

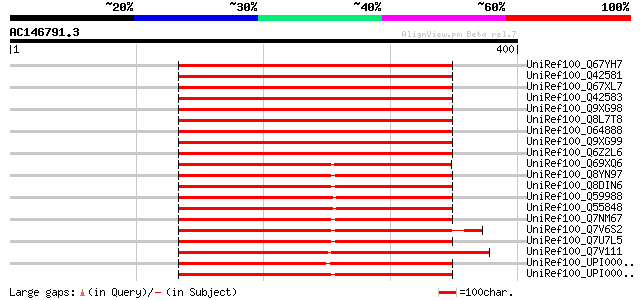
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146791.3 - phase: 0 /pseudo
(400 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q67YH7 Phosphoribosyl pyrophosphate synthetase [Arabid... 400 e-110
UniRef100_Q42581 Ribose-phosphate pyrophosphokinase 1 [Arabidops... 400 e-110
UniRef100_Q67XL7 Phosphoribosyl pyrophosphate synthetase [Arabid... 398 e-109
UniRef100_Q42583 Ribose-phosphate pyrophosphokinase 2 [Arabidops... 398 e-109
UniRef100_Q9XG98 Phosphoribosyl pyrophosphate synthase [Spinacia... 397 e-109
UniRef100_Q8L7T8 At2g44530/F16B22.2 [Arabidopsis thaliana] 395 e-109
UniRef100_O64888 Probable ribose-phosphate pyrophosphokinase [Ar... 395 e-109
UniRef100_Q9XG99 Phosphoribosyl pyrophosphate synthase isozyme 2... 394 e-108
UniRef100_Q6Z2L6 Putative phosphoribosyl pyrophosphate synthase ... 391 e-107
UniRef100_Q69XQ6 Putative phosphoribosyl pyrophosphate synthase ... 350 3e-95
UniRef100_Q8YN97 Ribose-phosphate pyrophosphokinase [Anabaena sp.] 335 2e-90
UniRef100_Q8DIN6 Ribose-phosphate pyrophosphokinase [Synechococc... 333 4e-90
UniRef100_Q59988 Ribose-phosphate pyrophosphokinase [Synechococc... 330 6e-89
UniRef100_Q55848 Ribose-phosphate pyrophosphokinase [Synechocyst... 325 1e-87
UniRef100_Q7NM67 Ribose-phosphate pyrophosphokinase [Gloeobacter... 314 2e-84
UniRef100_Q7V6S2 Ribose-phosphate pyrophosphokinase [Prochloroco... 313 7e-84
UniRef100_Q7U7L5 Ribose-phosphate pyrophosphokinase [Synechococc... 311 2e-83
UniRef100_Q7V111 Ribose-phosphate pyrophosphokinase [Prochloroco... 311 3e-83
UniRef100_UPI000031FFBA UPI000031FFBA UniRef100 entry 310 5e-83
UniRef100_UPI0000287166 UPI0000287166 UniRef100 entry 310 6e-83
>UniRef100_Q67YH7 Phosphoribosyl pyrophosphate synthetase [Arabidopsis thaliana]
Length = 352
Score = 400 bits (1027), Expect = e-110
Identities = 196/216 (90%), Positives = 208/216 (95%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGC+VYL+QPTC P NENLMEL +M+DACRRASAK +TAVIPYFGYARADR
Sbjct: 78 IYVQLQESVRGCDVYLVQPTCTPTNENLMELLIMVDACRRASAKKVTAVIPYFGYARADR 137
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLIT+AGADRVLACDLHSGQSMGYFDIPVDHV+CQPVILDYLASK+
Sbjct: 138 KTQGRESIAAKLVANLITEAGADRVLACDLHSGQSMGYFDIPVDHVYCQPVILDYLASKS 197
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
I S DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDV+GKVA+M
Sbjct: 198 IPSEDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVRGKVAIM 257
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTI KGAALLH+EGAREVYACCTH VFS
Sbjct: 258 VDDMIDTAGTIVKGAALLHQEGAREVYACCTHAVFS 293
>UniRef100_Q42581 Ribose-phosphate pyrophosphokinase 1 [Arabidopsis thaliana]
Length = 403
Score = 400 bits (1027), Expect = e-110
Identities = 196/216 (90%), Positives = 208/216 (95%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGC+VYL+QPTC P NENLMEL +M+DACRRASAK +TAVIPYFGYARADR
Sbjct: 129 IYVQLQESVRGCDVYLVQPTCTPTNENLMELLIMVDACRRASAKKVTAVIPYFGYARADR 188
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLIT+AGADRVLACDLHSGQSMGYFDIPVDHV+CQPVILDYLASK+
Sbjct: 189 KTQGRESIAAKLVANLITEAGADRVLACDLHSGQSMGYFDIPVDHVYCQPVILDYLASKS 248
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
