

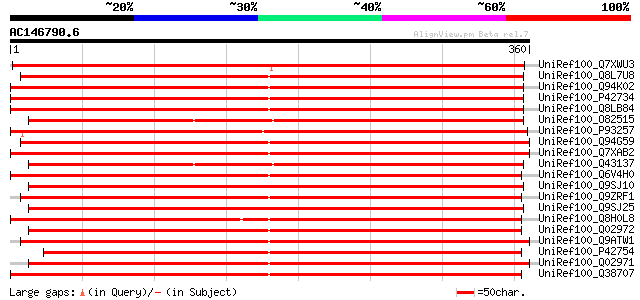
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146790.6 - phase: 0
(360 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XWU3 OSJNBa0065B15.8 protein [Oryza sativa] 541 e-152
UniRef100_Q8L7U8 Putative sinapyl alcohol dehydrogenase [Populus... 447 e-124
UniRef100_Q94K02 Cinnamyl-alcohol dehydrogenase CAD1 [Arabidopsi... 446 e-124
UniRef100_P42734 Probable mannitol dehydrogenase [Arabidopsis th... 446 e-124
UniRef100_Q8LB84 Cinnamyl-alcohol dehydrogenase CAD1 [Arabidopsi... 444 e-123
UniRef100_O82515 Probable mannitol dehydrogenase [Medicago sativa] 437 e-121
UniRef100_P93257 Probable mannitol dehydrogenase [Mesembryanthem... 437 e-121
UniRef100_Q94G59 Sinapyl alcohol dehydrogenase [Populus tremuloi... 433 e-120
UniRef100_Q7XAB2 10-hydroxygeraniol oxidoreductase [Camptotheca ... 433 e-120
UniRef100_Q43137 Probable mannitol dehydrogenase 1 [Stylosanthes... 431 e-119
UniRef100_Q6V4H0 10-hydroxygeraniol oxidoreductase [Catharanthus... 430 e-119
UniRef100_Q9SJ10 Cinnamyl alcohol dehydrogenase-like protein [Ar... 429 e-119
UniRef100_Q9ZRF1 Probable mannitol dehydrogenase [Fragaria anana... 428 e-118
UniRef100_Q9SJ25 Cinnamyl alcohol dehydrogenase-like protein [Ar... 428 e-118
UniRef100_Q8H0L8 Alcohol NADP+ oxidoreductase [Solanum tuberosum] 420 e-116
UniRef100_Q02972 Probable mannitol dehydrogenase 2 [Arabidopsis ... 418 e-115
UniRef100_Q9ATW1 Cinnamyl alcohol dehydrogenase [Fragaria ananassa] 417 e-115
UniRef100_P42754 Mannitol dehydrogenase [Petroselinum crispum] 415 e-114
UniRef100_Q02971 Probable mannitol dehydrogenase 1 [Arabidopsis ... 407 e-112
UniRef100_Q38707 Mannitol dehydrogenase [Apium graveolens] 405 e-111
>UniRef100_Q7XWU3 OSJNBa0065B15.8 protein [Oryza sativa]
Length = 360
Score = 541 bits (1394), Expect = e-152
Identities = 248/357 (69%), Positives = 305/357 (84%), Gaps = 2/357 (0%)
Query: 3 KTTPNHTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGI 62
+ TPNHTQTVSGWAA D SGKI P+ FKRRENG DVTIK+ YCG+CHTD+H +DWGI
Sbjct: 2 EVTPNHTQTVSGWAAMDESGKIVPFVFKRRENGVDDVTIKVKYCGMCHTDLHFIHNDWGI 61
Query: 63 TMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFV 122
TMYPVVPGHEITGV+TKVG++V GFK GD+VGVGC+AASCLDCE+C+ +ENYC+K+
Sbjct: 62 TMYPVVPGHEITGVVTKVGTNVAGFKVGDRVGVGCIAASCLDCEHCRRSEENYCDKVALT 121
Query: 123 YNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLV-- 180
YNGIFWDGSITYGGYS MLV R+VV IP++LP+DAAAPLLCAGITV+SP+K HG++
Sbjct: 122 YNGIFWDGSITYGGYSGMLVAHKRFVVRIPDTLPLDAAAPLLCAGITVYSPMKQHGMLQA 181
Query: 181 STAGKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQ 240
AG+R+GVVGLGGLGH+AVKFGKAFG HVTVISTSP+K+ EA+E L AD+F++ST+ Q
Sbjct: 182 DAAGRRLGVVGLGGLGHVAVKFGKAFGLHVTVISTSPAKEREARENLKADNFVVSTDQKQ 241
Query: 241 LQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKG 300
+QA RSLD+I+DTV+A H+L PILELLKVNG L +VGAP+KP++LP+FPLIFGKR++ G
Sbjct: 242 MQAMTRSLDYIIDTVAATHSLGPILELLKVNGKLVLVGAPEKPVELPSFPLIFGKRTVSG 301
Query: 301 GIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANAS 357
+ GG+KETQEM+D+CG+HNITCDIE++ D IN+A RL +NDVRYRFVI++ S
Sbjct: 302 SMTGGMKETQEMMDICGEHNITCDIEIVSTDRINDALARLARNDVRYRFVINVGGDS 358
>UniRef100_Q8L7U8 Putative sinapyl alcohol dehydrogenase [Populus tremula x Populus
tremuloides]
Length = 362
Score = 447 bits (1150), Expect = e-124
Identities = 206/349 (59%), Positives = 270/349 (77%), Gaps = 1/349 (0%)
Query: 8 HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPV 67
H GW A DSSG ++P+ F RR+NG DVTIKILYCG+CH+D+H AK++WG + YP+
Sbjct: 9 HQHKAFGWTAKDSSGVLSPFHFSRRDNGVEDVTIKILYCGVCHSDLHAAKNEWGFSRYPL 68
Query: 68 VPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIF 127
VPGHEI G++TK+GS+V+ FK D+VGVG + SC CEYC D ENYC K+ F YN
Sbjct: 69 VPGHEIVGIVTKIGSNVKKFKVDDQVGVGVMVNSCKSCEYCDQDSENYCPKMIFTYNAQN 128
Query: 128 WDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRI 187
+DG+ TYGGYS +VVD +V+HIP+S+P D AAPLLCAGITV+SP+K +G+ + GK +
