 e-156
UniRef100_UPI000021D62A UPI000021D62A UniRef100 entry 549 e-155
UniRef100_Q5XHK9 Hypothetical protein [Xenopus laevis] 549 e-155
UniRef100_Q9CWX9 DEAD-box protein 47 [Mus musculus] 549 e-155
UniRef100_Q9H4E3 DEAD box protein [Homo sapiens] 547 e-154
UniRef100_UPI000036F389 UPI000036F389 UniRef100 entry 545 e-153
UniRef100_Q9H0S4 DEAD-box protein 47 [Homo sapiens] 545 e-153
UniRef100_Q6INR0 MGC81303 protein [Xenopus laevis] 541 e-152
UniRef100_Q5ZLB0 Hypothetical protein [Gallus gallus] 534 e-150
UniRef100_UPI00003636F9 UPI00003636F9 UniRef100 entry 533 e-150
UniRef100_Q9VIF6 CG9253-PA [Drosophila melanogaster] 518 e-145
UniRef100_Q8MRB7 RE27528p [Drosophila melanogaster] 518 e-145
UniRef100_P34580 Putative ATP-dependent RNA helicase T26G10.1 in... 498 e-139
UniRef100_Q61IR7 Hypothetical protein CBG10097 [Caenorhabditis b... 493 e-138
UniRef100_Q9P6N8 SPAC823.08c protein [Schizosaccharomyces pombe] 493 e-138
UniRef100_Q6CH58 Yarrowia lipolytica chromosome A of strain CLIB... 486 e-136
>UniRef100_Q8LA24 Replication protein A1-like [Arabidopsis thaliana]
Length = 456
Score = 648 bits (1672), Expect = 0.0
Identities = 332/456 (72%), Positives = 386/456 (83%), Gaps = 18/456 (3%)
Query: 1 MEEENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKT 60
MEEEN+ +K+F +LG+ E LV+ACE++GWK P +IQ EA+P ALEGKD+IGLA+TGSGKT
Sbjct: 1 MEEENEVVKTFAELGVREELVKACERLGWKNPSKIQAEALPFALEGKDVIGLAQTGSGKT 60
Query: 61 GAFALPILHALLE---------APRPNH-FFACVMSPTRELAIQISEQFEALGSEIGVKC 110
GAFA+PIL ALLE RP+ FFACV+SPTRELAIQI+EQFEALG++I ++C
Sbjct: 61 GAFAIPILQALLEYVYDSEPKKGRRPDPAFFACVLSPTRELAIQIAEQFEALGADISLRC 120
Query: 111 AVLVGGIDMVQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNED 170
AVL GGID +QQ++ + K PH+IV TPGR+ DH+ +TKGFSL LKYLVLDEADRLLNED
Sbjct: 121 AVLFGGIDRMQQTIALGKRPHVIVATPGRLWDHMSDTKGFSLKSLKYLVLDEADRLLNED 180
Query: 171 FEESLNEILGMIPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYR 230
FE+SLN+IL IPRER+TFLFSATMT KV KLQR CLRNPVKIE +SKYSTVDTLKQQYR
Sbjct: 181 FEKSLNQILEEIPRERKTFLFSATMTKKVRKLQRACLRNPVKIEAASKYSTVDTLKQQYR 240
Query: 231 FLPAKHKDCYLVYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRL 290
F+ AK+KDCYLVYILSEM STSM+FTRTCD TR LAL+LR+LG +AIPI+G M+Q KRL
Sbjct: 241 FVAAKYKDCYLVYILSEMPESTSMIFTRTCDGTRFLALVLRSLGFRAIPISGQMTQSKRL 300
Query: 291 GALNKFKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVA 350
GALNKFK+G+CNIL+CTDVASRGLDIP+VD+VINYDIPTNSKDYIHRVGRTARAGRSGV
Sbjct: 301 GALNKFKAGECNILVCTDVASRGLDIPSVDVVINYDIPTNSKDYIHRVGRTARAGRSGVG 360
Query: 351 ISLVNQYELEWYVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRR 410
ISLVNQYELEWY+QIEKLIGKKLPEYPA E+EVL L ERV EAK+L+A MKESGG+KRR
Sbjct: 361 ISLVNQYELEWYIQIEKLIGKKLPEYPAEEDEVLSLLERVAEAKKLSAMNMKESGGRKRR 420
Query: 411 GEGDI-------GEEDDVDK-YFGLKDRKSSKKFRR 438
GE D G +D +K G KD+KSSKKF+R
Sbjct: 421 GEDDEESERFLGGNKDRGNKERGGNKDKKSSKKFKR 456
>UniRef100_Q9FME1 ATP-dependent RNA helicase-like protein [Arabidopsis thaliana]
Length = 456
Score = 647 bits (1670), Expect = 0.0
Identities = 332/456 (72%), Positives = 386/456 (83%), Gaps = 18/456 (3%)
Query: 1 MEEENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKT 60
MEEEN+ +K+F +LG+ E LV+ACE++GWK P +IQ EA+P ALEGKD+IGLA+TGSGKT
Sbjct: 1 MEEENEVVKTFAELGVREELVKACERLGWKNPSKIQAEALPFALEGKDVIGLAQTGSGKT 60
Query: 61 GAFALPILHALLE---------APRPNH-FFACVMSPTRELAIQISEQFEALGSEIGVKC 110
GAFA+PIL ALLE RP+ FFACV+SPTRELAIQI+EQFEALG++I ++C
Sbjct: 61 GAFAIPILQALLEYVYDSEPKKGRRPDPAFFACVLSPTRELAIQIAEQFEALGADISLRC 120
Query: 111 AVLVGGIDMVQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNED 170
AVLVGGID +QQ++ + K PH+IV TPGR+ DH+ +TKGFSL LKYLVLDEADRLLNED
Sbjct: 121 AVLVGGIDRMQQTIALGKRPHVIVATPGRLWDHMSDTKGFSLKSLKYLVLDEADRLLNED 180
Query: 171 FEESLNEILGMIPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYR 230
FE+SLN+IL IP ER+TFLFSATMT KV KLQR CLRNPVKIE +SKYSTVDTLKQQYR
Sbjct: 181 FEKSLNQILEEIPLERKTFLFSATMTKKVRKLQRACLRNPVKIEAASKYSTVDTLKQQYR 240
Query: 231 FLPAKHKDCYLVYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRL 290
F+ AK+KDCYLVYILSEM STSM+FTRTCD TR LAL+LR+LG +AIPI+G M+Q KRL
Sbjct: 241 FVAAKYKDCYLVYILSEMPESTSMIFTRTCDGTRFLALVLRSLGFRAIPISGQMTQSKRL 300
Query: 291 GALNKFKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVA 350
GALNKFK+G+CNIL+CTDVASRGLDIP+VD+VINYDIPTNSKDYIHRVGRTARAGRSGV
Sbjct: 301 GALNKFKAGECNILVCTDVASRGLDIPSVDVVINYDIPTNSKDYIHRVGRTARAGRSGVG 360
Query: 351 ISLVNQYELEWYVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRR 410
ISLVNQYELEWY+QIEKLIGKKLPEYPA E+EVL L ERV EAK+L+A MKESGG+KRR