I S DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDV+GKVA+M
Sbjct: 249 IPSEDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVRGKVAIM 308
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTI KGAALLH+EGAREVYACCTH VFS
Sbjct: 309 VDDMIDTAGTIVKGAALLHQEGAREVYACCTHAVFS 344
>UniRef100_Q67XL7 Phosphoribosyl pyrophosphate synthetase [Arabidopsis thaliana]
Length = 403
Score = 398 bits (1022), Expect = e-109
Identities = 195/216 (90%), Positives = 207/216 (95%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGC+VYL+QPTC P NENLMEL +M+DACRRASAK +TAV PYFGYARADR
Sbjct: 129 IYVQLQESVRGCDVYLVQPTCTPTNENLMELLIMVDACRRASAKKVTAVTPYFGYARADR 188
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLIT+AGADRVLACDLHSGQSMGYFDIPVDHV+CQPVILDYLASK+
Sbjct: 189 KTQGRESIAAKLVANLITEAGADRVLACDLHSGQSMGYFDIPVDHVYCQPVILDYLASKS 248
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
I S DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDV+GKVA+M
Sbjct: 249 IPSEDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVRGKVAIM 308
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTI KGAALLH+EGAREVYACCTH VFS
Sbjct: 309 VDDMIDTAGTIVKGAALLHQEGAREVYACCTHAVFS 344
>UniRef100_Q42583 Ribose-phosphate pyrophosphokinase 2 [Arabidopsis thaliana]
Length = 400
Score = 398 bits (1022), Expect = e-109
Identities = 196/216 (90%), Positives = 208/216 (95%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQL+ESVRGC+V+L+QPTC P NENLMEL +M+DACRRASAK +TAVIPYFGYARADR
Sbjct: 126 IYVQLKESVRGCDVFLVQPTCTPTNENLMELLIMVDACRRASAKKVTAVIPYFGYARADR 185
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLIT+AGADRVLACDLHSGQSMGYFDIPVDHV+CQPVILDYLASK+
Sbjct: 186 KTQGRESIAAKLVANLITEAGADRVLACDLHSGQSMGYFDIPVDHVYCQPVILDYLASKS 245
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
ISS DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRR GHNVAEVMNLIGDVKGKVAVM
Sbjct: 246 ISSEDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRHGHNVAEVMNLIGDVKGKVAVM 305
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDD+IDTAGTI KGAALLHEEGAREVYACCTH VFS
Sbjct: 306 VDDIIDTAGTIVKGAALLHEEGAREVYACCTHAVFS 341
>UniRef100_Q9XG98 Phosphoribosyl pyrophosphate synthase [Spinacia oleracea]
Length = 336
Score = 397 bits (1021), Expect = e-109
Identities = 199/216 (92%), Positives = 210/216 (97%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGC+V+L+QPTCPPANENLMEL +MIDACRRASAKNITAVIPYFGYARADR
Sbjct: 62 IYVQLQESVRGCDVFLVQPTCPPANENLMELLIMIDACRRASAKNITAVIPYFGYARADR 121
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLIT+AGA+RVLACDLHSGQSMGYFDIPVDHV+ QPVILDYLASKT
Sbjct: 122 KTQGRESIAAKLVANLITEAGANRVLACDLHSGQSMGYFDIPVDHVYGQPVILDYLASKT 181
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
I S+DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRR GHNVAEVMNLIGDVKGKVAVM
Sbjct: 182 ICSDDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRHGHNVAEVMNLIGDVKGKVAVM 241
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTI+KGAALLH+EGAREVYAC TH VFS
Sbjct: 242 VDDMIDTAGTISKGAALLHQEGAREVYACSTHAVFS 277
>UniRef100_Q8L7T8 At2g44530/F16B22.2 [Arabidopsis thaliana]
Length = 394
Score = 395 bits (1016), Expect = e-109