Sbjct: 129 YDGTKTYGGYSDTIVVDQHFVLHIPDSMPADGAAPLLCAGITVYSPMKYYGM-TEPGKHL 187
Query: 188 GVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRS 247
G+VGLGGLGH+AVK GKAFG VTVIS+S K+ EA +RLGAD F++S++P++++AA +
Sbjct: 188 GIVGLGGLGHVAVKIGKAFGLKVTVISSSSRKEREALDRLGADSFLVSSDPEKMKAAFGT 247
Query: 248 LDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIK 307
+D+I+DTVSA HAL P+L LLK NG L +G P+KPL+LP FPL+ G++ + G IGG+K
Sbjct: 248 MDYIIDTVSAVHALAPLLSLLKTNGKLVTLGLPEKPLELPIFPLVLGRKLVGGSDIGGMK 307
Query: 308 ETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANA 356
ETQEMLD C KHNI D+E+I+ D IN A RL K+DVRYRFVID+AN+
Sbjct: 308 ETQEMLDFCAKHNIIADVEVIRMDQINTAMDRLAKSDVRYRFVIDVANS 356
>UniRef100_Q94K02 Cinnamyl-alcohol dehydrogenase CAD1 [Arabidopsis thaliana]
Length = 360
Score = 446 bits (1148), Expect = e-124
Identities = 210/357 (58%), Positives = 271/357 (75%), Gaps = 2/357 (0%)
Query: 1 MAKTTPN-HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDD 59
MAK+ H V GW A D SG ++P+ F RR+NG DVT+KIL+CG+CHTD+H K+D
Sbjct: 1 MAKSPETEHPNKVFGWGARDKSGVLSPFHFSRRDNGENDVTVKILFCGVCHTDLHTIKND 60
Query: 60 WGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKL 119
WG + YPVVPGHEI G+ TKVG +V FK+GD+VGVG ++ SC CE C D ENYC ++
Sbjct: 61 WGYSYYPVVPGHEIVGIATKVGKNVTKFKEGDRVGVGVISGSCQSCESCDQDLENYCPQM 120
Query: 120 QFVYNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGL 179
F YN I DG+ YGGYS+ +VVD R+V+ PE+LP D+ APLLCAGITV+SP+K +G+
Sbjct: 121 SFTYNAIGSDGTKNYGGYSENIVVDQRFVLRFPENLPSDSGAPLLCAGITVYSPMKYYGM 180
Query: 180 VSTAGKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPD 239
+ AGK +GV GLGGLGH+AVK GKAFG VTVIS+S +K EA LGAD F+++T+P
Sbjct: 181 -TEAGKHLGVAGLGGLGHVAVKIGKAFGLKVTVISSSSTKAEEAINHLGADSFLVTTDPQ 239
Query: 240 QLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIK 299
+++AA ++D+I+DT+SA HAL P+L LLKVNG L +G P+KPL+LP FPL+ G++ +
Sbjct: 240 KMKAAIGTMDYIIDTISAVHALYPLLGLLKVNGKLIALGLPEKPLELPMFPLVLGRKMVG 299
Query: 300 GGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANA 356
G +GG+KETQEMLD C KHNIT DIELIK D IN A +RL K+DVRYRFVID+AN+
Sbjct: 300 GSDVGGMKETQEMLDFCAKHNITADIELIKMDEINTAMERLAKSDVRYRFVIDVANS 356
>UniRef100_P42734 Probable mannitol dehydrogenase [Arabidopsis thaliana]
Length = 360
Score = 446 bits (1146), Expect = e-124
Identities = 210/357 (58%), Positives = 271/357 (75%), Gaps = 2/357 (0%)
Query: 1 MAKTTPN-HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDD 59
MAK+ H V GW A D SG ++P+ F RR+NG DVT+KIL+CG+CHTD+H K+D
Sbjct: 1 MAKSPETEHPNKVFGWGARDKSGVLSPFHFSRRDNGENDVTVKILFCGVCHTDLHTIKND 60
Query: 60 WGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKL 119
WG + YPVVPGHEI G+ TKVG +V FK+GD+VGVG ++ SC CE C D ENYC ++
Sbjct: 61 WGYSYYPVVPGHEIVGIATKVGKNVTKFKEGDRVGVGVISGSCQSCESCDQDLENYCPQM 120
Query: 120 QFVYNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGL 179
F YN I DG+ YGGYS+ +VVD R+V+ PE+LP D+ APLLCAGITV+SP+K +G+
Sbjct: 121 SFTYNAIGSDGTKNYGGYSENIVVDQRFVLRFPENLPSDSGAPLLCAGITVYSPMKYYGM 180
Query: 180 VSTAGKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPD 239
+ AGK +GV GLGGLGH+AVK GKAFG VTVIS+S +K EA LGAD F+++T+P
Sbjct: 181 -TEAGKHLGVAGLGGLGHVAVKSGKAFGLKVTVISSSSTKAEEAINHLGADSFLVTTDPQ 239
Query: 240 QLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIK 299
+++AA ++D+I+DT+SA HAL P+L LLKVNG L +G P+KPL+LP FPL+ G++ +
Sbjct: 240 KMKAAIGTMDYIIDTISAVHALYPLLGLLKVNGKLIALGLPEKPLELPMFPLVLGRKMVG 299
Query: 300 GGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANA 356
G +GG+KETQEMLD C KHNIT DIELIK D IN A +RL K+DVRYRFVID+AN+
Sbjct: 300 GSDVGGMKETQEMLDFCAKHNITADIELIKMDEINTAMERLAKSDVRYRFVIDVANS 356
>UniRef100_Q8LB84 Cinnamyl-alcohol dehydrogenase CAD1 [Arabidopsis thaliana]
Length = 360
Score = 444 bits (1143), Expect = e-123
Identities = 209/357 (58%), Positives = 271/357 (75%), Gaps = 2/357 (0%)
Query: 1 MAKTTPN-HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDD 59