Sbjct: 361 ISLVNQYELEWYIQIEKLIGKKLPEYPAEEDEVLSLLERVAEAKKLSAMNMKESGGRKRR 420
Query: 411 GEGDI-------GEEDDVDK-YFGLKDRKSSKKFRR 438
GE D G +D +K G KD+KSSKKF+R
Sbjct: 421 GEDDEESERFLGGNKDRGNKERGGNKDKKSSKKFKR 456
>UniRef100_Q8GY84 Putative replication protein A1 [Arabidopsis thaliana]
Length = 456
Score = 647 bits (1668), Expect = 0.0
Identities = 332/456 (72%), Positives = 385/456 (83%), Gaps = 18/456 (3%)
Query: 1 MEEENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKT 60
MEEEN+ +K+F +LG+ E LV+ACE++GWK P +IQ EA+P ALEGKD+IGLA+TGSGKT
Sbjct: 1 MEEENEVVKTFAELGVREELVKACERLGWKNPSKIQAEALPFALEGKDVIGLAQTGSGKT 60
Query: 61 GAFALPILHALLE---------APRPNH-FFACVMSPTRELAIQISEQFEALGSEIGVKC 110
GAFA+PIL ALLE RP+ FFACV+SPTRELAIQI+EQFEALG++I ++C
Sbjct: 61 GAFAIPILQALLEYVYDSEPKKGRRPDPAFFACVLSPTRELAIQIAEQFEALGADISLRC 120
Query: 111 AVLVGGIDMVQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNED 170
AVLVGGID +QQ++ + K PH+IV TPGR+ DH+ +TKGFSL LKYLVLDEADRLLNED
Sbjct: 121 AVLVGGIDRMQQTIALGKRPHVIVATPGRLWDHMSDTKGFSLKSLKYLVLDEADRLLNED 180
Query: 171 FEESLNEILGMIPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYR 230
FE+SLN+IL IP ER TFLFSATMT KV KLQR CLRNPVKIE +SKYSTVDTLKQQYR
Sbjct: 181 FEKSLNQILEEIPLERETFLFSATMTKKVRKLQRACLRNPVKIEAASKYSTVDTLKQQYR 240
Query: 231 FLPAKHKDCYLVYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRL 290
F+ AK+KDCYLVYILSEM STSM+FTRTCD TR LAL+LR+LG +AIPI+G M+Q KRL
Sbjct: 241 FVAAKYKDCYLVYILSEMPESTSMIFTRTCDGTRFLALVLRSLGFRAIPISGQMTQSKRL 300
Query: 291 GALNKFKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVA 350
GALNKFK+G+CNIL+CTDVASRGLDIP+VD+VINYDIPTNSKDYIHRVGRTARAGRSGV
Sbjct: 301 GALNKFKAGECNILVCTDVASRGLDIPSVDVVINYDIPTNSKDYIHRVGRTARAGRSGVG 360
Query: 351 ISLVNQYELEWYVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRR 410
ISLVNQYELEWY+QIEKLIGKKLPEYPA E+EVL L ERV EAK+L+A MKESGG+KRR
Sbjct: 361 ISLVNQYELEWYIQIEKLIGKKLPEYPAEEDEVLSLLERVAEAKKLSAMNMKESGGRKRR 420
Query: 411 GEGDI-------GEEDDVDK-YFGLKDRKSSKKFRR 438
GE D G +D +K G KD+KSSKKF+R
Sbjct: 421 GEDDEESERFLGGNKDRGNKERGGNKDKKSSKKFKR 456
>UniRef100_Q7Y183 Putative RNA helicase [Oryza sativa]
Length = 472
Score = 568 bits (1464), Expect = e-160
Identities = 279/439 (63%), Positives = 349/439 (78%), Gaps = 2/439 (0%)
Query: 2 EEENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTG 61
E + +F +LG+ LV AC+ +GWK P IQ EAIP ALEG+DLIGL +TGSGKTG
Sbjct: 34 EPPARRPSTFAELGVVPELVAACDAMGWKEPTRIQAEAIPHALEGRDLIGLGQTGSGKTG 93
Query: 62 AFALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQ 121
AFALPI+ ALL+ +P FACV+SPTRELA QI +QFEALGS IG+ C VLVGG+D VQ
Sbjct: 94 AFALPIIQALLKQDKPQALFACVLSPTRELAFQIGQQFEALGSAIGLSCTVLVGGVDRVQ 153
Query: 122 QSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGM 181
Q+V +AK PHI+VGTPGR+LDHL +TKGFSL +LKYLVLDEAD+LLN +F+++L++IL +
Sbjct: 154 QAVSLAKRPHIVVGTPGRLLDHLTDTKGFSLNKLKYLVLDEADKLLNVEFQKALDDILNV 213
Query: 182 IPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYL 241
IP+ERRTFLFSATMTNKV KLQR CLRNPVK+E +SKYSTVDTL+Q++ F+PA +KDC+L
Sbjct: 214 IPKERRTFLFSATMTNKVSKLQRACLRNPVKVEVASKYSTVDTLRQEFYFVPADYKDCFL 273
Query: 242 VYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDC 301
V++L+E+ GS M+F RTC+STRLLAL LRNL KAI I+G MSQ KRLGALN+FK+ DC
Sbjct: 274 VHVLNELPGSMIMIFVRTCESTRLLALTLRNLRFKAISISGQMSQDKRLGALNRFKTKDC 333
Query: 302 NILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEW 361
NIL+CTDVASRGLDI VD+VINYDIP NSKDY+HRVGRTARAG +G A+SLVNQYE W
Sbjct: 334 NILICTDVASRGLDIQGVDVVINYDIPMNSKDYVHRVGRTARAGNTGYAVSLVNQYEAMW 393
Query: 362 YVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGG--KKRRGEGDIGEED 419
+ IEKL+G ++P+ + E+++L ER+ ++KR+A T MKE GG KKRR D EE+
Sbjct: 394 FKMIEKLLGYEIPDRKVDNAEIMILRERISDSKRIALTTMKEGGGHKKKRRKNEDDEEEE 453
Query: 420 DVDKYFGLKDRKSSKKFRR 438
+ + K + +K RR
Sbjct: 454 ERNAPVSRKSKSFNKSRRR 472
>UniRef100_Q9ZS10 RNA helicase [Arabidopsis thaliana]
Length = 376
Score = 553 bits (1425), Expect = e-156
Identities = 279/367 (76%), Positives = 319/367 (86%), Gaps = 8/367 (2%)
Query: 80 FFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIAKLPHIIVGTPGR 139
FFACV+SPTRELAIQI+EQFEALG++I ++CAVLVGGID +QQ++ + K PH+IV TPGR
Sbjct: 10 FFACVLSPTRELAIQIAEQFEALGADISLRCAVLVGGIDRMQQTIALGKRPHVIVATPGR 69
Query: 140 VLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRERRTFLFSATMTNKV 199
+ DH+ +TKGFSL LKYLVLDEADRLLNEDFE+SLN+IL IP ER+TFLFSATMT KV
Sbjct: 70 LWDHMSDTKGFSLKSLKYLVLDEADRLLNEDFEKSLNQILEEIPLERKTFLFSATMTKKV 129
Query: 200 EKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYILSEMAGSTSMVFTRT 259
KLQR CLRNPVKIE +SKYSTVDTLKQQYRF+ AK+KDCYLVYILSEM STSM+FTRT
