

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146784.3 - phase: 0
(348 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
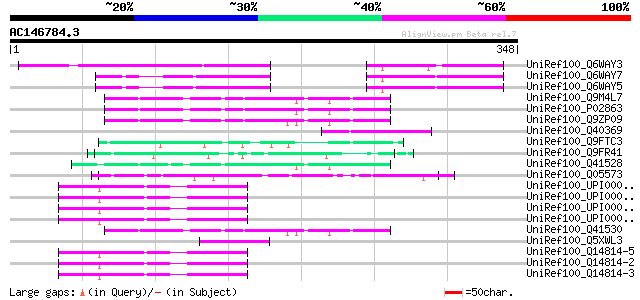
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum] 85 3e-15
UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum] 78 3e-13
UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum] 78 3e-13
UniRef100_Q9M4L7 Alpha-gliadin [Triticum aestivum] 56 1e-06
UniRef100_P02863 Alpha/beta-gliadin precursor [Triticum aestivum] 56 1e-06
UniRef100_Q9ZP09 Alpha-gliadin precursor [Triticum aestivum subs... 53 1e-05
UniRef100_Q40369 RPE15 protein [Medicago sativa] 53 1e-05
UniRef100_Q9FTC3 Gamma-gliadin [Aegilops sharonensis] 52 3e-05
UniRef100_Q9FR41 Secalin [Secale cereale] 51 4e-05
UniRef100_Q41528 Alpha-gliadin [Triticum aestivum] 51 4e-05
UniRef100_Q05573 Sec1 precursor [Secale cereale] 51 5e-05
UniRef100_UPI00003683EF UPI00003683EF UniRef100 entry 50 9e-05
UniRef100_UPI00003683EE UPI00003683EE UniRef100 entry 50 9e-05
UniRef100_UPI00003683ED UPI00003683ED UniRef100 entry 50 9e-05
UniRef100_UPI00003683EC UPI00003683EC UniRef100 entry 50 9e-05
UniRef100_Q41530 Alpha-gliadin storage protein [Triticum aestivum] 50 9e-05
UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum] 50 9e-05
UniRef100_Q14814-5 Splice isoform MEF2DA'0 of Q14814 [Homo sapiens] 50 9e-05
UniRef100_Q14814-2 Splice isoform MEF2DA'B of Q14814 [Homo sapiens] 50 9e-05
UniRef100_Q14814-3 Splice isoform MEF2D0B of Q14814 [Homo sapiens] 50 9e-05
>UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 85.1 bits (209), Expect = 3e-15
Identities = 54/173 (31%), Positives = 82/173 (47%), Gaps = 6/173 (3%)
Query: 7 GSHMQTLVQTGNMDQLEQEVQELRGEVTTLRAEVEKLASLVSSLMATNDPPLVQQRPQSP 66
G ++L++TG + + + R E E +++ D ++ R +
Sbjct: 319 GERTESLIKTGKIQDVGSSSSKKPFAGAPRRREGE-----TNAVQHRRDQNRIEYRQAAA 373
Query: 67 YQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQ 126
PQ +QQ R PQ Q Q+ Q + D +PM YA+LLP LL+ LV+
Sbjct: 374 VTIPAPQPRQQQQQRVQQPQQQQQQRPYQPRQRMPDRRFDSLPMSYAELLPELLRLGLVE 433
Query: 127 TLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSF 179
+ P LPP Y ++ C FH GAPGH TE+C L+ +VQ LI+ N +F
Sbjct: 434 LCTMAP-PTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDANAINF 485
Score = 75.9 bits (185), Expect = 2e-12
Identities = 44/98 (44%), Positives = 59/98 (59%), Gaps = 7/98 (7%)
Query: 246 QPQFQQYQQ--QPRQQAPRIKFDPIPMKYGELFPYLLERNLVQ--TRPPPPIPKKLPARW 301
QPQ QQ Q+ QPRQ+ P +FD +PM Y EL P LL LV+ T PP + LP +
Sbjct: 391 QPQQQQQQRPYQPRQRMPDRRFDSLPMSYAELLPELLRLGLVELCTMAPPTV---LPPGY 447
Query: 302 RPDLFCVFHQGAQGHDVERCFSLKIEVQKLIEDDLIPF 339
++ C FH GA GH E+C +L+ +VQ LI+ + I F
Sbjct: 448 DANVRCDFHSGAPGHHTEKCRALQHKVQDLIDANAINF 485
>UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 78.2 bits (191), Expect = 3e-13
Identities = 44/97 (45%), Positives = 57/97 (58%), Gaps = 4/97 (4%)
Query: 246 QPQFQQYQQ---QPRQQAPRIKFDPIPMKYGELFPYLLERNLVQTRPPPPIPKKLPARWR 302
QPQ QQ QQ QPRQ+ P +FD +PM Y EL P LL +V+ R P P LP +
Sbjct: 392 QPQQQQQQQRPYQPRQRMPDRRFDSLPMSYAELLPELLRLGMVELRTMAP-PTVLPPGYD 450
Query: 303 PDLFCVFHQGAQGHDVERCFSLKIEVQKLIEDDLIPF 339
++ C FH GA GH E+C +L+ +VQ LI+ I F
Sbjct: 451 ANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINF 487
Score = 77.4 bits (189), Expect = 5e-13
Identities = 47/120 (39%), Positives = 62/120 (51%), Gaps = 19/120 (15%)
Query: 60 QQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPIL 119
QQR Q P Q Q+P Q PRQ +P + D +PM YA+LLP L