MAK+ H V GW A D SG ++P+ F RR+NG DVT+KIL+CG+CHTD+H K+D
Sbjct: 1 MAKSPETEHPNKVFGWGARDKSGVLSPFHFSRRDNGENDVTVKILFCGVCHTDLHTIKND 60
Query: 60 WGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKL 119
WG + YPVVPGHEI G+ TKVG +V FK+GD+VGVG ++ SC CE C D ENYC ++
Sbjct: 61 WGYSYYPVVPGHEIVGIATKVGKNVTKFKEGDRVGVGVISGSCQSCESCDQDLENYCPQM 120
Query: 120 QFVYNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGL 179
F YN I DG+ YGGYS+ +VVD R+V+ PE+LP D+ APLLCAGITV+SP+K +G+
Sbjct: 121 SFTYNAIGSDGTKNYGGYSENIVVDQRFVLRFPENLPSDSGAPLLCAGITVYSPMKYYGM 180
Query: 180 VSTAGKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPD 239
+ AGK +GV GLGGLGH+AVK GKAFG VTVIS+S +K EA LGAD F+++T+P
Sbjct: 181 -TEAGKHLGVAGLGGLGHVAVKIGKAFGLKVTVISSSSTKAEEAINHLGADSFLVTTDPQ 239
Query: 240 QLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIK 299
+++AA ++D+I+DT+SA HAL P+L LLKVNG L +G P+KPL+LP FPL+ G++ +
Sbjct: 240 KMKAAIGTMDYIIDTISAVHALYPLLGLLKVNGKLIALGLPEKPLELPMFPLVLGRKMVG 299
Query: 300 GGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANA 356
G +GG+KETQEMLD C KHNIT DIELIK D IN A +RL K+DVRYRFVI++AN+
Sbjct: 300 GSDVGGMKETQEMLDFCAKHNITADIELIKMDEINTAMERLAKSDVRYRFVINVANS 356
>UniRef100_O82515 Probable mannitol dehydrogenase [Medicago sativa]
Length = 359
Score = 437 bits (1125), Expect = e-121
Identities = 207/343 (60%), Positives = 264/343 (76%), Gaps = 2/343 (0%)
Query: 14 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 73
GWAA D+SG ++P+ F RRENG DV++KILYCG+CH+D+H K+DWG T YPVVPGHEI
Sbjct: 15 GWAARDTSGTLSPFHFSRRENGDDDVSVKILYCGVCHSDLHTLKNDWGFTTYPVVPGHEI 74
Query: 74 TGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSIT 133
GV+TKVG +V+ F+ GD VGVG + SC CE C D E YC K F YN + G+ T
Sbjct: 75 VGVVTKVGINVKKFRVGDNVGVGVIVESCQTCENCNQDLEQYCPKPVFTYNSPY-KGTRT 133
Query: 134 YGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLG 193
YGGYS +VV RYVV P++LP+DA APLLCAGITV+SP+K +G+ GK +GV GLG
Sbjct: 134 YGGYSDFVVVHQRYVVQFPDNLPLDAGAPLLCAGITVYSPMKYYGMTEP-GKHLGVAGLG 192
Query: 194 GLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDFILD 253
GLGH+A+KFGKAFG VTVISTSP+K+ EA ++LGAD F++S +P++++AA ++D+I+D
Sbjct: 193 GLGHVAIKFGKAFGLKVTVISTSPNKETEAIDKLGADSFLVSKDPEKMKAAMGTMDYIID 252
Query: 254 TVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKETQEML 313
T+SA H+L+P+L LLK+NG L VG P KPL+L FPL+ G++ I G IGG+KETQEML
Sbjct: 253 TISAAHSLMPLLGLLKLNGKLVTVGLPSKPLELSVFPLVAGRKLIGGSNIGGMKETQEML 312
Query: 314 DVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANA 356
D CGKHNIT DIELIK IN A +RL K DV+YRFVID+AN+
Sbjct: 313 DFCGKHNITADIELIKMHEINTAMERLHKADVKYRFVIDVANS 355
>UniRef100_P93257 Probable mannitol dehydrogenase [Mesembryanthemum crystallinum]
Length = 361
Score = 437 bits (1124), Expect = e-121
Identities = 206/361 (57%), Positives = 265/361 (73%), Gaps = 3/361 (0%)
Query: 1 MAKTTPN--HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKD 58
MA + P H Q GWAA D+SG ++P F RR G DVT K+LYCGICH+D+H+ K+
Sbjct: 1 MANSAPENVHPQKAFGWAARDTSGTLSPLKFSRRATGEQDVTFKVLYCGICHSDLHYIKN 60
Query: 59 DWGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEK 118
+WG +YP +PGHEI GV+T+VG+ V+ FK GDKVGVGC+ SC CE C+ ENYC K
Sbjct: 61 EWGNAVYPAIPGHEIVGVVTEVGNKVQNFKVGDKVGVGCMVGSCRSCESCENHLENYCPK 120
Query: 119 LQFVYNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHG 178
+ Y ++DG++TYGGYS ++VV+ + V IP+++ +DA APLLCAG+TV+SPLK H
Sbjct: 121 MILTYGSTYYDGTLTYGGYSDIMVVEEHFAVRIPDNMALDATAPLLCAGVTVYSPLK-HF 179
Query: 179 LVSTAGKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNP 238
+ G IGVVGLGGLGHMAVKFGKAFG VTVISTSP+K+ EA RLGAD F++S P
Sbjct: 180 ELDKPGLHIGVVGLGGLGHMAVKFGKAFGAKVTVISTSPNKKDEAVNRLGADSFVVSREP 239
Query: 239 DQLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSI 298
+Q+Q+A +LD I+DTVSA H LLP+L LLK G + +VG PDKPL+LP FPL+ G++ +
Sbjct: 240 EQMQSAMGTLDGIIDTVSAAHPLLPLLGLLKSQGKMIMVGVPDKPLELPVFPLLQGRKIL 299
Query: 299 KGGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANASP 358