Sbjct: 130 RKLQRACLRNPVKIEAASKYSTVDTLKQQYRFVAAKYKDCYLVYILSEMPESTSMIFTRT 189
Query: 260 CDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILLCTDVASRGLDIPAV 319
CD TR LAL+LR+LG +AIPI+G M+Q KRLGALNKFK+G+CNIL+CTDVASRGLDIP+V
Sbjct: 190 CDGTRFLALVLRSLGFRAIPISGQMTQSKRLGALNKFKAGECNILVCTDVASRGLDIPSV 249
Query: 320 DMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQIEKLIGKKLPEYPAN 379
D+VINYDIPTNSKDYIHRVGRTARAGRSGV ISLVNQYELEWY+QIEKLIGKKLPEYPA
Sbjct: 250 DVVINYDIPTNSKDYIHRVGRTARAGRSGVGISLVNQYELEWYIQIEKLIGKKLPEYPAE 309
Query: 380 EEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDI-------GEEDDVDK-YFGLKDRK 431
E+EVL L ERV EAK+L+A MKESGG+KRRGE D G +D +K G KD+K
Sbjct: 310 EDEVLSLLERVAEAKKLSAMNMKESGGRKRRGEDDEESERFLGGNKDRGNKERGGNKDKK 369
Query: 432 SSKKFRR 438
SSKKF+R
Sbjct: 370 SSKKFKR 376
>UniRef100_UPI000021D62A UPI000021D62A UniRef100 entry
Length = 455
Score = 549 bits (1415), Expect = e-155
Identities = 270/435 (62%), Positives = 353/435 (81%), Gaps = 2/435 (0%)
Query: 4 ENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAF 63
E +E K+FKDLG+ + L EAC+++GW P +IQIEAIP AL+G+D+IGLA+TGSGKTGAF
Sbjct: 19 EEEETKTFKDLGVTDVLCEACDQLGWAKPTKIQIEAIPLALQGRDIIGLAETGSGKTGAF 78
Query: 64 ALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
ALPIL+ALLE P+ FA V++PTRELA QISEQFEALGS IGV+CAV+VGGID + QS
Sbjct: 79 ALPILNALLETPQ--RLFALVLTPTRELAFQISEQFEALGSSIGVQCAVIVGGIDSMSQS 136
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
+ +AK PHI++ TPGR++DHL+NTKGF+L LKYLV+DEADR+LN DFE +++IL +IP
Sbjct: 137 LALAKKPHIVIATPGRLIDHLENTKGFNLRALKYLVMDEADRILNMDFETEVDKILKVIP 196
Query: 184 RERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVY 243
R+R+TFLFSATMT KV+KLQR L+NPVK SSKY TV+ L+Q Y F+P+K KD YLVY
Sbjct: 197 RDRKTFLFSATMTKKVQKLQRAALKNPVKCAVSSKYQTVEKLQQYYLFIPSKFKDTYLVY 256
Query: 244 ILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNI 303
IL+E+AG++ M+F TC++T+ AL+LRNLG AIP++G MSQ KRLG+LNKFK+ +I
Sbjct: 257 ILNELAGNSFMIFCSTCNNTQRTALLLRNLGFTAIPLHGQMSQSKRLGSLNKFKAKARSI 316
Query: 304 LLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYV 363
LL TDVASRGLDIP VD+V+N+DIPT+SKDYIHRVGRTARAGRSG AI+ V QY++E +
Sbjct: 317 LLATDVASRGLDIPHVDVVVNFDIPTHSKDYIHRVGRTARAGRSGKAITFVTQYDVELFQ 376
Query: 364 QIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDDVDK 423
+IE LIGKKLP +P +EEV++L ERV EA+R A +++E G KK+R D G++DD +
Sbjct: 377 RIEHLIGKKLPVFPTQDEEVMMLTERVSEAQRFARMELREHGEKKKRKREDPGDDDDTEG 436
Query: 424 YFGLKDRKSSKKFRR 438
G++++ + K ++
Sbjct: 437 AIGVRNKVAGGKMKK 451
>UniRef100_Q5XHK9 Hypothetical protein [Xenopus laevis]
Length = 453
Score = 549 bits (1415), Expect = e-155
Identities = 270/435 (62%), Positives = 353/435 (81%), Gaps = 2/435 (0%)
Query: 4 ENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAF 63
E +E K+FKDLG+ + L EAC+++GW P +IQIEAIP AL+G+D+IGLA+TGSGKTGAF
Sbjct: 17 EEEETKTFKDLGVTDVLCEACDQLGWAKPTKIQIEAIPLALQGRDIIGLAETGSGKTGAF 76
Query: 64 ALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
ALPIL+ALLE P+ FA V++PTRELA QISEQFEALGS IGV+CAV+VGGID + QS
Sbjct: 77 ALPILNALLETPQ--RLFALVLTPTRELAFQISEQFEALGSSIGVQCAVIVGGIDSMSQS 134
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
+ +AK PHI++ TPGR++DHL+NTKGF+L LKYLV+DEADR+LN DFE +++IL +IP
Sbjct: 135 LALAKKPHIVIATPGRLIDHLENTKGFNLRALKYLVMDEADRILNMDFETEVDKILKVIP 194
Query: 184 RERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVY 243
R+R+TFLFSATMT KV+KLQR L+NPVK SSKY TV+ L+Q Y F+P+K KD YLVY
Sbjct: 195 RDRKTFLFSATMTKKVQKLQRAALKNPVKCAVSSKYQTVEKLQQYYLFIPSKFKDTYLVY 254
Query: 244 ILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNI 303
IL+E+AG++ M+F TC++T+ AL+LRNLG AIP++G MSQ KRLG+LNKFK+ +I
Sbjct: 255 ILNELAGNSFMIFCSTCNNTQRTALLLRNLGFTAIPLHGQMSQSKRLGSLNKFKAKARSI 314
Query: 304 LLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYV 363
LL TDVASRGLDIP VD+V+N+DIPT+SKDYIHRVGRTARAGRSG AI+ V QY++E +
Sbjct: 315 LLATDVASRGLDIPHVDVVVNFDIPTHSKDYIHRVGRTARAGRSGKAITFVTQYDVELFQ 374
Query: 364 QIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDDVDK 423
+IE LIGKKLP +P +EEV++L ERV EA+R A +++E G KK+R D G++DD +
Sbjct: 375 RIEHLIGKKLPVFPTQDEEVMMLTERVNEAQRFARMELREHGEKKKRKREDAGDDDDKEG 434
Query: 424 YFGLKDRKSSKKFRR 438
G++++ + K ++
Sbjct: 435 AIGVRNKVAGGKMKK 449
>UniRef100_Q9CWX9 DEAD-box protein 47 [Mus musculus]
Length = 455
Score = 549 bits (1415), Expect = e-155
Identities = 270/435 (62%), Positives = 353/435 (81%), Gaps = 2/435 (0%)
Query: 4 ENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAF 63
E +E K+FKDLG+ + L EAC+++GW P +IQIEAIP AL+G+D+IGLA+TGSGKTGAF