Sbjct: 387 QQRVQQPQQQQQQQRPYQ--PRQRMPDRRF----------------DSLPMSYAELLPEL 428
Query: 120 LKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSF 179
L+ +V+ L P LPP Y ++ C FH GAPGH TE+C L+ +VQ LI+ +F
Sbjct: 429 LRLGMVE-LRTMAPPTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINF 487
>UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum]
Length = 1105
Score = 78.2 bits (191), Expect = 3e-13
Identities = 44/97 (45%), Positives = 57/97 (58%), Gaps = 4/97 (4%)
Query: 246 QPQFQQYQQ---QPRQQAPRIKFDPIPMKYGELFPYLLERNLVQTRPPPPIPKKLPARWR 302
QPQ QQ QQ QPRQ+ P +FD +PM Y EL P LL +V+ R P P LP +
Sbjct: 392 QPQQQQQQQRPYQPRQRMPDRRFDSLPMSYAELLPELLRLGMVELRTMAP-PTVLPPGYD 450
Query: 303 PDLFCVFHQGAQGHDVERCFSLKIEVQKLIEDDLIPF 339
++ C FH GA GH E+C +L+ +VQ LI+ I F
Sbjct: 451 ANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINF 487
Score = 77.4 bits (189), Expect = 5e-13
Identities = 47/120 (39%), Positives = 62/120 (51%), Gaps = 19/120 (15%)
Query: 60 QQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPIL 119
QQR Q P Q Q+P Q PRQ +P + D +PM YA+LLP L
Sbjct: 387 QQRVQQPQQQQQQQRPYQ--PRQRMPDRRF----------------DSLPMSYAELLPEL 428
Query: 120 LKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSF 179
L+ +V+ L P LPP Y ++ C FH GAPGH TE+C L+ +VQ LI+ +F
Sbjct: 429 LRLGMVE-LRTMAPPTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINF 487
>UniRef100_Q9M4L7 Alpha-gliadin [Triticum aestivum]
Length = 269
Score = 56.2 bits (134), Expect = 1e-06
Identities = 60/213 (28%), Positives = 87/213 (40%), Gaps = 28/213 (13%)
Query: 66 PYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLV 125
P + PQ P QQ P++ +P Q Q+F+ Q Q Q P P + LP L +
Sbjct: 5 PVPQLQPQNPSQQQPQEQVPLVQ-QQQFLGQQQPFPPQQPYPQPQPFPSQLPYL----QL 59
Query: 126 QTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQDIK 185
Q P P++P S P +RP + Q P + Q P+ ++ Q+ + Q I
Sbjct: 60 QPFPQPQLPYSQPQPFRPQQ--PYPQPQPQYSQPQ-QPISQQQQQQQQQQQQQQQQQQIL 116
Query: 186 VLLQQQHLAP--------HSVAAVRPITNVVQDPGYQPQFR---------PSQQQYPDSH 228
+ QQ L P H++A R + V+Q YQ P Q Q H
Sbjct: 117 QQILQQQLIPCMDVVLQQHNIAHGR--SQVLQQSTYQLLQELCCQHLWQIPEQSQCQAIH 174
Query: 229 FVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAP 261
V + I++ + QP Q QQP QQ P
Sbjct: 175 NVVHAI-ILHQQQKQQQQPSSQVSFQQPLQQYP 206
Score = 37.0 bits (84), Expect = 0.80
Identities = 49/205 (23%), Positives = 76/205 (36%), Gaps = 28/205 (13%)
Query: 62 RPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLK 121
+PQ PY P +P+Q P Q PQ PQ+ I Q Q Q+ Q + +L +L+
Sbjct: 64 QPQLPYSQPQPFRPQQPYP-QPQPQYSQPQQPISQQQQQQQQQ-QQQQQQQQQILQQILQ 121
Query: 122 KNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLK--EEVQKLIENNVWSF 179
+ L+ P ++ V Q H Q + +Q+L ++W
Sbjct: 122 QQLI-----------------PCMDVVLQQHNIAHGRSQVLQQSTYQLLQELCCQHLWQI 164
Query: 180 DDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYP---DSHFVTTILPI 236
+Q Q H H++ + Q P Q F+ QQYP S + P
Sbjct: 165 PEQS---QCQAIHNVVHAI-ILHQQQKQQQQPSSQVSFQQPLQQYPLGQGSFRPSQQNPQ 220
Query: 237 MNVVRNPGYQPQFQQYQQQPRQQAP 261
P PQF++ + Q P
Sbjct: 221 AQGSVQPQQLPQFEEIRNLALQTLP 245
>UniRef100_P02863 Alpha/beta-gliadin precursor [Triticum aestivum]
Length = 286
Score = 56.2 bits (134), Expect = 1e-06
Identities = 60/213 (28%), Positives = 87/213 (40%), Gaps = 28/213 (13%)
Query: 66 PYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLV 125
P + PQ P QQ P++ +P Q Q+F+ Q Q Q P P + LP L +
Sbjct: 24 PVPQLQPQNPSQQLPQEQVPLVQ-QQQFLGQQQPFPPQQPYPQPQPFPSQLPYL----QL 78
Query: 126 QTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQDIK 185
Q P P++P S P +RP + Q P + Q P+ ++ Q+ + Q I