G IGG+KETQEM+D KH+I DIE++ D +N A +RLLK DVRYRFVID+AN
Sbjct: 300 AGSCIGGMKETQEMIDFAAKHDIKSDIEVVPMDYVNTAMERLLKGDVRYRFVIDVANTLK 359
Query: 359 A 359
A
Sbjct: 360 A 360
>UniRef100_Q94G59 Sinapyl alcohol dehydrogenase [Populus tremuloides]
Length = 362
Score = 433 bits (1114), Expect = e-120
Identities = 206/353 (58%), Positives = 257/353 (72%), Gaps = 1/353 (0%)
Query: 8 HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPV 67
H GWAA D SG ++P+ F RR G DV K+LYCG+CH+D+H K+DWG +MYP+
Sbjct: 9 HPVKAFGWAARDQSGHLSPFNFSRRATGEEDVRFKVLYCGVCHSDLHSIKNDWGFSMYPL 68
Query: 68 VPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIF 127
VPGHEI G +T+VGS V+ GDKVGVGCL +C CE C D ENYC K+ Y I+
Sbjct: 69 VPGHEIVGEVTEVGSKVKKVNVGDKVGVGCLVGACHSCESCANDLENYCPKMILTYASIY 128
Query: 128 WDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRI 187
DG+ITYGGYS +V + RY++ P+++P+D APLLCAGITV+SPLK GL GK I
Sbjct: 129 HDGTITYGGYSDHMVANERYIIRFPDNMPLDGGAPLLCAGITVYSPLKYFGL-DEPGKHI 187
Query: 188 GVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRS 247
G+VGLGGLGH+AVKF KAFG VTVISTSPSK+ EA + GAD F++S + +Q+QAA +
Sbjct: 188 GIVGLGGLGHVAVKFAKAFGSKVTVISTSPSKKEEALKNFGADSFLVSRDQEQMQAAAGT 247
Query: 248 LDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIK 307
LD I+DTVSA H LLP+ LLK +G L +VGAP+KPL+LPAF LI G++ + G IGG+K
Sbjct: 248 LDGIIDTVSAVHPLLPLFGLLKSHGKLILVGAPEKPLELPAFSLIAGRKIVAGSGIGGMK 307
Query: 308 ETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANASPAT 360
ETQEM+D KHNIT DIE+I D +N A +RL KNDVRYRFVID+ N AT
Sbjct: 308 ETQEMIDFAAKHNITADIEVISTDYLNTAMERLAKNDVRYRFVIDVGNTLAAT 360
>UniRef100_Q7XAB2 10-hydroxygeraniol oxidoreductase [Camptotheca acuminata]
Length = 360
Score = 433 bits (1113), Expect = e-120
Identities = 206/361 (57%), Positives = 267/361 (73%), Gaps = 2/361 (0%)
Query: 1 MAKT-TPNHTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDD 59
MAK+ H GWAA D+SG ++P TF RR G DV K++YCGICH+D+H +++
Sbjct: 1 MAKSPAEEHPVKAFGWAASDTSGVLSPLTFSRRATGEKDVKFKVMYCGICHSDLHCLRNE 60
Query: 60 WGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKL 119
WG T+YP++PGHEI G++T+VGS VE FK GDKVGVGC+ SC C C + ENYC K+
Sbjct: 61 WGSTIYPIIPGHEIAGIVTEVGSKVEKFKVGDKVGVGCMVGSCRSCNDCDNNIENYCPKM 120
Query: 120 QFVYNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGL 179
F Y+ I+ DG++TYGGYS ++V D +VV P++LP+D+ APLLCAGIT +SPL+ +GL
Sbjct: 121 MFTYSSIYHDGTVTYGGYSDIMVTDEHFVVRWPDNLPLDSGAPLLCAGITTYSPLRYYGL 180
Query: 180 VSTAGKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPD 239
G +GVVGLGGLGH+AVKF KA G VTVISTS +K+ EA ERLGAD F++S +PD
Sbjct: 181 -DKPGMNLGVVGLGGLGHVAVKFAKALGVKVTVISTSLNKKEEAIERLGADSFLVSRDPD 239
Query: 240 QLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIK 299
Q+QAA +LD ILDTVSA H LLP+L L+K +G L +VGAP+KPL+LP PL+ G++ +
Sbjct: 240 QMQAAAGTLDGILDTVSATHPLLPLLGLIKSHGKLVLVGAPEKPLELPVMPLLMGRKIVG 299
Query: 300 GGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANASPA 359
G +GG+KETQEMLD KHNIT D+E++ D +N A +RL K DVRYRFV+DIAN A
Sbjct: 300 GSNMGGMKETQEMLDFAAKHNITADVEVVPMDYVNTAMKRLEKGDVRYRFVLDIANTLKA 359
Query: 360 T 360
T
Sbjct: 360 T 360
>UniRef100_Q43137 Probable mannitol dehydrogenase 1 [Stylosanthes humilis]
Length = 354
Score = 431 bits (1107), Expect = e-119
Identities = 202/343 (58%), Positives = 267/343 (76%), Gaps = 2/343 (0%)
Query: 14 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 73
GWAA D+SG ++P+ F RR+N DVT+KILYCG+CH+D+H K+DWG T YPVVPGHEI
Sbjct: 8 GWAAKDASGHLSPFHFTRRQNEADDVTLKILYCGVCHSDLHTVKNDWGFTTYPVVPGHEI 67
Query: 74 TGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSIT 133
G++TKVGS+V FK+GD+VGVG + SC +CE C+ D E+YC K F YN + G+ T
Sbjct: 68 AGIVTKVGSNVTKFKEGDRVGVGVIVDSCQECECCQQDLESYCPKPVFTYNSPY-KGTRT 126
Query: 134 YGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLG 193
GGYS +VV R+V+ P++LP+DA APLLCAGITV+SP+K +G+ GK +GV GLG
Sbjct: 127 QGGYSDFVVVHQRFVLQFPDNLPLDAGAPLLCAGITVYSPMKYYGMTEP-GKHLGVAGLG 185