Sbjct: 19 EEEETKTFKDLGVTDVLCEACDQLGWAKPTKIQIEAIPLALQGRDIIGLAETGSGKTGAF 78
Query: 64 ALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
ALPIL+ALLE P+ FA V++PTRELA QISEQFEALGS IGV+CAV+VGGID + QS
Sbjct: 79 ALPILNALLETPQ--RLFALVLTPTRELAFQISEQFEALGSSIGVQCAVIVGGIDSMSQS 136
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
+ +AK PHI++ TPGR++DHL+NTKGF+L LKYLV+DEADR+LN DFE +++IL +IP
Sbjct: 137 LALAKKPHIVIATPGRLIDHLENTKGFNLRALKYLVMDEADRILNMDFETEVDKILKVIP 196
Query: 184 RERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVY 243
R+R+TFLFSATMT KV+KLQR L+NPVK SSKY TV+ L+Q Y F+P+K KD YLVY
Sbjct: 197 RDRKTFLFSATMTKKVQKLQRAALKNPVKCAVSSKYQTVEKLQQYYLFIPSKFKDTYLVY 256
Query: 244 ILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNI 303
IL+E+AG++ M+F TC++T+ AL+LRNLG AIP++G MSQ KRLG+LNKFK+ +I
Sbjct: 257 ILNELAGNSFMIFCSTCNNTQRTALLLRNLGFTAIPLHGQMSQSKRLGSLNKFKAKARSI 316
Query: 304 LLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYV 363
LL TDVASRGLDIP VD+V+N+DIPT+SKDYIHRVGRTARAGRSG AI+ V QY++E +
Sbjct: 317 LLATDVASRGLDIPHVDVVVNFDIPTHSKDYIHRVGRTARAGRSGKAITFVTQYDVELFQ 376
Query: 364 QIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDDVDK 423
+IE LIGKKLP +P +EEV++L ERV EA+R A +++E G KK+R D G++DD +
Sbjct: 377 RIEHLIGKKLPVFPTQDEEVMMLTERVNEAQRFARMELREHGEKKKRKREDAGDDDDKEG 436
Query: 424 YFGLKDRKSSKKFRR 438
G++++ + K ++
Sbjct: 437 AIGVRNKVAGGKMKK 451
>UniRef100_Q9H4E3 DEAD box protein [Homo sapiens]
Length = 455
Score = 547 bits (1410), Expect = e-154
Identities = 269/435 (61%), Positives = 352/435 (80%), Gaps = 2/435 (0%)
Query: 4 ENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAF 63
E +E K+FKDLG+ + L EAC+++GW P +IQIEAIP AL+G+D+IGLA+TGSGKTGAF
Sbjct: 19 EEEETKTFKDLGVTDVLCEACDQLGWTKPTKIQIEAIPLALQGRDIIGLAETGSGKTGAF 78
Query: 64 ALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
ALPIL+ALLE P+ FA V++PTRELA QISEQFEALGS IGV+ AV+VGGID + QS
Sbjct: 79 ALPILNALLETPQ--RLFALVLTPTRELAFQISEQFEALGSSIGVQSAVIVGGIDSMSQS 136
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
+ +AK PHII+ TPGR++DHL+NTKGF+L LKYLV+DEADR+LN DFE +++IL +IP
Sbjct: 137 LALAKKPHIIIATPGRLIDHLENTKGFNLRALKYLVMDEADRILNMDFETEVDKILKVIP 196
Query: 184 RERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVY 243
R+R+TFLFSATMT KV+KLQR L+NPVK SSKY TV+ L+Q Y F+P+K KD YLVY
Sbjct: 197 RDRKTFLFSATMTKKVQKLQRAALKNPVKCAVSSKYQTVEKLQQYYIFIPSKFKDTYLVY 256
Query: 244 ILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNI 303
IL+E+AG++ M+F TC++T+ AL+LRNLG AIP++G MSQ KRLG+LNKFK+ + +I
Sbjct: 257 ILNELAGNSFMIFCSTCNNTQRTALLLRNLGFTAIPLHGQMSQSKRLGSLNKFKAKESSI 316
Query: 304 LLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYV 363
LL TDVASRGLDIP VD+V+N+DIPT+SKDYIHRVGRTARAGRSG AI+ V QY++E +
Sbjct: 317 LLATDVASRGLDIPHVDVVVNFDIPTHSKDYIHRVGRTARAGRSGKAITFVTQYDVELFQ 376
Query: 364 QIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDDVDK 423
+IE LIGKKLP +P ++EV++L ERV EA+R A +++E G KK+R D G+ DD +
Sbjct: 377 RIEHLIGKKLPGFPTQDDEVMMLTERVAEAQRFARMELREHGEKKKRSREDAGDNDDTEG 436
Query: 424 YFGLKDRKSSKKFRR 438
G++++ + K ++
Sbjct: 437 AIGVRNKVAGGKMKK 451
>UniRef100_UPI000036F389 UPI000036F389 UniRef100 entry
Length = 455
Score = 545 bits (1404), Expect = e-153
Identities = 269/435 (61%), Positives = 351/435 (79%), Gaps = 2/435 (0%)
Query: 4 ENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAF 63
E +E K+FKDLG+ + L EAC+++GW P +IQIEAIP AL+G+D+IGLA+TGSGKTGAF
Sbjct: 19 EKEETKTFKDLGVTDVLCEACDQLGWTKPTKIQIEAIPLALQGRDIIGLAETGSGKTGAF 78
Query: 64 ALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
ALPIL+ALLE P+ FA V++PTRELA QISEQFEALGS IGV+ AV+VGGID + QS
Sbjct: 79 ALPILNALLETPQ--RLFALVLTPTRELAFQISEQFEALGSSIGVQSAVIVGGIDSMSQS 136
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
+ +AK PHII+ TPGR++DHL+NTKGF+L LKYLV+DEADR+LN DFE +++IL +IP
Sbjct: 137 LALAKKPHIIIATPGRLIDHLENTKGFNLRALKYLVMDEADRILNMDFETEVDKILKVIP 196
Query: 184 RERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVY 243
R+R+TFLFSATMT KV+KLQR L+NPVK SSKY TV+ L+Q Y F+P+K KD YLVY
Sbjct: 197 RDRKTFLFSATMTKKVQKLQRAALKNPVKCAVSSKYQTVEKLQQYYIFIPSKFKDTYLVY 256
Query: 244 ILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNI 303
IL+E+AG++ M+F TC++T+ AL+LRNLG AIP++G MSQ KRLG+LNKFK+ +I
Sbjct: 257 ILNELAGNSFMIFCSTCNNTQRTALLLRNLGFTAIPLHGQMSQSKRLGSLNKFKAKARSI 316
Query: 304 LLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYV 363
LL TDVASRGLDIP VD+V+N+DIPT+SKDYIHRVGRTARAGRSG AI+ V QY++E +
Sbjct: 317 LLATDVASRGLDIPHVDVVVNFDIPTHSKDYIHRVGRTARAGRSGKAITFVTQYDVELFQ 376
Query: 364 QIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDDVDK 423