Sbjct: 79 QPFPQPQLPYSQPQPFRPQQ--PYPQPQPQYSQPQ-QPISQQQQQQQQQQQQQQQQQQIL 135
Query: 186 VLLQQQHLAP--------HSVAAVRPITNVVQDPGYQPQFR---------PSQQQYPDSH 228
+ QQ L P H++A R + V+Q YQ P Q Q H
Sbjct: 136 QQILQQQLIPCMDVVLQQHNIAHGR--SQVLQQSTYQLLQELCCQHLWQIPEQSQCQAIH 193
Query: 229 FVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAP 261
V + I++ + QP Q QQP QQ P
Sbjct: 194 NVVHAI-ILHQQQKQQQQPSSQVSFQQPLQQYP 225
Score = 37.0 bits (84), Expect = 0.80
Identities = 49/205 (23%), Positives = 76/205 (36%), Gaps = 28/205 (13%)
Query: 62 RPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLK 121
+PQ PY P +P+Q P Q PQ PQ+ I Q Q Q+ Q + +L +L+
Sbjct: 83 QPQLPYSQPQPFRPQQPYP-QPQPQYSQPQQPISQQQQQQQQQ-QQQQQQQQQILQQILQ 140
Query: 122 KNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLK--EEVQKLIENNVWSF 179
+ L+ P ++ V Q H Q + +Q+L ++W
Sbjct: 141 QQLI-----------------PCMDVVLQQHNIAHGRSQVLQQSTYQLLQELCCQHLWQI 183
Query: 180 DDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYP---DSHFVTTILPI 236
+Q Q H H++ + Q P Q F+ QQYP S + P
Sbjct: 184 PEQS---QCQAIHNVVHAI-ILHQQQKQQQQPSSQVSFQQPLQQYPLGQGSFRPSQQNPQ 239
Query: 237 MNVVRNPGYQPQFQQYQQQPRQQAP 261
P PQF++ + Q P
Sbjct: 240 AQGSVQPQQLPQFEEIRNLALQTLP 264
>UniRef100_Q9ZP09 Alpha-gliadin precursor [Triticum aestivum subsp. spelta]
Length = 288
Score = 52.8 bits (125), Expect = 1e-05
Identities = 60/215 (27%), Positives = 88/215 (40%), Gaps = 30/215 (13%)
Query: 66 PYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLV 125
P + PQ P QQ P++ +P Q Q+F+ Q Q Q P P + P L +
Sbjct: 24 PVPQLQPQNPSQQQPQEQVPLVQ-QQQFLGQQQPFPPQQPYPQPQPFPSQQPYL----QL 78
Query: 126 QTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQDIK 185
Q P P++P S P +RP + Q P + Q P+ ++ Q+ + Q +
Sbjct: 79 QPFPQPQLPYSQPQPFRPQQ--PYPQPQPQYSQPQ-QPISQQQQQQQQQQQQQQQQQQQQ 135
Query: 186 VLLQ--QQHLAP--------HSVAAVRPITNVVQDPGYQPQFR---------PSQQQYPD 226
+L Q QQ L P H++A R + V+Q YQ P Q Q
Sbjct: 136 ILQQILQQQLIPCMDVVLQQHNIAHGR--SQVLQQSTYQLLQELCCQHLWQIPEQSQCQA 193
Query: 227 SHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAP 261
H V + I++ + QP Q QQP QQ P
Sbjct: 194 IHKVVHAI-ILHQQQKQQQQPSSQVSFQQPLQQYP 227
Score = 37.0 bits (84), Expect = 0.80
Identities = 50/209 (23%), Positives = 78/209 (36%), Gaps = 34/209 (16%)
Query: 62 RPQSPY---QPMCPQKP-RQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLP 117
+PQ PY QP PQ+P Q P+ S PQ + Q+ Q Q Q+ Q + +L
Sbjct: 83 QPQLPYSQPQPFRPQQPYPQPQPQYSQPQQPISQQQQQQQQQQQQQQ----QQQQQQILQ 138
Query: 118 ILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLK--EEVQKLIENN 175
+L++ L+ P ++ V Q H Q + +Q+L +
Sbjct: 139 QILQQQLI-----------------PCMDVVLQQHNIAHGRSQVLQQSTYQLLQELCCQH 181
Query: 176 VWSFDDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYP---DSHFVTT 232
+W +Q Q H H++ + Q P Q F+ QQYP S +
Sbjct: 182 LWQIPEQS---QCQAIHKVVHAI-ILHQQQKQQQQPSSQVSFQQPLQQYPLGQGSFRPSQ 237
Query: 233 ILPIMNVVRNPGYQPQFQQYQQQPRQQAP 261
P P PQF++ + Q P
Sbjct: 238 QNPQAQGSVQPQQLPQFEEIRNLALQTLP 266
>UniRef100_Q40369 RPE15 protein [Medicago sativa]
Length = 219
Score = 52.8 bits (125), Expect = 1e-05
Identities = 30/75 (40%), Positives = 39/75 (52%), Gaps = 1/75 (1%)
Query: 215 PQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDPIPMKYGE 274
P F P QQYP + P+ + + +QP QQ QQQ + P+ F PIPM+Y E
Sbjct: 126 PFFPPFYQQYPLPPGQPPV-PVNAMAQQVQHQPPAQQQQQQQQGPRPKTYFPPIPMRYAE 184
Query: 275 LFPYLLERNLVQTRP 289
L P LL + TRP
Sbjct: 185 LLPTLLAKGHCVTRP 199
Score = 45.1 bits (105), Expect = 0.003
Identities = 28/74 (37%), Positives = 34/74 (45%), Gaps = 3/74 (4%)