Query: 194 GLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDFILD 253
GLGH+A+KFGKAFG VTVIS+SP+K++EA + LGAD F++S++P++++AA ++D+I+D
Sbjct: 186 GLGHVAIKFGKAFGLKVTVISSSPNKESEAIDVLGADSFLLSSDPEKMKAATGTMDYIID 245
Query: 254 TVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKETQEML 313
T+SA H+L+ +L LLK+NG L VG P KPLQLP FPL+ G++ I G GG+KETQEML
Sbjct: 246 TISAVHSLVSLLGLLKLNGKLVTVGLPSKPLQLPIFPLVAGRKLIGGSNFGGLKETQEML 305
Query: 314 DVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANA 356
D CGKHNI +IELIK D IN A +RL K DV+YRFVID+AN+
Sbjct: 306 DFCGKHNIAANIELIKMDEINTAIERLSKADVKYRFVIDVANS 348
>UniRef100_Q6V4H0 10-hydroxygeraniol oxidoreductase [Catharanthus roseus]
Length = 360
Score = 430 bits (1106), Expect = e-119
Identities = 205/356 (57%), Positives = 261/356 (72%), Gaps = 2/356 (0%)
Query: 1 MAKTTP-NHTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDD 59
MAK+ H GWAA D+SG ++P+ F RR G DV K+LYCGICH+D+H K++
Sbjct: 1 MAKSPEVEHPVKAFGWAARDTSGHLSPFHFSRRATGEHDVQFKVLYCGICHSDLHMIKNE 60
Query: 60 WGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKL 119
WG T YP+VPGHEI G++T+VGS VE FK GDKVGVGCL SC C+ C D ENYC
Sbjct: 61 WGFTKYPIVPGHEIVGIVTEVGSKVEKFKVGDKVGVGCLVGSCRKCDMCTKDLENYCPGQ 120
Query: 120 QFVYNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGL 179
Y+ + DG+ TYGGYS ++V D +V+ PE+LPMD APLLCAGIT +SPL+ GL
Sbjct: 121 ILTYSATYTDGTTTYGGYSDLMVADEHFVIRWPENLPMDIGAPLLCAGITTYSPLRYFGL 180
Query: 180 VSTAGKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPD 239
G +GVVGLGGLGH+AVKF KAFG VTVISTS SK+ EA E+LGAD F++S +P+
Sbjct: 181 -DKPGTHVGVVGLGGLGHVAVKFAKAFGAKVTVISTSESKKQEALEKLGADSFLVSRDPE 239
Query: 240 QLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIK 299
Q++AA SLD I+DTVSA H ++P+L +LK +G L +VGAP+KPL+LP+FPLI G++ I
Sbjct: 240 QMKAAAASLDGIIDTVSAIHPIMPLLSILKSHGKLILVGAPEKPLELPSFPLIAGRKIIA 299
Query: 300 GGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIAN 355
G IGG+KETQEM+D KHN+ D+EL+ D +N A +RLLK DV+YRFVID+AN
Sbjct: 300 GSAIGGLKETQEMIDFAAKHNVLPDVELVSMDYVNTAMERLLKADVKYRFVIDVAN 355
>UniRef100_Q9SJ10 Cinnamyl alcohol dehydrogenase-like protein [Arabidopsis thaliana]
Length = 375
Score = 429 bits (1104), Expect = e-119
Identities = 198/343 (57%), Positives = 259/343 (74%)
Query: 14 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 73
GWAA+D SG ++P+ F RRENG DVT+KIL+CG+CH+D+H K+ WG + YP++PGHEI
Sbjct: 9 GWAANDESGVLSPFHFSRRENGENDVTVKILFCGVCHSDLHTIKNHWGFSRYPIIPGHEI 68
Query: 74 TGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSIT 133
G+ TKVG +V FK+GD+VGVG + SC CE C D ENYC K+ F YN DG+
Sbjct: 69 VGIATKVGKNVTKFKEGDRVGVGVIIGSCQSCESCNQDLENYCPKVVFTYNSRSSDGTRN 128
Query: 134 YGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLG 193
GGYS ++VVD+R+V+ IP+ LP D+ APLLCAGITV+SP+K +G+ +GKR+GV GLG
Sbjct: 129 QGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGMTKESGKRLGVNGLG 188
Query: 194 GLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDFILD 253
GLGH+AVK GKAFG VTVIS S K+ EA +RLGAD F+++T+ +++ A ++DFI+D
Sbjct: 189 GLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDSQKMKEAVGTMDFIID 248
Query: 254 TVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKETQEML 313
TVSA+HALLP+ LLKV+G L +G +KPL LP FPL+ G++ + G IGG+KETQEML
Sbjct: 249 TVSAEHALLPLFSLLKVSGKLVALGLLEKPLDLPIFPLVLGRKMVGGSQIGGMKETQEML 308
Query: 314 DVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANA 356
+ C KH I DIELIK IN A RL+K+DVRYRFVID+AN+
Sbjct: 309 EFCAKHKIVSDIELIKMSDINSAMDRLVKSDVRYRFVIDVANS 351
>UniRef100_Q9ZRF1 Probable mannitol dehydrogenase [Fragaria ananassa]
Length = 359
Score = 428 bits (1101), Expect = e-118
Identities = 202/348 (58%), Positives = 257/348 (73%), Gaps = 1/348 (0%)
Query: 8 HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPV 67
H + SGWAA DSSG ++P+ F RRE G DVT K+LYCGICH+D+H K++WG + YP+
Sbjct: 7 HRKKASGWAARDSSGVLSPFNFYRRETGEKDVTFKVLYCGICHSDLHMVKNEWGFSTYPL 66