+IE LIGKKLP +P ++EV++L ERV EA+R A +++E G KK+R D G+ DD +
Sbjct: 377 RIEHLIGKKLPGFPTQDDEVMMLTERVAEAQRFARMELREHGEKKKRSREDAGDNDDTEG 436
Query: 424 YFGLKDRKSSKKFRR 438
G++++ + K ++
Sbjct: 437 AIGVRNKVAGGKMKK 451
>UniRef100_Q9H0S4 DEAD-box protein 47 [Homo sapiens]
Length = 455
Score = 545 bits (1404), Expect = e-153
Identities = 269/435 (61%), Positives = 351/435 (79%), Gaps = 2/435 (0%)
Query: 4 ENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAF 63
E +E K+FKDLG+ + L EAC+++GW P +IQIEAIP AL+G+D+IGLA+TGSGKTGAF
Sbjct: 19 EEEETKTFKDLGVTDVLCEACDQLGWTKPTKIQIEAIPLALQGRDIIGLAETGSGKTGAF 78
Query: 64 ALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
ALPIL+ALLE P+ FA V++PTRELA QISEQFEALGS IGV+ AV+VGGID + QS
Sbjct: 79 ALPILNALLETPQ--RLFALVLTPTRELAFQISEQFEALGSSIGVQSAVIVGGIDSMSQS 136
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
+ +AK PHII+ TPGR++DHL+NTKGF+L LKYLV+DEADR+LN DFE +++IL +IP
Sbjct: 137 LALAKKPHIIIATPGRLIDHLENTKGFNLRALKYLVMDEADRILNMDFETEVDKILKVIP 196
Query: 184 RERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVY 243
R+R+TFLFSATMT KV+KLQR L+NPVK SSKY TV+ L+Q Y F+P+K KD YLVY
Sbjct: 197 RDRKTFLFSATMTKKVQKLQRAALKNPVKCAVSSKYQTVEKLQQYYIFIPSKFKDTYLVY 256
Query: 244 ILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNI 303
IL+E+AG++ M+F TC++T+ AL+LRNLG AIP++G MSQ KRLG+LNKFK+ +I
Sbjct: 257 ILNELAGNSFMIFCSTCNNTQRTALLLRNLGFTAIPLHGQMSQSKRLGSLNKFKAKARSI 316
Query: 304 LLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYV 363
LL TDVASRGLDIP VD+V+N+DIPT+SKDYIHRVGRTARAGRSG AI+ V QY++E +
Sbjct: 317 LLATDVASRGLDIPHVDVVVNFDIPTHSKDYIHRVGRTARAGRSGKAITFVTQYDVELFQ 376
Query: 364 QIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDDVDK 423
+IE LIGKKLP +P ++EV++L ERV EA+R A +++E G KK+R D G+ DD +
Sbjct: 377 RIEHLIGKKLPGFPTQDDEVMMLTERVAEAQRFARMELREHGEKKKRSREDAGDNDDTEG 436
Query: 424 YFGLKDRKSSKKFRR 438
G++++ + K ++
Sbjct: 437 AIGVRNKVAGGKMKK 451
>UniRef100_Q6INR0 MGC81303 protein [Xenopus laevis]
Length = 448
Score = 541 bits (1395), Expect = e-152
Identities = 270/438 (61%), Positives = 352/438 (79%), Gaps = 4/438 (0%)
Query: 1 MEEENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKT 60
+E +E K+F+DLG+ + L EACE++GWK P +IQIEAIP AL+G+D+IGLA+TGSGKT
Sbjct: 11 VENGEEEPKTFRDLGVTDVLCEACEQLGWKQPTKIQIEAIPMALQGRDIIGLAETGSGKT 70
Query: 61 GAFALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMV 120
GAFALPIL LLE+P+ +A V++PTRELA QISEQFEALGS IGVK AV+VGGIDM+
Sbjct: 71 GAFALPILQTLLESPQ--RLYALVLTPTRELAFQISEQFEALGSSIGVKSAVIVGGIDMM 128
Query: 121 QQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILG 180
QS+ +AK PHI++ TPGR++DHL+NTKGF+L +KYLV+DEADR+LN DFE +++IL
Sbjct: 129 SQSLALAKKPHIVIATPGRLIDHLENTKGFNLRAIKYLVMDEADRILNMDFETEVDKILK 188
Query: 181 MIPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCY 240
+IPR+R+TFLFSATMT KV KLQR L++PVK SSKY TV+ L+Q Y F+P+K KD Y
Sbjct: 189 VIPRDRKTFLFSATMTKKVHKLQRAALKDPVKCAVSSKYQTVEKLQQFYVFIPSKFKDSY 248
Query: 241 LVYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGD 300
LVYIL+E+AG++ M+F TC++T+ +AL+LRNLG AIP++G M Q KRLGALNKFK+
Sbjct: 249 LVYILNELAGNSFMIFCSTCNNTQRVALLLRNLGFTAIPLHGQMGQNKRLGALNKFKAKS 308
Query: 301 CNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELE 360
+ILL TDVASRGLDIP VD+VIN+DIPT+SKDYIHRVGRTARAGRSG AI+ V+QY++E
Sbjct: 309 RSILLATDVASRGLDIPHVDVVINFDIPTHSKDYIHRVGRTARAGRSGKAITFVSQYDVE 368
Query: 361 WYVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDD 420
+ +IE LIGKKLP +P EEEV++L ERV EA+R A +++E G KK+R D +DD
Sbjct: 369 LFQRIEHLIGKKLPAFPTQEEEVMMLNERVSEAQRFARIELREHGEKKKRPRND--ADDD 426
Query: 421 VDKYFGLKDRKSSKKFRR 438
+ G++ + + KF++
Sbjct: 427 KEGSLGVRKKVTGGKFKK 444
>UniRef100_Q5ZLB0 Hypothetical protein [Gallus gallus]
Length = 453
Score = 534 bits (1375), Expect = e-150
Identities = 267/433 (61%), Positives = 348/433 (79%), Gaps = 5/433 (1%)
Query: 6 KEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFAL 65
+E +SFKDLG+ + L EAC+++GWK P +IQ+EAIP AL+G+D+IGLA+TGSGKTGAFAL
Sbjct: 22 EEARSFKDLGVTDVLCEACDQLGWKVPTKIQVEAIPVALQGRDIIGLAETGSGKTGAFAL 81
Query: 66 PILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVK 125
PIL ALL+AP+ FA V++PTRELA QISEQFEALGS IGV AV+VGGID + QS+
Sbjct: 82 PILQALLDAPQ--RLFALVLTPTRELAFQISEQFEALGSSIGVHSAVIVGGIDSMSQSLA 139
Query: 126 IAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRE 185
+AK PHII+ TPGR++DHL+NTKGF+L LK+LV+DEADR+LN DFE +++IL +IPR+
Sbjct: 140 LAKKPHIIIATPGRLVDHLENTKGFNLRALKFLVMDEADRILNMDFETEVDKILKVIPRD 199
Query: 186 RRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYIL 245