Query: 56 PPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADL 115
PP QQ P P QP P Q + P Q Q+ Q + + PIPM+YA+L
Sbjct: 129 PPFYQQYPLPPGQPPVPVNAMAQQVQHQPPAQQQQQQ---QQGPRPKTYFPPIPMRYAEL 185
Query: 116 LPILLKKNLVQTLP 129
LP LL K T P
Sbjct: 186 LPTLLAKGHCVTRP 199
>UniRef100_Q9FTC3 Gamma-gliadin [Aegilops sharonensis]
Length = 234
Score = 51.6 bits (122), Expect = 3e-05
Identities = 62/224 (27%), Positives = 88/224 (38%), Gaps = 51/224 (22%)
Query: 62 RPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKA----SQCDPIPMKYADLLP 117
+PQ P+ P PQ+P Q P+Q PQ Q PQ+ PQ+Q + P P
Sbjct: 41 QPQQPF-PQQPQQPYPQQPQQPFPQTQQPQQLFPQSQQPQQPYPQQPQQPFPQTQQPQQQ 99
Query: 118 ILLKKNLVQTLPLPR-----VPNSLPPWYRPDLNCVFHQGAPGHDT--EQCYPLKEEVQK 170
+ Q P P+ P PP+ +P L Q P + +QC P+
Sbjct: 100 FPQSQQPQQPFPQPQQPQQSFPRQQPPFIQPSLQ---QQLNPCKNLLLQQCRPVS----- 151
Query: 171 LIENNVWS--FDDQDIKVLLQQ--QHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYPD 226
+ +++WS + D +V+ QQ Q LA P Q Q
Sbjct: 152 -LVSSLWSMIWPQSDCQVMRQQCCQQLAQI----------------------PQQLQCAA 188
Query: 227 SHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDPIPM 270
H V + IM + Q Q QQ QQQ +QQ RI P+P+
Sbjct: 189 IHSVVHSI-IMQQEQQQQQQQQQQQQQQQRQQQGLRI---PLPL 228
>UniRef100_Q9FR41 Secalin [Secale cereale]
Length = 455
Score = 51.2 bits (121), Expect = 4e-05
Identities = 63/215 (29%), Positives = 85/215 (39%), Gaps = 42/215 (19%)
Query: 59 VQQRPQSPYQPMCPQKPRQQAPRQ-SIPQNQVPQKFIPQN------QVQKASQCDPIPMK 111
+QQ PQ P QP PQ+P QQ P+Q P Q PQ+ +PQ Q Q+ Q P P +
Sbjct: 125 LQQFPQQPQQPF-PQQPLQQFPQQPQQPFPQQPQQPVPQQSQQPFPQTQQPQQPFPQPQQ 183
Query: 112 YADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKL 171
L P + + Q + P P +P Q +P ++Q YP +E Q+L
Sbjct: 184 PQQLFPQTQQSSPQQPQQVTSQPQQPFPQAQPP-----QQSSP--QSQQPYP--QEPQQL 234
Query: 172 IENNVWSFDDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQ--FRPSQQQYPDSHF 229
+ Q Q Q P + Q P QPQ F SQ+Q+P H
Sbjct: 235 FPQS--QQPQQPFPQPQQPQQPFPQPQPQTQQSIPQPQQPFPQPQQPFPQSQEQFPQVH- 291
Query: 230 VTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIK 264
QPQ Q P+QQ P I+
Sbjct: 292 ----------------QPQ----QPSPQQQQPSIQ 306
Score = 49.7 bits (117), Expect = 1e-04
Identities = 64/230 (27%), Positives = 87/230 (37%), Gaps = 14/230 (6%)
Query: 54 NDPPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQN-QVPQKFIPQNQVQKASQCDPIPMKY 112
+ P Q PQ P+ P PQ+P Q P+Q PQ Q P PQ Q + Q P+P +
Sbjct: 67 SSPQPQQPYPQQPF-PQQPQQPYPQQPQQPFPQQPQQPYPQQPQQQFPQQPQ-QPVPQQP 124
Query: 113 ADLLPILLKKNLVQTLPLPRVPN--SLPPWYRPDLNCVFHQGAPGHDT---EQCYPLKEE 167
P ++ Q PL + P P +P P T +Q +P ++
Sbjct: 125 LQQFPQQPQQPFPQQ-PLQQFPQQPQQPFPQQPQQPVPQQSQQPFPQTQQPQQPFPQPQQ 183
Query: 168 VQKLIENNVWSFDDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYPDS 227
Q+L S Q +V Q Q P + + Q +PQ Q Q P
Sbjct: 184 PQQLFPQTQQSSPQQPQQVTSQPQQPFPQAQPPQQSSPQSQQPYPQEPQQLFPQSQQPQQ 243
Query: 228 HFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDPIPMKYGELFP 277
F P P QPQ QQ QP+Q P+ + P P E FP
Sbjct: 244 PFPQPQQPQQPF---PQPQPQTQQSIPQPQQPFPQPQ-QPFPQSQ-EQFP 288
Score = 45.1 bits (105), Expect = 0.003
Identities = 73/295 (24%), Positives = 114/295 (37%), Gaps = 40/295 (13%)
Query: 57 PLVQQRPQSPYQPMCP-----QKPRQQAPRQSIPQNQ--VPQKFIPQNQVQKASQ--CDP 107
P Q + SP QP P Q+P Q P+QS PQ Q PQ+ PQ Q Q P
Sbjct: 36 PFPQPQQSSPQQPQQPFPQQSQQPFPQQPQQSSPQPQQPYPQQPFPQQPQQPYPQQPQQP 95
Query: 108 IPMKYADLLPILLKKNLVQ--TLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLK 165
P + P ++ Q P+P+ P P +Q +P +