Query: 68 VPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIF 127
VPGHEI G +T+VGS V+ FK GD+VGVGC+ SC CE C ENYC K Y +
Sbjct: 67 VPGHEIVGEVTEVGSKVQKFKVGDRVGVGCIVGSCRSCENCTDHLENYCPKQILTYGAKY 126
Query: 128 WDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRI 187
+DGS TYGGYS ++V D ++V IP++LP+D AAPLLCAGIT +SPL+ GL G +
Sbjct: 127 YDGSTTYGGYSDIMVADEHFIVRIPDNLPLDGAAPLLCAGITTYSPLRYFGL-DKPGMHV 185
Query: 188 GVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRS 247
GVVGLGGLGH+AVKF KA G VTVISTSP K+ EA + LGAD F++S + DQ+QAA +
Sbjct: 186 GVVGLGGLGHVAVKFAKAMGVKVTVISTSPKKEEEALKHLGADSFLVSRDQDQMQAAIGT 245
Query: 248 LDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIK 307
+D I+DTVSA H LLP++ LL +G L +VGAP+KPL+LP FPL+ G++ + G IGG+K
Sbjct: 246 MDGIIDTVSAQHPLLPLIGLLNSHGKLVMVGAPEKPLELPVFPLLMGRKMVAGSGIGGMK 305
Query: 308 ETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIAN 355
ETQEM+D +HNIT DIE+I D +N A +RL+K DVRYRFVIDI N
Sbjct: 306 ETQEMIDFAARHNITADIEVIPIDYLNTAMERLVKADVRYRFVIDIGN 353
>UniRef100_Q9SJ25 Cinnamyl alcohol dehydrogenase-like protein [Arabidopsis thaliana]
Length = 376
Score = 428 bits (1100), Expect = e-118
Identities = 200/344 (58%), Positives = 258/344 (74%), Gaps = 1/344 (0%)
Query: 14 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 73
GWAA+D SG ++P+ F RRENG DVT+KIL+CG+CH+D+H K+ WG + YP++PGHEI
Sbjct: 9 GWAANDESGVLSPFHFSRRENGENDVTVKILFCGVCHSDLHTIKNHWGFSRYPIIPGHEI 68
Query: 74 TGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDG-SI 132
G+ TKVG +V FK+GD+VGVG + SC CE C D ENYC K+ F YN DG S
Sbjct: 69 VGIATKVGKNVTKFKEGDRVGVGVIIGSCQSCESCNQDLENYCPKVVFTYNSRSSDGTSR 128
Query: 133 TYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGL 192
GGYS ++VVD+R+V+ IP+ LP D+ APLLCAGITV+SP+K +G+ +GKR+GV GL
Sbjct: 129 NQGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGMTKESGKRLGVNGL 188
Query: 193 GGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDFIL 252
GGLGH+AVK GKAFG VTVIS S K+ EA +RLGAD F+++T+ +++ A ++DFI+
Sbjct: 189 GGLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDSQKMKEAVGTMDFII 248
Query: 253 DTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKETQEM 312
DTVSA+HALLP+ LLKVNG L +G P+KPL LP F L+ G++ + G IGG+KETQEM
Sbjct: 249 DTVSAEHALLPLFSLLKVNGKLVALGLPEKPLDLPIFSLVLGRKMVGGSQIGGMKETQEM 308
Query: 313 LDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANA 356
L+ C KH I DIELIK IN A RL K+DVRYRFVID+AN+
Sbjct: 309 LEFCAKHKIVSDIELIKMSDINSAMDRLAKSDVRYRFVIDVANS 352
>UniRef100_Q8H0L8 Alcohol NADP+ oxidoreductase [Solanum tuberosum]
Length = 359
Score = 420 bits (1080), Expect = e-116
Identities = 207/356 (58%), Positives = 261/356 (73%), Gaps = 3/356 (0%)
Query: 1 MAKTTPN-HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDD 59
MAK++ N H GWA +SG ++P+ F RR G V K++YCGICH+D+H K++
Sbjct: 1 MAKSSENEHPIKAFGWATRHTSGVLSPFNFSRRVTGEKHVQFKVMYCGICHSDLHQLKNE 60
Query: 60 WGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKL 119
WG T YP+VPGHE+ GV+ +VGS VE FK GDKVGVGC+ SC CE C D ENYC +
Sbjct: 61 WGNTKYPMVPGHEVVGVVIEVGSKVEKFKVGDKVGVGCMVGSCRKCENCTVDLENYCPRQ 120
Query: 120 QFVYNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGL 179
YNG DG++T+GGYS M+V D +VV PE+L MDAA PLLCAGIT +SPLK GL
Sbjct: 121 IPTYNGYSLDGTLTFGGYSDMMVSDEHFVVRWPENLSMDAA-PLLCAGITTYSPLKYFGL 179
Query: 180 VSTAGKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPD 239
G IGVVGLGGLGHMAVKF KAFG VTVISTS +K+ EA ERLGAD F+IS +P+
Sbjct: 180 -DKPGMHIGVVGLGGLGHMAVKFAKAFGTKVTVISTSANKKQEAIERLGADSFLISRDPE 238
Query: 240 QLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIK 299
Q++AA +LD I+DTVSA H +LP+L L+K +G L +VGAP+KP++LP FPL+ G++ +
Sbjct: 239 QMKAAMNTLDGIIDTVSAVHPILPLLMLMKSHGKLVMVGAPEKPVELPVFPLLMGRKLVA 298
Query: 300 GGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIAN 355
G IGG+KETQEMLD KHNIT DIE++ D +N A +RLLK+DV+YRFV+DI N
Sbjct: 299 GSCIGGMKETQEMLDFAAKHNITPDIEVVPMDYVNTALERLLKSDVKYRFVLDIGN 354