Identities = 198/216 (91%), Positives = 208/216 (95%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGC+V+L+QPTCPPANENLMEL VMIDACRRASAK ITAVIPYFGYARADR
Sbjct: 114 IYVQLQESVRGCDVFLVQPTCPPANENLMELLVMIDACRRASAKTITAVIPYFGYARADR 173
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLIT++GADRVLACDLHSGQSMGYFDIPVDHV+ QPVILDYLASK
Sbjct: 174 KTQGRESIAAKLVANLITQSGADRVLACDLHSGQSMGYFDIPVDHVYGQPVILDYLASKA 233
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
ISS DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRR GHNVAEVMNLIGDVKGKVA+M
Sbjct: 234 ISSEDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRHGHNVAEVMNLIGDVKGKVAIM 293
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTI+KGAALLH+EGAREVYAC TH VFS
Sbjct: 294 VDDMIDTAGTISKGAALLHQEGAREVYACTTHAVFS 329
>UniRef100_O64888 Probable ribose-phosphate pyrophosphokinase [Arabidopsis thaliana]
Length = 386
Score = 395 bits (1016), Expect = e-109
Identities = 198/216 (91%), Positives = 208/216 (95%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGC+V+L+QPTCPPANENLMEL VMIDACRRASAK ITAVIPYFGYARADR
Sbjct: 114 IYVQLQESVRGCDVFLVQPTCPPANENLMELLVMIDACRRASAKTITAVIPYFGYARADR 173
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLIT++GADRVLACDLHSGQSMGYFDIPVDHV+ QPVILDYLASK
Sbjct: 174 KTQGRESIAAKLVANLITQSGADRVLACDLHSGQSMGYFDIPVDHVYGQPVILDYLASKA 233
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
ISS DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRR GHNVAEVMNLIGDVKGKVA+M
Sbjct: 234 ISSEDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRHGHNVAEVMNLIGDVKGKVAIM 293
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTI+KGAALLH+EGAREVYAC TH VFS
Sbjct: 294 VDDMIDTAGTISKGAALLHQEGAREVYACTTHAVFS 329
>UniRef100_Q9XG99 Phosphoribosyl pyrophosphate synthase isozyme 2 precursor [Spinacia
oleracea]
Length = 395
Score = 394 bits (1012), Expect = e-108
Identities = 196/216 (90%), Positives = 208/216 (95%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQL+ESVRGC+V++IQPT PPANENLMEL +MIDACRRASAKNITAVIPYFGYARADR
Sbjct: 121 IYVQLEESVRGCDVFIIQPTSPPANENLMELLIMIDACRRASAKNITAVIPYFGYARADR 180
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLIT+AGA+RVLACDLHSGQSMGYFDIPVDHV+CQPVILDYLASK
Sbjct: 181 KTQGRESIAAKLVANLITEAGANRVLACDLHSGQSMGYFDIPVDHVYCQPVILDYLASKG 240
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
I+S+DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRR HNVAEVMNLIGDVKGKVAVM
Sbjct: 241 IASSDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRHAHNVAEVMNLIGDVKGKVAVM 300
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
+DDMIDTAGTI KGA LLHEEGAREVYACCTH VFS
Sbjct: 301 LDDMIDTAGTITKGAELLHEEGAREVYACCTHAVFS 336
>UniRef100_Q6Z2L6 Putative phosphoribosyl pyrophosphate synthase [Oryza sativa]
Length = 396
Score = 391 bits (1004), Expect = e-107
Identities = 193/216 (89%), Positives = 208/216 (95%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGC+V+L+QPTCPPANENLMEL +MIDACRRASAKNITAVIPYFGYARADR
Sbjct: 116 IYVQLQESVRGCDVFLVQPTCPPANENLMELLIMIDACRRASAKNITAVIPYFGYARADR 175
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
K+QGRESIAAKLVAN+IT+AGA+RVL CDLHS Q+MGYFDIPVDHV+ QPVILDYLASKT
Sbjct: 176 KSQGRESIAAKLVANMITEAGANRVLVCDLHSSQAMGYFDIPVDHVYGQPVILDYLASKT 235