Sbjct: 96 FPQQPQQPYPQQPQQQFPQQPQQPVPQQPLQQFP----------------QQPQQPFP-Q 138
Query: 166 EEVQKLIENNVWSFDDQDIKVLLQQ-QHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQY 224
+ +Q+ + F Q + + QQ Q P + +P Q PQ + S Q
Sbjct: 139 QPLQQFPQQPQQPFPQQPQQPVPQQSQQPFPQTQQPQQPFPQPQQPQQLFPQTQQSSPQQ 198
Query: 225 PDSHFVTTILPIMNVVRNPGYQPQFQQ-YQQQPRQ-----QAPRIKFDPIPMKYGELFPY 278
P P PQ QQ Y Q+P+Q Q P+ F P P + + FP
Sbjct: 199 PQQVTSQPQQPFPQAQPPQQSSPQSQQPYPQEPQQLFPQSQQPQQPF-PQPQQPQQPFPQ 257
Query: 279 ---LLERNLVQTRPPPPIPKKLPARWRPDLFCVFHQGAQGHDVERCFSLKIEVQK 330
++++ Q + P P P++ P + F HQ Q ++ S+++ +Q+
Sbjct: 258 PQPQTQQSIPQPQQPFPQPQQ-PFPQSQEQFPQVHQPQQPSPQQQQPSIQLSLQQ 311
Score = 37.7 bits (86), Expect = 0.47
Identities = 58/235 (24%), Positives = 81/235 (33%), Gaps = 22/235 (9%)
Query: 68 QPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQ----VQKASQCDPIPMKYADLLPILLKKN 123
Q CPQ+ P+QS PQ PQ+ PQ Q+ Q P P + P +
Sbjct: 28 QVQCPQQQPFPQPQQSSPQQ--PQQPFPQQSQQPFPQQPQQSSPQPQQPYPQQPFPQQPQ 85
Query: 124 LVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQD 183
P P+ P P Q +Q P ++ +Q+ + F Q
Sbjct: 86 ----QPYPQQPQQPFPQQPQQPYPQQPQQQFPQQPQQPVP-QQPLQQFPQQPQQPFPQQP 140
Query: 184 IKVLLQQQHLAPHSVAAVRPITNVVQDP---GYQPQFRPSQQQYPDSHFVTTILPIMNVV 240
++ QQ P +P+ Q P QPQ Q Q P F T
Sbjct: 141 LQQFPQQPQ-QPFPQQPQQPVPQQSQQPFPQTQQPQQPFPQPQQPQQLFPQTQQSSPQQP 199
Query: 241 RNPGYQPQFQQYQQQPRQQAPRIKFDPIPMKYGELFPYLLERNLVQTRPPPPIPK 295
+ QPQ Q QP QQ+ P P + +LFP +P P P+
Sbjct: 200 QQVTSQPQQPFPQAQPPQQSSPQSQQPYPQEPQQLFPQ-------SQQPQQPFPQ 247
Score = 35.4 bits (80), Expect = 2.3
Identities = 41/140 (29%), Positives = 58/140 (41%), Gaps = 13/140 (9%)
Query: 54 NDPPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQN---QVQKASQCDPIPM 110
+ P QQ P QP P+ Q P+QS PQ+Q P PQ Q Q+ Q P P
Sbjct: 194 SSPQQPQQVTSQPQQPF----PQAQPPQQSSPQSQQPYPQEPQQLFPQSQQPQQPFPQPQ 249
Query: 111 KYADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQK 170
+ P + Q++P P+ P P P F Q H +Q P ++ Q
Sbjct: 250 QPQQPFP-QPQPQTQQSIPQPQQPFPQPQQPFPQSQEQFPQ---VHQPQQ--PSPQQQQP 303
Query: 171 LIENNVWSFDDQDIKVLLQQ 190
I+ ++ + VLLQQ
Sbjct: 304 SIQLSLQQQLNPCKNVLLQQ 323
>UniRef100_Q41528 Alpha-gliadin [Triticum aestivum]
Length = 287
Score = 51.2 bits (121), Expect = 4e-05
Identities = 63/236 (26%), Positives = 95/236 (39%), Gaps = 31/236 (13%)
Query: 43 LASLVSSLMATNDPPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKA 102
L ++ +++AT P V+ P + PQ P QQ P++ +P Q Q+F+ Q Q
Sbjct: 5 LILVLLAIVATTAPTAVR----FPVPQLQPQNPSQQLPQEQVPLVQ-QQQFLGQQQPFPP 59
Query: 103 SQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCY 162
Q P P ++ LP L +Q P P++P S P +RP + Q P + Q
Sbjct: 60 QQPYPQP-QFPSQLPYL----QLQPFPQPQLPYSQPQPFRPQQ--PYPQPQPQYSQPQQP 112
Query: 163 PLKEEVQKLIENNVWSFDDQDIKVLLQQQHLAP--------HSVAAVRPITNVVQDPGYQ 214
+++ Q+ + Q I + QQ L P H+ A R + V+Q YQ
Sbjct: 113 ISQQQQQQQQQQQQQQQQQQQILQQILQQQLIPCMDVVLQQHNKAHGR--SQVLQQSTYQ 170
Query: 215 PQFR---------PSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAP 261
P Q Q H V + + + QP Q QQP QQ P
Sbjct: 171 LLRELCCQHLWQIPEQSQCQAIHNVVHAIILHQQQKQQQQQPSSQVSFQQPLQQYP 226
Score = 43.1 bits (100), Expect = 0.011
Identities = 49/207 (23%), Positives = 77/207 (36%), Gaps = 30/207 (14%)
Query: 62 RPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLK 121
+PQ PY P +P+Q P Q PQ PQ+ I Q Q Q+ Q + +L +L+
Sbjct: 82 QPQLPYSQPQPFRPQQPYP-QPQPQYSQPQQPISQQQQQQQQQQQQQQQQQQQILQQILQ 140
Query: 122 KNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIE----NNVW 177
+ L+ P ++ V Q H Q L++ +L+ ++W