>UniRef100_Q02972 Probable mannitol dehydrogenase 2 [Arabidopsis thaliana]
Length = 359
Score = 418 bits (1075), Expect = e-115
Identities = 202/342 (59%), Positives = 258/342 (75%), Gaps = 1/342 (0%)
Query: 14 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 73
G AA D+SG ++P++F RRE G DV K+L+CGICH+D+H K++WG++ YP+VPGHEI
Sbjct: 11 GLAAKDNSGVLSPFSFTRRETGEKDVRFKVLFCGICHSDLHMVKNEWGMSTYPLVPGHEI 70
Query: 74 TGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSIT 133
GV+T+VG+ V FK G+KVGVGCL +SC C+ C ENYC K Y ++D +IT
Sbjct: 71 VGVVTEVGAKVTKFKTGEKVGVGCLVSSCGSCDSCTEGMENYCPKSIQTYGFPYYDNTIT 130
Query: 134 YGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLG 193
YGGYS +V + +V+ IP++LP+DAAAPLLCAGITV+SP+K HGL G IGVVGLG
Sbjct: 131 YGGYSDHMVCEEGFVIRIPDNLPLDAAAPLLCAGITVYSPMKYHGL-DKPGMHIGVVGLG 189
Query: 194 GLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDFILD 253
GLGH+ VKF KA G VTVISTS K+ EA RLGAD F++S +P Q++ A ++D I+D
Sbjct: 190 GLGHVGVKFAKAMGTKVTVISTSEKKRDEAINRLGADAFLVSRDPKQIKDAMGTMDGIID 249
Query: 254 TVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKETQEML 313
TVSA H+LLP+L LLK G L +VGAP+KPL+LP PLIF ++ + G +IGGIKETQEM+
Sbjct: 250 TVSATHSLLPLLGLLKHKGKLVMVGAPEKPLELPVMPLIFERKMVMGSMIGGIKETQEMI 309
Query: 314 DVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIAN 355
D+ GKHNIT DIELI AD +N A +RL K DVRYRFVID+AN
Sbjct: 310 DMAGKHNITADIELISADYVNTAMERLEKADVRYRFVIDVAN 351
>UniRef100_Q9ATW1 Cinnamyl alcohol dehydrogenase [Fragaria ananassa]
Length = 359
Score = 417 bits (1073), Expect = e-115
Identities = 200/353 (56%), Positives = 253/353 (71%), Gaps = 1/353 (0%)
Query: 8 HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPV 67
H SGWAA DSSG ++P+ F RRE G DV K+LYCGICH+D H K++WG + YP+
Sbjct: 7 HPNKASGWAARDSSGVLSPFNFSRRETGEKDVMFKVLYCGICHSDHHMVKNEWGFSTYPL 66
Query: 68 VPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIF 127
VPGHEI G +T+VGS V+ FK GD+VGVGC+ SC CE C ENYC K Y +
Sbjct: 67 VPGHEIVGEVTEVGSKVQKFKVGDRVGVGCIVGSCRSCENCTDHLENYCPKQILTYGANY 126
Query: 128 WDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRI 187
+DG+ TYGG S ++V +VV IP++LP+D AAPLLCAGIT +SPL+ GL G +
Sbjct: 127 YDGTTTYGGCSDIMVAHEHFVVRIPDNLPLDGAAPLLCAGITTYSPLRYFGL-DKPGMHV 185
Query: 188 GVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRS 247
GVVGLGGLGH+AVKF KA G VTVISTSP K+ EA + LGAD F++S + D +QAA +
Sbjct: 186 GVVGLGGLGHVAVKFAKAMGVKVTVISTSPKKEEEALKHLGADSFLVSRDQDHMQAAIGT 245
Query: 248 LDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIK 307
+D I+DTVSA H LLP++ LLK +G L +VGAP+KPL+LP FPL+ G++ + G IGG+
Sbjct: 246 MDGIIDTVSAQHPLLPLIGLLKSHGKLVMVGAPEKPLELPVFPLLMGRKMVAGSGIGGMM 305
Query: 308 ETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANASPAT 360
ETQEM+D KHNIT DIE+I D +N A +RL+K DVRYRFVIDI N A+
Sbjct: 306 ETQEMIDFAAKHNITADIEVIPIDYLNTAMERLVKADVRYRFVIDIGNTLKAS 358
>UniRef100_P42754 Mannitol dehydrogenase [Petroselinum crispum]
Length = 337
Score = 415 bits (1066), Expect = e-114
Identities = 197/332 (59%), Positives = 251/332 (75%), Gaps = 1/332 (0%)
Query: 24 ITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGSD 83
++P+ F RR G DV K+LYCG+CH+D+H K++WG+T YP+VPGHEI G +T+VGS
Sbjct: 2 LSPFKFSRRATGDNDVRFKVLYCGVCHSDLHMVKNEWGMTTYPIVPGHEIVGRVTEVGSK 61
Query: 84 VEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSITYGGYSQMLVV 143
VE FK GD VGVGCL SCL CE C D EN C K Y DGSITYGGY+ +V
Sbjct: 62 VEKFKVGDAVGVGCLVGSCLSCENCDDDSENNCAKQVQTYAFTNVDGSITYGGYADSMVA 121
Query: 144 DYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLGGLGHMAVKFG 203
D +V+ PE+LP+D+ APLLCAGIT +SPL+ HGL G ++GVVGLGGLGH+AVK
Sbjct: 122 DQHFVLRWPENLPLDSGAPLLCAGITTYSPLRYHGL-DKPGTKVGVVGLGGLGHVAVKMA 180
Query: 204 KAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDFILDTVSADHALLP 263
KAFG HVTVISTS SK+ EA E+LGAD+F++S++ DQ+QAA +L I+DTVSA H ++P
Sbjct: 181 KAFGAHVTVISTSESKKQEALEKLGADEFLVSSDSDQMQAATGTLHGIIDTVSALHPVVP 240
