

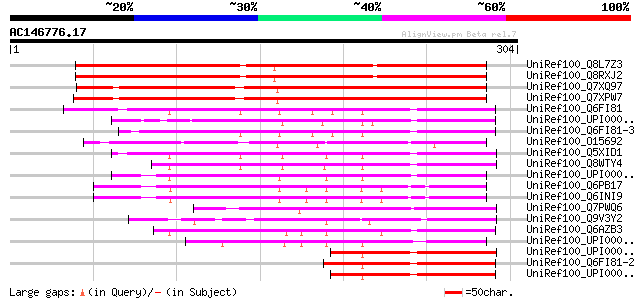
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146776.17 - phase: 0
(304 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8L7Z3 AT5g18400/F20L16_120 [Arabidopsis thaliana] 267 3e-70
UniRef100_Q8RXJ2 Hypothetical protein At5g18400 [Arabidopsis tha... 265 9e-70
UniRef100_Q7XQ97 OSJNBa0018M05.11 protein [Oryza sativa] 249 8e-65
UniRef100_Q7XPW7 OSJNBa0032F06.14 protein [Oryza sativa] 248 1e-64
UniRef100_Q6FI81 Anamorsin [Homo sapiens] 131 2e-29
UniRef100_UPI00002F984C UPI00002F984C UniRef100 entry 128 2e-28
UniRef100_Q6FI81-3 Splice isoform 3 of Q6FI81 [Homo sapiens] 128 2e-28
UniRef100_O15692 Hypothetical protein rsc43 [Dictyostelium disco... 126 8e-28
UniRef100_Q5XID1 Similar to RIKEN cDNA 2810413N20 [Rattus norveg... 125 1e-27
UniRef100_Q8WTY4 Anamorsin [Mus musculus] 125 2e-27
UniRef100_UPI000001AB38 UPI000001AB38 UniRef100 entry 120 6e-26
UniRef100_Q6PB17 LOC398841 protein [Xenopus laevis] 119 1e-25
UniRef100_Q6INI9 LOC398841 protein [Xenopus laevis] 119 1e-25
UniRef100_Q7PWQ6 ENSANGP00000013898 [Anopheles gambiae str. PEST] 119 1e-25
UniRef100_Q9V3Y2 CG4180-PA (L(2)35Bg protein) [Drosophila melano... 118 2e-25
UniRef100_Q6AZB3 Zgc:92009 [Brachydanio rerio] 117 4e-25
UniRef100_UPI00003AA558 UPI00003AA558 UniRef100 entry 117 5e-25
UniRef100_UPI000021E8FD UPI000021E8FD UniRef100 entry 111 3e-23
UniRef100_Q6FI81-2 Splice isoform 2 of Q6FI81 [Homo sapiens] 110 6e-23
UniRef100_UPI000036A7F3 UPI000036A7F3 UniRef100 entry 109 1e-22
>UniRef100_Q8L7Z3 AT5g18400/F20L16_120 [Arabidopsis thaliana]
Length = 272
Score = 267 bits (682), Expect = 3e-70
Identities = 144/259 (55%), Positives = 181/259 (69%), Gaps = 17/259 (6%)
Query: 40 VLACTDEAVLPVSQVFDAIRELGNEGVEKLDPLVITSASSLSKFPVESSSVDLVVLIWKS 99
VLA TD+ VLPVS V ++ELG E +E DPL+IT AS++++FP+++SSV+ V+ I K+
Sbjct: 10 VLAVTDDVVLPVSSVLAIMKELGKEVIESFDPLIITQASTINQFPLDASSVEAVLAISKT 69
Query: 100 LDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQALQ-- 157
DFP D++ E R+LK GGT + K + G + I + ++ LAGF E Q L
Sbjct: 70 SDFPSDKICGEFSRILKPGGTVSVCKVLEGETGEIQQTI---QRRVTLAGFLEPQCLDLK 126
Query: 158 ---------SSVIKAKKPSWKIGSSFALKKFVKSSPKVQIDFDSDLIDKNSLLSEEDLKK 208
S IKAKKPSWKIGSSFALKK V + K+ +D D DLID++SLL+EEDL K
Sbjct: 127 SIKLSTFSLSFGIKAKKPSWKIGSSFALKKPVTNLFKIDLDDDVDLIDEDSLLTEEDLMK 186