Sbjct: 141 QQLI-----------------PCMDVVLQQHNKAHGRSQV--LQQSTYQLLRELCCQHLW 181
Query: 178 SFDDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYP---DSHFVTTIL 234
+Q Q H H++ + Q P Q F+ QQYP S +
Sbjct: 182 QIPEQS---QCQAIHNVVHAIILHQQQKQQQQQPSSQVSFQQPLQQYPLGQGSFRPSQQN 238
Query: 235 PIMNVVRNPGYQPQFQQYQQQPRQQAP 261
P P PQF++ + Q P
Sbjct: 239 PQTQGSVQPQQLPQFEEIRNLALQTLP 265
>UniRef100_Q05573 Sec1 precursor [Secale cereale]
Length = 357
Score = 50.8 bits (120), Expect = 5e-05
Identities = 62/241 (25%), Positives = 96/241 (39%), Gaps = 42/241 (17%)
Query: 62 RPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLK 121
+PQ P P+ PQ+P Q P+Q PQ Q PQ + Q PIP + P +
Sbjct: 70 QPQQP-APIQPQQPFPQQPQQPFPQPQQQLPLQPQQPFPQPQQ--PIPQQPQQSFPQQPQ 126
Query: 122 KNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDD 181
+ Q P+ P + P T+Q +PL+ + Q + SF
Sbjct: 127 RPEQQ---FPQQPQQIIP----------------QQTQQPFPLQPQ-QPFPQQPQRSFAQ 166
Query: 182 QDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQP-QFRPSQQQYPDSHFVTTILPIMNVV 240
Q +++ QQ P + +P + Q QP Q P Q Q P + + P
Sbjct: 167 QPKQIISQQ----PFPLQPQQPFSQPQQPFPQQPGQIIPQQPQQP-----SPLQPQQPFS 217
Query: 241 RNPGYQPQFQQYQQQPRQQAPRIKFDPIPMKYGELFPYLLER-------NLVQTRPPPPI 293
+ P +PQ Q + QQP+Q P+ P P++ + P +R ++ RP P
Sbjct: 218 QQP-QRPQ-QPFPQQPQQIIPQQPQQPFPLQPQQPVPQQPQRPFGQQPEQIISQRPQQPF 275
Query: 294 P 294
P
Sbjct: 276 P 276
Score = 49.7 bits (117), Expect = 1e-04
Identities = 71/265 (26%), Positives = 116/265 (42%), Gaps = 35/265 (13%)
Query: 57 PLVQQRP-QSPYQPMCPQKPRQQAPRQ-SIPQNQVPQKFIPQNQVQKASQCDPIPMKYAD 114
PL Q+P P QP+ PQ+P+Q P+Q P+ Q PQ+ PQ + + +Q P P++
Sbjct: 99 PLQPQQPFPQPQQPI-PQQPQQSFPQQPQRPEQQFPQQ--PQQIIPQQTQ-QPFPLQPQQ 154
Query: 115 LLPILLKKNLV---------QTLPL-PRVPNSLP--PWYRPDLNCVFHQGAPGHDTEQCY 162
P +++ Q PL P+ P S P P+ + + Q +
Sbjct: 155 PFPQQPQRSFAQQPKQIISQQPFPLQPQQPFSQPQQPFPQQPGQIIPQQPQQPSPLQPQQ 214
Query: 163 PLKEEVQKLIENNVWSFDDQDIKVLLQQQHLAPHSVAAVRPITNVVQDP-GYQPQFRPSQ 221
P ++ Q+ + F Q +++ QQ P + +P+ Q P G QP+ SQ
Sbjct: 215 PFSQQPQRPQQ----PFPQQPQQIIPQQPQ-QPFPLQPQQPVPQQPQRPFGQQPEQIISQ 269
Query: 222 QQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDPIPMKYGELFPYLLE 281
+ P F P+ + P QPQ Q QQP Q P+ P P++ + FP E
Sbjct: 270 R--PQQPF-----PLQ--PQQPFSQPQ-QPLPQQPGQIIPQQPQQPFPLQPQQPFPQQSE 319
Query: 282 RNL-VQTRPPPPIPKKLPARWRPDL 305
+ + Q + P P+ + P+ +P L
Sbjct: 320 QIIPQQPQQPFPLQPQQPSPQQPQL 344
Score = 34.3 bits (77), Expect = 5.2
Identities = 31/104 (29%), Positives = 43/104 (40%), Gaps = 8/104 (7%)
Query: 189 QQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYPDSHFVTTILPIMNVVRNPGY--- 245
QQ P A ++P Q P Q F QQQ P P + + P
Sbjct: 65 QQPFPQPQQPAPIQPQQPFPQQP--QQPFPQPQQQLPLQPQQPFPQPQQPIPQQPQQSFP 122
Query: 246 -QPQF--QQYQQQPRQQAPRIKFDPIPMKYGELFPYLLERNLVQ 286
QPQ QQ+ QQP+Q P+ P P++ + FP +R+ Q
Sbjct: 123 QQPQRPEQQFPQQPQQIIPQQTQQPFPLQPQQPFPQQPQRSFAQ 166
>UniRef100_UPI00003683EF UPI00003683EF UniRef100 entry
Length = 519
Score = 50.1 bits (118), Expect = 9e-05
Identities = 38/137 (27%), Positives = 58/137 (41%), Gaps = 20/137 (14%)
Query: 34 TTLRAEVEKLASLVSSLMATNDPPLVQ-------QRPQSPYQPMCPQKPRQQAPRQSIPQ 86
T + + ++ +SSL A + P + Q+PQ P QP PQ P+QQ P+ PQ
Sbjct: 330 TAYNTDYQLTSAELSSLPAFSSPGGLSLGNVTAWQQPQQPQQPQQPQPPQQQPPQPQQPQ 389
Query: 87 NQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLN 146
Q PQ+ PQ Q+ S P+ + NL+ PLP V +L P ++
Sbjct: 390 PQQPQQ--PQQPPQQQSHLVPVSL-----------SNLIPGSPLPHVGAALTVTTHPHIS 436
Query: 147 CVFHQGAPGHDTEQCYP 163
+P + P
Sbjct: 437 IKSEPVSPSRERSPAPP 453
>UniRef100_UPI00003683EE UPI00003683EE UniRef100 entry