R+TFLFSATMT +V+KLQR L+NPVK SSKY TV+ L+Q Y F+P+K KD YLVYIL
Sbjct: 200 RKTFLFSATMTKQVQKLQRAALKNPVKCAVSSKYQTVEKLQQYYIFIPSKFKDSYLVYIL 259
Query: 246 SEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILL 305
+E+AG++ M+F TC++T+ AL+LRNLG AIP++G MSQ KRLG+LNKFK+ +ILL
Sbjct: 260 NELAGNSFMIFCSTCNNTQRTALLLRNLGFTAIPLHGQMSQNKRLGSLNKFKAKARSILL 319
Query: 306 CTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQI 365
TDVASRGLDIP VD+VIN+DIPT+SKDYIHRVGRTARAGRSG +I+ V QY++E + +I
Sbjct: 320 ATDVASRGLDIPHVDVVINFDIPTHSKDYIHRVGRTARAGRSGKSITFVTQYDVELFQRI 379
Query: 366 EKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDDVDKYF 425
E LIGKKLP +P EEEV++L ERV EA+R A +++E G KKR D +DD ++
Sbjct: 380 EHLIGKKLPAFPMQEEEVMMLTERVAEAQRFARMELREQGEKKRSRNAD---DDDTEEAL 436
Query: 426 GLKDRKSSKKFRR 438
G++ + + K ++
Sbjct: 437 GVRKKVAGGKKKK 449
>UniRef100_UPI00003636F9 UPI00003636F9 UniRef100 entry
Length = 418
Score = 533 bits (1374), Expect = e-150
Identities = 263/416 (63%), Positives = 342/416 (81%), Gaps = 2/416 (0%)
Query: 15 GLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILHALLEA 74
G+ E L EAC+++GWK+P +IQIEA+P AL+GKD+IGLA+TGSGKTGAFALPIL +LL +
Sbjct: 1 GVTEVLCEACDQLGWKSPTKIQIEAVPVALQGKDVIGLAETGSGKTGAFALPILQSLLAS 60
Query: 75 PRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIAKLPHIIV 134
P+ H V++PTRELA QISEQFEALGS IGVKCAV+VGGIDM+ QS+ +AK PHI++
Sbjct: 61 PQRLH--TLVLTPTRELAFQISEQFEALGSSIGVKCAVIVGGIDMMSQSLVLAKKPHIVI 118
Query: 135 GTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRERRTFLFSAT 194
TPGR++DHL+NTKGF+L LK+LV+DEADR+LN DFE +++IL +IPRERRTFLFSAT
Sbjct: 119 ATPGRLIDHLENTKGFTLRALKFLVMDEADRILNMDFETEVDKILKVIPRERRTFLFSAT 178
Query: 195 MTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYILSEMAGSTSM 254
MT KV+KLQR L++PVK S+KYSTVD L+Q Y F+P+K+KDCYLV IL+++AG++ +
Sbjct: 179 MTKKVQKLQRAALKDPVKCAVSTKYSTVDKLQQYYIFIPSKYKDCYLVSILNDLAGNSFI 238
Query: 255 VFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILLCTDVASRGL 314
+F TC++ + +AL+LRNLG+ AI ++G MSQ KRLGALNKFKS ++LL TDVASRGL
Sbjct: 239 IFCSTCNNAQRVALLLRNLGITAISLHGQMSQNKRLGALNKFKSKSRSVLLATDVASRGL 298
Query: 315 DIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQIEKLIGKKLP 374
DIP VD VINYDIPT+SKDYIHRVGRTARAGRSG +I+ V QY++E + +IE LIGKKLP
Sbjct: 299 DIPHVDCVINYDIPTHSKDYIHRVGRTARAGRSGKSITFVTQYDVELFQRIESLIGKKLP 358
Query: 375 EYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDDVDKYFGLKDR 430
+P EEEV++L ERV EA+R A +MKE G K++R +G ++DD ++ G++ +
Sbjct: 359 AFPTQEEEVMMLVERVSEAQRFARLEMKEQGEKRKRPKGGDRDDDDTEQSSGVRKK 414
>UniRef100_Q9VIF6 CG9253-PA [Drosophila melanogaster]
Length = 507
Score = 518 bits (1333), Expect = e-145
Identities = 262/425 (61%), Positives = 340/425 (79%), Gaps = 10/425 (2%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
++KDLGL E+L +AC+++ WK P +IQ EAIP AL+GKD+IGLA+TGSGKTGAFALPILH
Sbjct: 62 TWKDLGLNEALCQACDELKWKAPSKIQREAIPVALQGKDVIGLAETGSGKTGAFALPILH 121
Query: 70 ALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIAKL 129
ALLE P +FA V++PTRELA QI EQFEALGS IG+KC V+VGG+DMV Q +++AK
Sbjct: 122 ALLE--NPQRYFALVLTPTRELAFQIGEQFEALGSGIGIKCCVVVGGMDMVAQGLQLAKK 179
Query: 130 PHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRERRTF 189
PHII+ TPGR++DHL+N KGF+L +KYLV+DEADR+LN DFE L++IL ++PRERRTF
Sbjct: 180 PHIIIATPGRLVDHLENMKGFNLKAIKYLVMDEADRILNMDFEVELDKILKVLPRERRTF 239
Query: 190 LFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYILSEMA 249
LFSATMT KV+KLQR L++PVK+E S+KY TV+ L+Q Y F+P K+KD YLV+IL+E+A
Sbjct: 240 LFSATMTKKVKKLQRASLKDPVKVEVSNKYQTVEQLQQSYLFIPVKYKDVYLVHILNELA 299
Query: 250 GSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILLCTDV 309
G++ M+F TC++T AL+LR LGL AIP++G MSQ KRL ALNKFK+ + +IL+ TDV
Sbjct: 300 GNSFMIFCSTCNNTVKTALMLRALGLAAIPLHGQMSQNKRLAALNKFKAKNRSILISTDV 359
Query: 310 ASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQIEKLI 369
ASRGLDIP VD+V+N+DIPT+SKDYIHRVGRTARAGRSG AI+LV+QY++E Y +IE L+
Sbjct: 360 ASRGLDIPHVDVVVNFDIPTHSKDYIHRVGRTARAGRSGKAITLVSQYDIELYQRIEHLL 419
Query: 370 GKKLPEYPANEEEVLLLEERVGEAKRLAATKMKE----SGGKKRRGEGDIGEEDDVDKYF 425
GK+L Y E+EV+ L+ERV EA+R A ++K+ GG KR G+ DD + +
Sbjct: 420 GKQLTLYKCEEDEVMALQERVAEAQRTAKLELKDLEDTRGGHKRGGD----THDDSENFT 475
Query: 426 GLKDR 430
G + R
Sbjct: 476 GARKR 480
>UniRef100_Q8MRB7 RE27528p [Drosophila melanogaster]
Length = 507
Score = 518 bits (1333), Expect = e-145
Identities = 262/425 (61%), Positives = 340/425 (79%), Gaps = 10/425 (2%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
++KDLGL E+L +AC+++ WK P +IQ EAIP AL+GKD+IGLA+TGSGKTGAFALPILH