Query: 209 PELP-SGDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNPQSACGSCGLGDAFRC 267
P+LP + CE T+KACKNC CGRAE EEK +KLGLT +QI NPQS+CGSCGLGDAFRC
Sbjct: 187 PQLPVASGCET--TKKACKNCVCGRAEIEEKAVKLGLTEDQIENPQSSCGSCGLGDAFRC 244
Query: 268 STCPYKGLPAFKMGETVLL 286
TCPYKGLP FK+GE V L
Sbjct: 245 GTCPYKGLPPFKLGEKVTL 263
>UniRef100_Q8RXJ2 Hypothetical protein At5g18400 [Arabidopsis thaliana]
Length = 269
Score = 265 bits (678), Expect = 9e-70
Identities = 144/259 (55%), Positives = 180/259 (68%), Gaps = 17/259 (6%)
Query: 40 VLACTDEAVLPVSQVFDAIRELGNEGVEKLDPLVITSASSLSKFPVESSSVDLVVLIWKS 99
VLA TD+ VLPVS V ++ELG E +E DPL+IT AS++++FP+++SSV+ V+ I K+
Sbjct: 7 VLAVTDDVVLPVSSVLAIMKELGKEVIESFDPLIITQASTINQFPLDASSVEAVLAISKT 66
Query: 100 LDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQALQ-- 157
DFP D+ E R+LK GGT + K + G + I + ++ LAGF E Q L
Sbjct: 67 SDFPSDKKCGEFSRILKPGGTVSVCKVLEGETGEIQQTI---QRRVTLAGFLEPQCLDLK 123
Query: 158 ---------SSVIKAKKPSWKIGSSFALKKFVKSSPKVQIDFDSDLIDKNSLLSEEDLKK 208
S IKAKKPSWKIGSSFALKK V + K+ +D D DLID++SLL+EEDL K
Sbjct: 124 SIKLSTFSLSFGIKAKKPSWKIGSSFALKKPVTNLFKIDLDDDVDLIDEDSLLTEEDLMK 183
Query: 209 PELP-SGDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNPQSACGSCGLGDAFRC 267
P+LP + CE T+KACKNC CGRAE EEK +KLGLT +QI NPQS+CGSCGLGDAFRC
Sbjct: 184 PQLPVASGCET--TKKACKNCVCGRAEIEEKAVKLGLTEDQIENPQSSCGSCGLGDAFRC 241
Query: 268 STCPYKGLPAFKMGETVLL 286
TCPYKGLP FK+GE V L
Sbjct: 242 GTCPYKGLPPFKLGEKVTL 260
>UniRef100_Q7XQ97 OSJNBa0018M05.11 protein [Oryza sativa]
Length = 265
Score = 249 bits (635), Expect = 8e-65
Identities = 139/256 (54%), Positives = 170/256 (66%), Gaps = 18/256 (7%)
Query: 41 LACTDEAVLPVSQVFDAIRELGNEGVEKLDPLVITSASSLS-KFPVESSSVDLVVLIWKS 99
LA TDE LP+ V D G V + + +VIT +SL K P + +SV V+ + K
Sbjct: 9 LAVTDELALPLRAVGDLAAAAG---VSREEVVVITQCASLGGKLPFDDASVGSVLAVIKK 65
Query: 100 LDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQALQSS 159
++ D E+ RVLKAGG LI S + ++ KLLL GF ++QA +S
Sbjct: 66 VENLGDLFITEISRVLKAGGMVLIQSSPSDQDPNNS-----IQRKLLLGGFVDVQASAAS 120
Query: 160 --------VIKAKKPSWKIGSSFALKKFVKSSPKVQIDFDSDLIDKNSLLSEEDLKKPEL 211
IKAKK SW +GSSF LKK K PK+QID DS+LID++SLL+E+DLKKPEL
Sbjct: 121 SQDSEHSVTIKAKKVSWSLGSSFPLKKATKGLPKIQIDDDSELIDEDSLLTEDDLKKPEL 180
Query: 212 PS-GDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNPQSACGSCGLGDAFRCSTC 270