Length = 512
Score = 50.1 bits (118), Expect = 9e-05
Identities = 38/137 (27%), Positives = 58/137 (41%), Gaps = 20/137 (14%)
Query: 34 TTLRAEVEKLASLVSSLMATNDPPLVQ-------QRPQSPYQPMCPQKPRQQAPRQSIPQ 86
T + + ++ +SSL A + P + Q+PQ P QP PQ P+QQ P+ PQ
Sbjct: 323 TAYNTDYQLTSAELSSLPAFSSPGGLSLGNVTAWQQPQQPQQPQQPQPPQQQPPQPQQPQ 382
Query: 87 NQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLN 146
Q PQ+ PQ Q+ S P+ + NL+ PLP V +L P ++
Sbjct: 383 PQQPQQ--PQQPPQQQSHLVPVSL-----------SNLIPGSPLPHVGAALTVTTHPHIS 429
Query: 147 CVFHQGAPGHDTEQCYP 163
+P + P
Sbjct: 430 IKSEPVSPSRERSPAPP 446
>UniRef100_UPI00003683ED UPI00003683ED UniRef100 entry
Length = 474
Score = 50.1 bits (118), Expect = 9e-05
Identities = 38/137 (27%), Positives = 58/137 (41%), Gaps = 20/137 (14%)
Query: 34 TTLRAEVEKLASLVSSLMATNDPPLVQ-------QRPQSPYQPMCPQKPRQQAPRQSIPQ 86
T + + ++ +SSL A + P + Q+PQ P QP PQ P+QQ P+ PQ
Sbjct: 285 TAYNTDYQLTSAELSSLPAFSSPGGLSLGNVTAWQQPQQPQQPQQPQPPQQQPPQPQQPQ 344
Query: 87 NQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLN 146
Q PQ+ PQ Q+ S P+ + NL+ PLP V +L P ++
Sbjct: 345 PQQPQQ--PQQPPQQQSHLVPVSL-----------SNLIPGSPLPHVGAALTVTTHPHIS 391
Query: 147 CVFHQGAPGHDTEQCYP 163
+P + P
Sbjct: 392 IKSEPVSPSRERSPAPP 408
>UniRef100_UPI00003683EC UPI00003683EC UniRef100 entry
Length = 520
Score = 50.1 bits (118), Expect = 9e-05
Identities = 38/137 (27%), Positives = 58/137 (41%), Gaps = 20/137 (14%)
Query: 34 TTLRAEVEKLASLVSSLMATNDPPLVQ-------QRPQSPYQPMCPQKPRQQAPRQSIPQ 86
T + + ++ +SSL A + P + Q+PQ P QP PQ P+QQ P+ PQ
Sbjct: 330 TAYNTDYQLTSAELSSLPAFSSPGGLSLGNVTAWQQPQQPQQPQQPQPPQQQPPQPQQPQ 389
Query: 87 NQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLN 146
Q PQ+ PQ Q+ S P+ + NL+ PLP V +L P ++
Sbjct: 390 PQQPQQ--PQQPPQQQSHLVPVSL-----------SNLIPGSPLPHVGAALTVTTHPHIS 436
Query: 147 CVFHQGAPGHDTEQCYP 163
+P + P
Sbjct: 437 IKSEPVSPSRERSPAPP 453
>UniRef100_Q41530 Alpha-gliadin storage protein [Triticum aestivum]
Length = 288
Score = 50.1 bits (118), Expect = 9e-05
Identities = 58/215 (26%), Positives = 89/215 (40%), Gaps = 30/215 (13%)
Query: 66 PYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLV 125
P + PQ P QQ P++ +P Q Q+F+ Q Q Q P P + P L +
Sbjct: 24 PVPQLQPQNPSQQQPQEQVPLVQ-QQQFLGQQQPFPPQQPYPQPQPFPSQQPYL----QL 78
Query: 126 QTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQDIK 185
Q P++P S P +RP + Q P + Q P+ ++ Q+ + Q+ +
Sbjct: 79 QPFSQPQLPYSQPQPFRPQQ--PYPQPQPQYSQPQ-QPISQQQQQQQQQQQQQQQQQEQQ 135
Query: 186 VLLQ--QQHLAP--------HSVAAVRPITNVVQDPGYQPQFR---------PSQQQYPD 226
+L Q QQ L P H++A R + V+Q YQ P + Q
Sbjct: 136 ILQQILQQQLTPCMDVVLQQHNIARGR--SQVLQQSTYQLLQELCCQHLWQIPEKLQCQA 193
Query: 227 SHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAP 261
H V + I++ + QP Q QQP+QQ P
Sbjct: 194 IHNVVHAI-ILHQQQQKQQQPSSQVSFQQPQQQYP 227
Score = 37.7 bits (86), Expect = 0.47
Identities = 48/208 (23%), Positives = 75/208 (35%), Gaps = 31/208 (14%)
Query: 57 PLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLL 116
P Q +P P QP +P+ P+Q I Q Q Q+ Q Q Q+ Q +L
Sbjct: 87 PYSQPQPFRPQQPYPQPQPQYSQPQQPISQQQQQQQQQQQQQQQQQEQ---------QIL 137
Query: 117 PILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNV 176
+L++ L + D+ H A G + +Q+L ++
Sbjct: 138 QQILQQQLTPCM---------------DVVLQQHNIARGRSQVLQQSTYQLLQELCCQHL 182
Query: 177 WSFDDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYP---DSHFVTTI 233
W + K+ Q H H++ + Q P Q F+ QQQYP S +
Sbjct: 183 WQIPE---KLQCQAIHNVVHAI-ILHQQQQKQQQPSSQVSFQQPQQQYPLGQGSFRPSQQ 238
Query: 234 LPIMNVVRNPGYQPQFQQYQQQPRQQAP 261