Query: 264 ILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKETQEMLDVCGKHNITC 323
+L LLKVNG L +VGAP+KPL+LP FPL+ G++ + G IGG+KETQEMLD +HNIT
Sbjct: 241 LLGLLKVNGKLVMVGAPEKPLELPVFPLLMGRKVLAGSNIGGLKETQEMLDFAAQHNITA 300
Query: 324 DIELIKADTINEAFQRLLKNDVRYRFVIDIAN 355
D+E+I D +N A +RL+K+DVRYRFVID+AN
Sbjct: 301 DVEVIPVDYVNTAMERLVKSDVRYRFVIDVAN 332
>UniRef100_Q02971 Probable mannitol dehydrogenase 1 [Arabidopsis thaliana]
Length = 357
Score = 407 bits (1045), Expect = e-112
Identities = 197/347 (56%), Positives = 254/347 (72%), Gaps = 1/347 (0%)
Query: 14 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 73
G AA D SG ++P++F RR G DV K+L+CGICHTD+ AK++WG+T YP+VPGHEI
Sbjct: 11 GLAAKDESGILSPFSFSRRATGEKDVRFKVLFCGICHTDLSMAKNEWGLTTYPLVPGHEI 70
Query: 74 TGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSIT 133
GV+T+VG+ V+ F GDKVGVG +A SC C+ C ENYC K+ +D ++T
Sbjct: 71 VGVVTEVGAKVKKFNAGDKVGVGYMAGSCRSCDSCNDGDENYCPKMILTSGAKNFDDTMT 130
Query: 134 YGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLG 193
+GGYS +V +++ IP++LP+D AAPLLCAG+TV+SP+K HGL G IGVVGLG
Sbjct: 131 HGGYSDHMVCAEDFIIRIPDNLPLDGAAPLLCAGVTVYSPMKYHGL-DKPGMHIGVVGLG 189
Query: 194 GLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDFILD 253
GLGH+AVKF KA G VTVISTS K+ EA RLGAD F++S +P Q++ A ++D I+D
Sbjct: 190 GLGHVAVKFAKAMGTKVTVISTSERKRDEAVTRLGADAFLVSRDPKQMKDAMGTMDGIID 249
Query: 254 TVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKETQEML 313
TVSA H LLP+L LLK G L +VGAP +PL+LP FPLIFG++ + G ++GGIKETQEM+
Sbjct: 250 TVSATHPLLPLLGLLKNKGKLVMVGAPAEPLELPVFPLIFGRKMVVGSMVGGIKETQEMV 309
Query: 314 DVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANASPAT 360
D+ GKHNIT DIELI AD +N A +RL K DV+YRFVID+AN T
Sbjct: 310 DLAGKHNITADIELISADYVNTAMERLAKADVKYRFVIDVANTMKPT 356
>UniRef100_Q38707 Mannitol dehydrogenase [Apium graveolens]
Length = 365
Score = 405 bits (1040), Expect = e-111
Identities = 192/356 (53%), Positives = 259/356 (71%), Gaps = 2/356 (0%)
Query: 1 MAKTTP-NHTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDD 59
MAK++ H GWAA D++G ++P+ F RR G DV +K+L+CG+CH+D H ++
Sbjct: 1 MAKSSEIEHPVKAFGWAARDTTGLLSPFKFSRRATGEKDVRLKVLFCGVCHSDHHMIHNN 60
Query: 60 WGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKL 119
WG T YP+VPGHEI GV+T+VGS VE K GD VG+GCL SC CE C ++E++CE
Sbjct: 61 WGFTTYPIVPGHEIVGVVTEVGSKVEKVKVGDNVGIGCLVGSCRSCESCCDNRESHCENT 120
Query: 120 QFVYNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGL 179
Y I++DG++T+GGYS +V D +++ P++LP+D+ APLLCAGIT +SPLK +GL
Sbjct: 121 IDTYGSIYFDGTMTHGGYSDTMVADEHFILRWPKNLPLDSGAPLLCAGITTYSPLKYYGL 180
Query: 180 VSTAGKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPD 239
G +IGVVGLGGLGH+AVK KAFG VTVI S SK+ EA E+LGAD F+++++ +
Sbjct: 181 -DKPGTKIGVVGLGGLGHVAVKMAKAFGAQVTVIDISESKRKEALEKLGADSFLLNSDQE 239
Query: 240 QLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIK 299
Q++ A+ SLD I+DTV +H L P+ +LLK NG L +VGAP+KP +LP F L+ G++ +
Sbjct: 240 QMKGARSSLDGIIDTVPVNHPLAPLFDLLKPNGKLVMVGAPEKPFELPVFSLLKGRKLLG 299
Query: 300 GGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIAN 355
G I GGIKETQEMLD KHNIT D+E+I D +N A +RL+K+DVRYRFVIDIAN
Sbjct: 300 GTINGGIKETQEMLDFAAKHNITADVEVIPMDYVNTAMERLVKSDVRYRFVIDIAN 355
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 633,428,791
Number of Sequences: 2790947
Number of extensions: 27763468
Number of successful extensions: 76569
Number of sequences better than 10.0: 4035
Number of HSP's better than 10.0 without gapping: 1774
Number of HSP's successfully gapped in prelim test: 2262
Number of HSP's that attempted gapping in prelim test: 69759
Number of HSP's gapped (non-prelim): 4843
length of query: 360
length of database: 848,049,833
effective HSP length: 128
effective length of query: 232
effective length of database: 490,808,617
effective search space: 113867599144
effective search space used: 113867599144
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146790.6