Sbjct: 62 TWKDLGLNEALCQACDELKWKAPSKIQREAIPVALQGKDVIGLAETGSGKTGAFALPILH 121
Query: 70 ALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIAKL 129
ALLE P +FA V++PTRELA QI EQFEALGS IG+KC V+VGG+DMV Q +++AK
Sbjct: 122 ALLE--NPQRYFALVLTPTRELAFQIGEQFEALGSGIGIKCCVVVGGMDMVAQGLQLAKK 179
Query: 130 PHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRERRTF 189
PHII+ TPGR++DHL+N KGF+L +KYLV+DEADR+LN DFE L++IL ++PRERRTF
Sbjct: 180 PHIIIATPGRLVDHLENMKGFNLKAIKYLVMDEADRILNMDFEVELDKILKVLPRERRTF 239
Query: 190 LFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYILSEMA 249
LFSATMT KV+KLQR L++PVK+E S+KY TV+ L+Q Y F+P K+KD YLV+IL+E+A
Sbjct: 240 LFSATMTKKVKKLQRASLKDPVKVEVSNKYQTVEQLQQSYLFIPVKYKDVYLVHILNELA 299
Query: 250 GSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILLCTDV 309
G++ M+F TC++T AL+LR LGL AIP++G MSQ KRL ALNKFK+ + +IL+ TDV
Sbjct: 300 GNSFMIFCSTCNNTVKTALMLRALGLAAIPLHGQMSQNKRLAALNKFKAKNRSILISTDV 359
Query: 310 ASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQIEKLI 369
ASRGLDIP VD+V+N+DIPT+SKDYIHRVGRTARAGRSG AI+LV+QY++E Y +IE L+
Sbjct: 360 ASRGLDIPHVDVVVNFDIPTHSKDYIHRVGRTARAGRSGKAITLVSQYDIELYQRIEHLL 419
Query: 370 GKKLPEYPANEEEVLLLEERVGEAKRLAATKMKE----SGGKKRRGEGDIGEEDDVDKYF 425
GK+L Y E+EV+ L+ERV EA+R A ++K+ GG KR G+ DD + +
Sbjct: 420 GKQLTLYKCEEDEVMALQERVAEAQRTAKLELKDLEDTRGGHKRGGD----THDDSENFT 475
Query: 426 GLKDR 430
G + R
Sbjct: 476 GARKR 480
>UniRef100_P34580 Putative ATP-dependent RNA helicase T26G10.1 in chromosome III
[Caenorhabditis elegans]
Length = 489
Score = 498 bits (1282), Expect = e-139
Identities = 247/424 (58%), Positives = 331/424 (77%), Gaps = 7/424 (1%)
Query: 2 EEENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTG 61
+EE+ + KSF +LG+ + L +AC+++GW P +IQ A+P AL+GKD+IGLA+TGSGKTG
Sbjct: 37 DEEDVKEKSFAELGVSQPLCDACQRLGWMKPSKIQQAALPHALQGKDVIGLAETGSGKTG 96
Query: 62 AFALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQ 121
AFA+P+L +LL+ P+ FF V++PTRELA QI +QFEALGS IG+ AV+VGG+DM
Sbjct: 97 AFAIPVLQSLLDHPQA--FFCLVLTPTRELAFQIGQQFEALGSGIGLIAAVIVGGVDMAA 154
Query: 122 QSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGM 181
Q++ +A+ PHIIV TPGR++DHL+NTKGF+L LK+L++DEADR+LN DFE L++IL +
Sbjct: 155 QAMALARRPHIIVATPGRLVDHLENTKGFNLKALKFLIMDEADRILNMDFEVELDKILKV 214
Query: 182 IPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYL 241
IPRERRT+LFSATMT KV KL+R LR+P ++ SS+Y TVD LKQ Y F+P K+K+ YL
Sbjct: 215 IPRERRTYLFSATMTKKVSKLERASLRDPARVSVSSRYKTVDNLKQHYIFVPNKYKETYL 274
Query: 242 VYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDC 301
VY+L+E AG++++VF TC +T +A++LR LG++A+P++G MSQ KRLG+LNKFKS
Sbjct: 275 VYLLNEHAGNSAIVFCATCATTMQIAVMLRQLGMQAVPLHGQMSQEKRLGSLNKFKSKAR 334
Query: 302 NILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEW 361
IL+CTDVA+RGLDIP VDMVINYD+P+ SKDY+HRVGRTARAGRSG+AI++V QY++E
Sbjct: 335 EILVCTDVAARGLDIPHVDMVINYDMPSQSKDYVHRVGRTARAGRSGIAITVVTQYDVEA 394
Query: 362 YVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKE-----SGGKKRRGEGDIG 416
Y +IE +GKKL EY E EV++L ER EA A +MKE GKKRR D G
Sbjct: 395 YQKIEANLGKKLDEYKCVENEVMVLVERTQEATENARIEMKEMDEKKKSGKKRRQNDDFG 454
Query: 417 EEDD 420
+ ++
Sbjct: 455 DTEE 458
>UniRef100_Q61IR7 Hypothetical protein CBG10097 [Caenorhabditis briggsae]
Length = 486
Score = 493 bits (1269), Expect = e-138
Identities = 246/423 (58%), Positives = 329/423 (77%), Gaps = 7/423 (1%)
Query: 3 EENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGA 62
EE +E K+F +LG+ + L +AC ++GW P +IQ A+P ALEGKD+IGLA+TGSGKTGA
Sbjct: 38 EEIEEEKTFAELGVSQPLCDACLRLGWTKPSKIQQAALPHALEGKDVIGLAETGSGKTGA 97
Query: 63 FALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQ 122
FA+P+L +LL+ P+ FF V++PTRELA QI +QFEALGS IG+ AV+VGG+DM Q
Sbjct: 98 FAIPVLQSLLDHPQA--FFCLVLTPTRELAFQIGQQFEALGSGIGLIVAVIVGGVDMAAQ 155
Query: 123 SVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMI 182
++ +A+ PHIIV TPGR++DHL+NTKGF+L LK+L++DEADR+LN DFE L++IL +I
Sbjct: 156 AMALARRPHIIVATPGRLVDHLENTKGFNLKALKFLIMDEADRILNMDFEVELDKILKVI 215
Query: 183 PRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLV 242
P+ERRT+LFSATMT KV KL+R LR+P ++ S++Y TVD LKQ Y F+P K+K+ YLV
Sbjct: 216 PKERRTYLFSATMTKKVSKLERASLRDPARVSISTRYKTVDNLKQHYIFIPNKYKETYLV 275
Query: 243 YILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCN 302
Y+L+E AG++++VF TC +T +A++LR LG++A+P++G MSQ KRLG+LNKFKS
Sbjct: 276 YLLNEHAGNSAIVFCATCATTMQVAVMLRQLGMQAVPLHGQMSQEKRLGSLNKFKSKARE 335
Query: 303 ILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWY 362
IL+CTDVA+RGLDIP VDMVINYD+P+ SKDY+HRVGRTARAGRSG+AI++V QY++E Y
Sbjct: 336 ILVCTDVAARGLDIPHVDMVINYDMPSQSKDYVHRVGRTARAGRSGLAITVVTQYDVEAY 395