P GDCE+G TRKACKNC+CGRAE EEKV KL LT+EQINNPQSACG+CGLGDAFRC TC
Sbjct: 181 PVVGDCEVGATRKACKNCTCGRAEAEEKVEKLNLTSEQINNPQSACGNCGLGDAFRCGTC 240
Query: 271 PYKGLPAFKMGETVLL 286
PY+GLPAFK GE + L
Sbjct: 241 PYRGLPAFKPGEKIAL 256
>UniRef100_Q7XPW7 OSJNBa0032F06.14 protein [Oryza sativa]
Length = 264
Score = 248 bits (633), Expect = 1e-64
Identities = 139/258 (53%), Positives = 170/258 (65%), Gaps = 18/258 (6%)
Query: 39 AVLACTDEAVLPVSQVFDAIRELGNEGVEKLDPLVITSASSLS-KFPVESSSVDLVVLIW 97
A LA TDE LP+ V D G + + +VIT +SL K P + +SV V+ +
Sbjct: 6 AALAVTDELALPLRAVGDLAAAAG---FSREEVVVITQCASLGGKLPFDDASVGSVLAVI 62
Query: 98 KSLDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQALQ 157
K ++ D E+ RVLKAGG LI S + ++ KLLL GF ++QA
Sbjct: 63 KKVENLGDLFITEISRVLKAGGMVLIQSSPSDQDPNNS-----IQRKLLLGGFVDVQASA 117
Query: 158 SS--------VIKAKKPSWKIGSSFALKKFVKSSPKVQIDFDSDLIDKNSLLSEEDLKKP 209
+S IKAKK SW +GSSF LKK K PK+QID DS+LID++SLL+E+DLKKP
Sbjct: 118 ASSQDSEHSVTIKAKKVSWSMGSSFPLKKATKGLPKIQIDDDSELIDEDSLLTEDDLKKP 177
Query: 210 ELPS-GDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNPQSACGSCGLGDAFRCS 268
ELP GDCE+G TRKACKNC+CGRAE EEKV KL LT+EQINNPQSACG+CGLGDAFRC
Sbjct: 178 ELPVVGDCEVGATRKACKNCTCGRAEAEEKVEKLNLTSEQINNPQSACGNCGLGDAFRCG 237
Query: 269 TCPYKGLPAFKMGETVLL 286
TCPY+GLPAFK GE + L
Sbjct: 238 TCPYRGLPAFKPGEKIAL 255
>UniRef100_Q6FI81 Anamorsin [Homo sapiens]
Length = 312
Score = 131 bits (330), Expect = 2e-29
Identities = 104/312 (33%), Positives = 155/312 (49%), Gaps = 62/312 (19%)
Query: 33 AAKMYGAVLACTDEAVLPVSQVFDAIREL-GNEGVEKLDPLVITSASSLSKFPVESSSVD 91
+A + AV+ V + + D ++ L GNEG + + + L + + SS D
Sbjct: 7 SAGQFVAVVWDKSSPVEALKGLVDKLQALTGNEG-----RVSVENIKQLLQSAHKESSFD 61
Query: 92 LVV--LIWKSLDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKM--IPDLENKLLL 147
+++ L+ S ++ E+ R+L+ GG + + ++AV + K+ L + L L
Sbjct: 62 IILSGLVPGSTTLHSAEILAEIARILRPGGCLFLKEPVETAVDNNSKVKTASKLCSALTL 121
Query: 148 AGFSEIQALQSSV------------------------IKAKKPSWKIGSSFALKKFV--K 181
+G E++ LQ I KKP++++GSS LK + K
Sbjct: 122 SGLVEVKELQREPLTPEEVQSVREHLGHESDNLLFVQITGKKPNFEVGSSRQLKLSITKK 181
Query: 182 SSPKVQIDFDS------------------DLIDKNSLLSEEDLKKPE---LPSGDCEIGP 220
SSP V+ D DLID + LL EDLKKP+ L + C G
Sbjct: 182 SSPSVKPAVDPAAAKLWTLSANDMEDDSMDLIDSDELLDPEDLKKPDPASLRAASCGEGK 241