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
I S+DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRR GHNVAEVMNLIGDV+GKVAVM
Sbjct: 236 ICSDDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRHGHNVAEVMNLIGDVRGKVAVM 295
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
+DDMIDTAGTIAKGA LLH+EGAREVYACCTH VFS
Sbjct: 296 MDDMIDTAGTIAKGAELLHQEGAREVYACCTHAVFS 331
>UniRef100_Q69XQ6 Putative phosphoribosyl pyrophosphate synthase [Oryza sativa]
Length = 387
Score = 350 bits (899), Expect = 3e-95
Identities = 173/215 (80%), Positives = 193/215 (89%), Gaps = 1/215 (0%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGC+V+L+QPTC P NENLMEL VMIDACRRASA++IT VIPYFGYARADR
Sbjct: 126 IYVQLQESVRGCDVFLVQPTCSPVNENLMELFVMIDACRRASARSITVVIPYFGYARADR 185
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
K QGRE+I AKL ANL+T+AG+DRV+ CD+HS Q++GYFDIPVDH+H QPVILDYLASKT
Sbjct: 186 KAQGREAITAKLSANLLTEAGSDRVIVCDIHSTQALGYFDIPVDHIHGQPVILDYLASKT 245
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
I S DLVVVSPDVGGV RARAFAKKLSDAPLAIVDKRR GHN++EVM+LIGDVKGKVA+M
Sbjct: 246 I-SKDLVVVSPDVGGVVRARAFAKKLSDAPLAIVDKRRQGHNMSEVMHLIGDVKGKVAIM 304
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVF 348
VDDMIDTAGTI AALL +EGA VYAC TH VF
Sbjct: 305 VDDMIDTAGTITSAAALLKQEGAEAVYACSTHAVF 339
>UniRef100_Q8YN97 Ribose-phosphate pyrophosphokinase [Anabaena sp.]
Length = 330
Score = 335 bits (858), Expect = 2e-90
Identities = 164/216 (75%), Positives = 189/216 (86%), Gaps = 2/216 (0%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQ+QES+RGC+VYLIQP C P N++LMEL +M+DACRRASA+ +TAVIPY+GYARADR
Sbjct: 58 LYVQIQESIRGCDVYLIQPCCQPVNDHLMELLIMVDACRRASARQVTAVIPYYGYARADR 117
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KT GRESI AKLVANLIT+AGA+RVLA DLH+ Q GYFDIP DHV+ PV+LDYL SK
Sbjct: 118 KTAGRESITAKLVANLITQAGANRVLAMDLHAAQIQGYFDIPFDHVYGSPVLLDYLTSKQ 177
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
+ +D+VVVSPDVGGVARARAFAKKL+DAPLAI+DKRR HNVAEV+N+IGDVKGK AV+
Sbjct: 178 L--HDIVVVSPDVGGVARARAFAKKLNDAPLAIIDKRRQAHNVAEVLNVIGDVKGKTAVL 235
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDT GTIA GA LL EEGAR+VYAC TH VFS
Sbjct: 236 VDDMIDTGGTIAAGAKLLREEGARQVYACATHAVFS 271
>UniRef100_Q8DIN6 Ribose-phosphate pyrophosphokinase [Synechococcus elongatus]
Length = 396
Score = 333 bits (855), Expect = 4e-90
Identities = 164/216 (75%), Positives = 186/216 (85%), Gaps = 2/216 (0%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQ+QES+RGC+VYLIQP C P N++LMEL +M+DACRRASA+ +TAVIPY+GYARADR
Sbjct: 124 LYVQIQESIRGCDVYLIQPCCRPVNDHLMELLIMVDACRRASARQVTAVIPYYGYARADR 183
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KT GRESI AKLVANLIT+AGA RVLA DLHS Q GYFDIPVDHV+ PV+LDYL SK
Sbjct: 184 KTAGRESITAKLVANLITQAGASRVLAMDLHSAQIQGYFDIPVDHVYGSPVLLDYLRSKN 243
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
+ D+VVVSPDVGGVARARAFA KL DAPLAI+DKRR HNVAEVMN++GDVKGK AV+
Sbjct: 244 L--EDIVVVSPDVGGVARARAFANKLDDAPLAIIDKRRQAHNVAEVMNVVGDVKGKTAVL 301
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTI +GA LL EGA+EVYAC TH VFS
Sbjct: 302 VDDMIDTAGTILEGARLLRREGAKEVYACATHAVFS 337
>UniRef100_Q59988 Ribose-phosphate pyrophosphokinase [Synechococcus sp.]