P P PQF++ + Q P
Sbjct: 239 NPQAQGSVQPQQLPQFEEIRNLALQTLP 266
>UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum]
Length = 1716
Score = 50.1 bits (118), Expect = 9e-05
Identities = 21/48 (43%), Positives = 29/48 (59%)
Query: 131 PRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWS 178
P+ ++ +YRPD C +H + GHDTE C LK ++Q LI NV S
Sbjct: 448 PKPVDTSSKFYRPDQRCAYHSNSVGHDTEDCINLKHKIQDLINQNVVS 495
Score = 45.8 bits (107), Expect = 0.002
Identities = 23/83 (27%), Positives = 39/83 (46%), Gaps = 1/83 (1%)
Query: 258 QQAPRIKFDPIPMKYGELFPYLLERNLVQTRPPPPIPKKLPARWRPDLFCVFHQGAQGHD 317
++ P F P+ +LF L + P P+ +RPD C +H + GHD
Sbjct: 416 EKKPARNFTPLIKSRTKLFERLTVAGYIHPVGPKPVDTSSKF-YRPDQRCAYHSNSVGHD 474
Query: 318 VERCFSLKIEVQKLIEDDLIPFE 340
E C +LK ++Q LI +++ +
Sbjct: 475 TEDCINLKHKIQDLINQNVVSLQ 497
>UniRef100_Q14814-5 Splice isoform MEF2DA'0 of Q14814 [Homo sapiens]
Length = 513
Score = 50.1 bits (118), Expect = 9e-05
Identities = 38/137 (27%), Positives = 58/137 (41%), Gaps = 20/137 (14%)
Query: 34 TTLRAEVEKLASLVSSLMATNDPPLVQ-------QRPQSPYQPMCPQKPRQQAPRQSIPQ 86
T + + ++ +SSL A + P + Q+PQ P QP PQ P+QQ P+ PQ
Sbjct: 323 TAYNTDYQLTSAELSSLPAFSSPGGLSLGNVTAWQQPQQPQQPQQPQPPQQQPPQPQQPQ 382
Query: 87 NQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLN 146
Q PQ+ PQ Q+ S P+ + NL+ PLP V +L P ++
Sbjct: 383 PQQPQQ--PQQPPQQQSHLVPVSL-----------SNLIPGSPLPHVGAALTVTTHPHIS 429
Query: 147 CVFHQGAPGHDTEQCYP 163
+P + P
Sbjct: 430 IKSEPVSPSRERSPAPP 446
>UniRef100_Q14814-2 Splice isoform MEF2DA'B of Q14814 [Homo sapiens]
Length = 520
Score = 50.1 bits (118), Expect = 9e-05
Identities = 38/137 (27%), Positives = 58/137 (41%), Gaps = 20/137 (14%)
Query: 34 TTLRAEVEKLASLVSSLMATNDPPLVQ-------QRPQSPYQPMCPQKPRQQAPRQSIPQ 86
T + + ++ +SSL A + P + Q+PQ P QP PQ P+QQ P+ PQ
Sbjct: 330 TAYNTDYQLTSAELSSLPAFSSPGGLSLGNVTAWQQPQQPQQPQQPQPPQQQPPQPQQPQ 389
Query: 87 NQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLN 146
Q PQ+ PQ Q+ S P+ + NL+ PLP V +L P ++
Sbjct: 390 PQQPQQ--PQQPPQQQSHLVPVSL-----------SNLIPGSPLPHVGAALTVTTHPHIS 436
Query: 147 CVFHQGAPGHDTEQCYP 163
+P + P
Sbjct: 437 IKSEPVSPSRERSPAPP 453
>UniRef100_Q14814-3 Splice isoform MEF2D0B of Q14814 [Homo sapiens]
Length = 475
Score = 50.1 bits (118), Expect = 9e-05
Identities = 38/137 (27%), Positives = 58/137 (41%), Gaps = 20/137 (14%)
Query: 34 TTLRAEVEKLASLVSSLMATNDPPLVQ-------QRPQSPYQPMCPQKPRQQAPRQSIPQ 86
T + + ++ +SSL A + P + Q+PQ P QP PQ P+QQ P+ PQ
Sbjct: 285 TAYNTDYQLTSAELSSLPAFSSPGGLSLGNVTAWQQPQQPQQPQQPQPPQQQPPQPQQPQ 344
Query: 87 NQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLN 146
Q PQ+ PQ Q+ S P+ + NL+ PLP V +L P ++
Sbjct: 345 PQQPQQ--PQQPPQQQSHLVPVSL-----------SNLIPGSPLPHVGAALTVTTHPHIS 391
Query: 147 CVFHQGAPGHDTEQCYP 163
+P + P
Sbjct: 392 IKSEPVSPSRERSPAPP 408
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 622,528,348
Number of Sequences: 2790947
Number of extensions: 28188885
Number of successful extensions: 144466
Number of sequences better than 10.0: 661
Number of HSP's better than 10.0 without gapping: 127
Number of HSP's successfully gapped in prelim test: 593
Number of HSP's that attempted gapping in prelim test: 135696
Number of HSP's gapped (non-prelim): 4481
length of query: 348
length of database: 848,049,833
effective HSP length: 128
effective length of query: 220
effective length of database: 490,808,617
effective search space: 107977895740
effective search space used: 107977895740
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146784.3