Query: 363 VQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKE-----SGGKKRRGEGDIGE 417
+IE +GKKL EY E EV++L ER EA A +MKE GKKRR D G+
Sbjct: 396 QKIEANLGKKLDEYKCVENEVMVLVERTQEAMENAKVEMKEMDEKRKSGKKRRQNDDFGD 455
Query: 418 EDD 420
++
Sbjct: 456 TEE 458
>UniRef100_Q9P6N8 SPAC823.08c protein [Schizosaccharomyces pombe]
Length = 465
Score = 493 bits (1268), Expect = e-138
Identities = 252/424 (59%), Positives = 330/424 (77%), Gaps = 6/424 (1%)
Query: 3 EENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGA 62
+ N+ K+FK+LG+ + L EACEK+G+KTP IQ EAIP L +D+IGLA+TGSGKT A
Sbjct: 40 QNNEAPKTFKELGVIDELCEACEKLGFKTPTPIQQEAIPVVLNKRDVIGLAQTGSGKTAA 99
Query: 63 FALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQ 122
FALP++ L P P FFA V++PTRELA QISEQFEA+G IGV+ V+VGG+DMV Q
Sbjct: 100 FALPVIQELWNNPSP--FFAVVLAPTRELAYQISEQFEAIGGSIGVRSVVIVGGMDMVTQ 157
Query: 123 SVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMI 182
+V ++K PH++V TPGR++DHL+NTKGFSL LKYL++DEADRLL+ DF +++IL +I
Sbjct: 158 AVALSKKPHVLVCTPGRLMDHLENTKGFSLKNLKYLIMDEADRLLDMDFGPIIDKILKII 217
Query: 183 PRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLV 242
P ERRT LFSATMT+KVEKLQR L PV++ SSK+STVDTL Q+Y F P KHKD YLV
Sbjct: 218 PHERRTLLFSATMTSKVEKLQRASLHQPVRVAVSSKFSTVDTLIQRYLFFPFKHKDTYLV 277
Query: 243 YILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCN 302
Y+++E+AG++ ++F RT + T+ LA++LR LG AIP++G +SQ RLGALNKFKSG +
Sbjct: 278 YLVNELAGNSIIIFARTVNDTQRLAILLRTLGFSAIPLHGQLSQSNRLGALNKFKSGARS 337
Query: 303 ILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWY 362
L+ TDVA+RGLDIP VD+VINYDIPT+SK YIHRVGRTARAGR+G +I+LV QY+LE +
Sbjct: 338 TLVATDVAARGLDIPLVDVVINYDIPTDSKAYIHRVGRTARAGRAGKSIALVTQYDLEPF 397
Query: 363 VQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDI---GEED 419
++IE IGKK+ EY ++E V LL ERVGEA+R A +MKE +R+ +G + + D
Sbjct: 398 LRIEATIGKKMQEYEIDKEGVFLLSERVGEAQREAIIQMKEI-HDRRKSKGKLHTKRKRD 456
Query: 420 DVDK 423
D+D+
Sbjct: 457 DLDR 460
>UniRef100_Q6CH58 Yarrowia lipolytica chromosome A of strain CLIB99 of Yarrowia
lipolytica [Yarrowia lipolytica]
Length = 480
Score = 486 bits (1250), Expect = e-136
Identities = 246/427 (57%), Positives = 328/427 (76%), Gaps = 11/427 (2%)
Query: 1 MEEENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKT 60
++E ++ K+FKDLG+ +S+ E CE++ + P IQ ++IP ALEG+D+IGLA+TGSGKT
Sbjct: 56 VDESEEQTKTFKDLGVIDSICETCEELKFTKPTPIQAQSIPYALEGRDIIGLAQTGSGKT 115
Query: 61 GAFALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMV 120
AFA+P+L +L E P+P + V++PTRELA QISE FEALGS +G++ AV+VGG++M+
Sbjct: 116 AAFAIPVLQSLYENPQP--LYCVVLAPTRELAYQISETFEALGSAMGLRTAVVVGGMNMM 173
Query: 121 QQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILG 180
Q+V ++K PH+IV TPGR++DHL+NTKGFSL LK+LV+DEADRLL+ +F SL++IL
Sbjct: 174 TQAVALSKKPHVIVATPGRLVDHLENTKGFSLRTLKFLVMDEADRLLDMEFGPSLDKILK 233
Query: 181 MIPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCY 240
+IPR+R T+LFSATMT+KVEKLQR L +PV++ S+KY T D L Q F P KHKD +
Sbjct: 234 VIPRQRNTYLFSATMTSKVEKLQRASLVDPVRVAVSTKYQTADNLLQYMVFCPFKHKDTH 293
Query: 241 LVYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGD 300
LVY++SE AG++ ++F RT T+ ++L+LRNLG AIP++G +SQ RLGALNKFKSG
Sbjct: 294 LVYLVSENAGNSMIIFARTKSDTQRISLLLRNLGYGAIPLHGDLSQTARLGALNKFKSGS 353
Query: 301 CNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELE 360
NIL+ TDVASRGLDIPAVD+VINYDIP++SK YIHRVGRTARAGR+G +++LV+QY+LE
Sbjct: 354 RNILIATDVASRGLDIPAVDLVINYDIPSDSKSYIHRVGRTARAGRAGKSVALVSQYDLE 413
Query: 361 WYVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGG---------KKRRG 411
Y++IE +GKKL YP E V+L ERV EA R A +MK G K+RR
Sbjct: 414 LYLRIEGALGKKLDSYPLESEAVMLFSERVAEASRAAIQEMKGEDGTKKRSKFDKKRRRD 473
Query: 412 EGDIGEE 418
E DIGE+
Sbjct: 474 EMDIGEQ 480
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.137 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 713,826,655
Number of Sequences: 2790947
Number of extensions: 30340973
Number of successful extensions: 96890
Number of sequences better than 10.0: 4361
Number of HSP's better than 10.0 without gapping: 3603
Number of HSP's successfully gapped in prelim test: 758
Number of HSP's that attempted gapping in prelim test: 84305
Number of HSP's gapped (non-prelim): 5506
length of query: 438
length of database: 848,049,833
effective HSP length: 130
effective length of query: 308
effective length of database: 485,226,723
effective search space: 149449830684
effective search space used: 149449830684
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146793.2