Length = 331
Score = 330 bits (845), Expect = 6e-89
Identities = 162/216 (75%), Positives = 189/216 (87%), Gaps = 2/216 (0%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQ+QES+RGC+VYL+QPTC P N++LMEL +MIDACRRASA+ ITAV+PY+GYARADR
Sbjct: 58 LYVQIQESIRGCDVYLMQPTCWPVNDHLMELLIMIDACRRASARQITAVLPYYGYARADR 117
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KT GRESI AKLVANLIT+AGA+RVLA DLHS Q GYFDIP DHV+ PV++DYL SK
Sbjct: 118 KTAGRESITAKLVANLITQAGANRVLAMDLHSAQIQGYFDIPFDHVYGSPVLIDYLRSKN 177
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
++ DLVVVSPDVGGVARARAFAKKL DAPLAI+DKRR HNVAEV+N+IGDV+GK AV+
Sbjct: 178 LA--DLVVVSPDVGGVARARAFAKKLDDAPLAIIDKRRQAHNVAEVLNVIGDVQGKTAVL 235
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTI +GA LL ++GA +VYAC TH VFS
Sbjct: 236 VDDMIDTAGTICEGARLLRKQGASQVYACATHAVFS 271
>UniRef100_Q55848 Ribose-phosphate pyrophosphokinase [Synechocystis sp.]
Length = 333
Score = 325 bits (833), Expect = 1e-87
Identities = 161/216 (74%), Positives = 187/216 (86%), Gaps = 2/216 (0%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+Y+Q+QES+RG +VYLIQP C P N+NLMEL +MIDACRRASA+ ITAV+PY+GYARADR
Sbjct: 61 LYIQIQESIRGGDVYLIQPCCHPVNDNLMELLIMIDACRRASARQITAVLPYYGYARADR 120
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KT GRESI+AKLVANLIT AGA RVLA DLHS Q GYFDIP DH++ PVI+DYL SK
Sbjct: 121 KTAGRESISAKLVANLITGAGAQRVLAMDLHSAQIQGYFDIPCDHMYGSPVIIDYLKSKQ 180
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
++ DLVVVSPDVGGVARARAFAKKL+DAPLAI+DKRR HNVAEV+NLIGDV GK AV+
Sbjct: 181 LT--DLVVVSPDVGGVARARAFAKKLNDAPLAIIDKRRQSHNVAEVLNLIGDVDGKTAVL 238
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGT+++G+ LL +GAR+VYAC TH VFS
Sbjct: 239 VDDMIDTAGTLSEGSRLLRAQGARQVYACATHAVFS 274
>UniRef100_Q7NM67 Ribose-phosphate pyrophosphokinase [Gloeobacter violaceus]
Length = 332
Score = 314 bits (805), Expect = 2e-84
Identities = 153/216 (70%), Positives = 181/216 (82%), Gaps = 2/216 (0%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQ+QES+RGC+VYL+QPTC P N++LMEL ++IDACRRASA+ ITAV+PY+GYARADR
Sbjct: 60 LYVQIQESIRGCDVYLVQPTCSPVNDSLMELLILIDACRRASARQITAVLPYYGYARADR 119
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KT GRESI AKLVANLIT AG DRVLA DLHS Q YFDIP+DHV+ PV+L Y+ K
Sbjct: 120 KTAGRESITAKLVANLITAAGVDRVLAMDLHSAQIQAYFDIPLDHVYGSPVLLQYIKEKQ 179
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
+ D+V+VSPDVGGV+RARAFAKKL DAPLAIVDKRR N EVMN+IGDVKGK A++
Sbjct: 180 L--GDMVIVSPDVGGVSRARAFAKKLDDAPLAIVDKRRQAPNEVEVMNVIGDVKGKTAIL 237
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTI++ A +L +GA+EVYAC TH VFS
Sbjct: 238 VDDMIDTAGTISEAAKVLLRQGAKEVYACATHAVFS 273
>UniRef100_Q7V6S2 Ribose-phosphate pyrophosphokinase [Prochlorococcus marinus]
Length = 332
Score = 313 bits (801), Expect = 7e-84
Identities = 151/240 (62%), Positives = 191/240 (78%), Gaps = 11/240 (4%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQ+QES+RGC+V+LIQPTC P N+NLMEL +M+DAC+RASA+ ITAV+PY+GYARADR
Sbjct: 58 LYVQIQESIRGCDVFLIQPTCAPVNDNLMELLIMVDACQRASARQITAVVPYYGYARADR 117
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KT GRESI AKL ANL+ K+G +RVLA DLHS Q GYFDIP DH++ PV++DYLA++
Sbjct: 118 KTAGRESITAKLTANLLVKSGVNRVLAMDLHSAQIQGYFDIPCDHIYGSPVLVDYLAAQE 177
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
+ N++VVVSPDVGGVARARAFAK++ DAPLAI+DKRRSGHNVAE + +IGDV GK A++
Sbjct: 178 L--NEVVVVSPDVGGVARARAFAKQMRDAPLAIIDKRRSGHNVAESLTVIGDVAGKTAIL 235
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFSLIMFVLLYLQPSCNREAVQRLISR 373
+DDMIDT GTI GA LL ++GA+ V AC +H VFS P+C R + + L +
Sbjct: 236 IDDMIDTGGTICSGARLLRQQGAKRVIACASHAVFS---------PPACERLSEEGLFEQ 286
>UniRef100_Q7U7L5 Ribose-phosphate pyrophosphokinase [Synechococcus sp.]
Length = 331
Score = 311 bits (798), Expect = 2e-83
Identities = 145/216 (67%), Positives = 184/216 (85%), Gaps = 2/216 (0%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQ+QES+RGC+V+LIQPTC P N++LMEL +M+DACRRASA+ ITAV+PY+GYARADR
Sbjct: 58 LYVQIQESIRGCDVFLIQPTCAPVNDHLMELLIMVDACRRASARQITAVVPYYGYARADR 117
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KT GRESI AKL ANL+ K+G DRVLA DLHS Q GYFDIP DH++ PV++DYL+++
Sbjct: 118 KTAGRESITAKLTANLLVKSGVDRVLAMDLHSAQIQGYFDIPCDHIYGSPVLVDYLSTQN 177
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
+ +D+VVVSPDVGGVARARAFAK+++DAPLAI+DKRR+GHN+AE + +IGDV G+ A++
Sbjct: 178 L--DDIVVVSPDVGGVARARAFAKQMNDAPLAIIDKRRTGHNLAESLTVIGDVSGRTAIL 235
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
+DDMIDT GTI GA LL ++GA+ V AC TH VFS
Sbjct: 236 IDDMIDTGGTICAGARLLRQQGAKRVIACATHAVFS 271
>UniRef100_Q7V111 Ribose-phosphate pyrophosphokinase [Prochlorococcus marinus subsp.
pastoris]
Length = 331
Score = 311 bits (796), Expect = 3e-83
Identities = 152/245 (62%), Positives = 191/245 (77%), Gaps = 2/245 (0%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQ+Q+S+RGC+V+LIQPTC P N++LMEL +M+DAC+RASA+ ITAVIPYFGYARADR
Sbjct: 58 LYVQIQQSIRGCDVFLIQPTCAPVNDSLMELMIMVDACKRASARQITAVIPYFGYARADR 117
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KT GRESI AKL ANL+ K+G DRVLA DLHS Q GYFDIP DH++ PV++DYL S
Sbjct: 118 KTSGRESITAKLTANLLEKSGVDRVLAMDLHSAQIQGYFDIPCDHIYGSPVLIDYLES-- 175
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
+ ++VVVSPDVGGVARARAFAK +SDAPLAI+DKRRS HN+AE + +IG+VKGK A++
Sbjct: 176 LKLEEVVVVSPDVGGVARARAFAKLMSDAPLAIIDKRRSAHNIAESLTVIGEVKGKTAIL 235
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFSLIMFVLLYLQPSCNREAVQRLISR 373
+DDMIDT GTI GA LL +EGA+ ++AC +H VFS + L + + V I
Sbjct: 236 IDDMIDTGGTICSGANLLKKEGAKRIFACASHAVFSPPSYERLSSKDLFEQVIVTNSIPV 295
Query: 374 GDNHE 378
DN+E
Sbjct: 296 IDNYE 300
>UniRef100_UPI000031FFBA UPI000031FFBA UniRef100 entry
Length = 288
Score = 310 bits (794), Expect = 5e-83
Identities = 146/216 (67%), Positives = 182/216 (83%), Gaps = 2/216 (0%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQ+QES+RGC+V+LIQPTC P N++LMEL +M+DAC+RASA+ ITAVIPYFGYARADR
Sbjct: 20 LYVQIQESIRGCDVFLIQPTCAPVNDSLMELMIMVDACKRASARQITAVIPYFGYARADR 79
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KT GRESI AKL AN++ K+G DRVLA DLHS Q GYFDIP DH++ PV++DYL+ T
Sbjct: 80 KTAGRESITAKLTANILVKSGVDRVLAMDLHSAQIQGYFDIPCDHIYGSPVLVDYLS--T 137
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
+ ++VVVSPDVGGVARARAFAK++ D+PLAI+DKRRSGHNVAE + +IG+V GK A++
Sbjct: 138 MELEEIVVVSPDVGGVARARAFAKQMKDSPLAIIDKRRSGHNVAESLTVIGEVSGKTAIL 197
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
+DDMIDT GTI GA LL +EGA++V AC +H VFS
Sbjct: 198 IDDMIDTGGTICAGAELLRKEGAKKVIACASHAVFS 233
>UniRef100_UPI0000287166 UPI0000287166 UniRef100 entry
Length = 327
Score = 310 bits (793), Expect = 6e-83
Identities = 145/216 (67%), Positives = 183/216 (84%), Gaps = 2/216 (0%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQ+QES+RGC+V+LIQPTC P N++LMEL +M+DACRRASA+ ITAV+PY+GYARADR
Sbjct: 54 LYVQIQESIRGCDVFLIQPTCAPVNDHLMELLIMVDACRRASARQITAVVPYYGYARADR 113
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KT GRESI AKL ANL+ K+G +RVLA DLHS Q GYFDIP DH++ PV++DYL+++
Sbjct: 114 KTAGRESITAKLTANLLVKSGVNRVLAMDLHSSQIQGYFDIPCDHIYGSPVLVDYLSTQN 173
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
+ D+VVVSPDVGGVARARAFAK+++DAPLAI+DKRR+GHN+AE + +IGDV G+ AV+
Sbjct: 174 L--GDIVVVSPDVGGVARARAFAKQMNDAPLAIIDKRRTGHNMAESLTVIGDVSGRTAVL 231
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
+DDMIDT GTI GA LL ++GA+ V AC TH VFS
Sbjct: 232 IDDMIDTGGTICAGARLLRQQGAKRVIACATHAVFS 267
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.337 0.146 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 568,104,730
Number of Sequences: 2790947
Number of extensions: 20787186
Number of successful extensions: 111728
Number of sequences better than 10.0: 1023
Number of HSP's better than 10.0 without gapping: 947
Number of HSP's successfully gapped in prelim test: 76
Number of HSP's that attempted gapping in prelim test: 109124
Number of HSP's gapped (non-prelim): 1287
length of query: 400
length of database: 848,049,833
effective HSP length: 129
effective length of query: 271
effective length of database: 488,017,670
effective search space: 132252788570
effective search space used: 132252788570
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146791.3