Query: 221 TRKACKNCSCGRAEEEEKVLKLGLTAEQINN-PQSACGSCGLGDAFRCSTCPYKGLPAFK 279
RKACKNC+CG AEE EK + EQ+++ P+SACG+C LGDAFRC++CPY G+PAFK
Sbjct: 242 KRKACKNCTCGLAEELEKEK----SREQMSSQPKSACGNCYLGDAFRCASCPYLGMPAFK 297
Query: 280 MGETVLLLSPSL 291
GE VLL +L
Sbjct: 298 PGEKVLLSDSNL 309
>UniRef100_UPI00002F984C UPI00002F984C UniRef100 entry
Length = 313
Score = 128 bits (322), Expect = 2e-28
Identities = 97/280 (34%), Positives = 137/280 (48%), Gaps = 59/280 (21%)
Query: 62 GNEGVEKLDPLVITSASSLSKFPVESSSVDLVVLIWKSLDFPIDQLTQEVLRVLKAGGTT 121
G VE +D L+++S S+ S F S V L S +D L E+ RVLK GG
Sbjct: 40 GKVSVENVDRLLLSSHSA-STFDCAVSCV----LADSSAVHSLDTLA-ELARVLKPGGKL 93
Query: 122 LIHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQALQSSVIK------------------- 162
++ + A ++ L + L L+GF+ + + + +
Sbjct: 94 ILEEVVTGAEAQRERTSEKLVSTLKLSGFTSVTEISKAELSPDALSAIRTATGYQGNALF 153
Query: 163 -----AKKPSWKIGSSFALK-KFVKSSPKV--------------------QIDFDSDLID 196
A KP +++GSS +K F +P+ D D DL+D
Sbjct: 154 RIRMSASKPDFEVGSSSQIKLSFGNKAPRPADKPAPDPNTVKMWMLSANDMNDDDLDLVD 213
Query: 197 KNSLLSEEDLKKPE---LPSGDC--EIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINN 251
+SLL EEDLKKP+ L + C G +KACKNC+CG AEE E+ K E+ N
Sbjct: 214 SDSLLDEEDLKKPDPSSLKASTCGEAAGKKKKACKNCTCGLAEELEQESK---EKEKTNL 270
Query: 252 PQSACGSCGLGDAFRCSTCPYKGLPAFKMGETVLLLSPSL 291
P+SACGSC LGDAFRC++CPY G+PAFK GE +LL + +L
Sbjct: 271 PKSACGSCYLGDAFRCASCPYLGMPAFKPGEKILLDNKTL 310
>UniRef100_Q6FI81-3 Splice isoform 3 of Q6FI81 [Homo sapiens]
Length = 299
Score = 128 bits (322), Expect = 2e-28
Identities = 97/276 (35%), Positives = 137/276 (49%), Gaps = 58/276 (21%)
Query: 66 VEKLDPLVITSASSLSKFPVESSSVDLVVLIWKSLDFPIDQLTQEVLRVLKAGGTTLIHK 125
V+KL L + + VE+ L L+ S ++ E+ R+L+ GG + +
Sbjct: 29 VDKLQALT----GNEGRVSVENIKQLLQCLVPGSTTLHSAEILAEIARILRPGGCLFLKE 84
Query: 126 SSQSAVGSGDKM--IPDLENKLLLAGFSEIQALQSSV----------------------- 160
++AV + K+ L + L L+G E++ LQ
Sbjct: 85 PVETAVDNNSKVKTASKLCSALTLSGLVEVKELQREPLTPEEVQSVREHLGHESDNLLFV 144
Query: 161 -IKAKKPSWKIGSSFALKKFV--KSSPKVQIDFDS------------------DLIDKNS 199
I KKP++++GSS LK + KSSP V+ D DLID +
Sbjct: 145 QITGKKPNFEVGSSRQLKLSITKKSSPSVKPAVDPAAAKLWTLSANDMEDDSMDLIDSDE 204
Query: 200 LLSEEDLKKPE---LPSGDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINN-PQSA 255
LL EDLKKP+ L + C G RKACKNC+CG AEE EK + EQ+++ P+SA
Sbjct: 205 LLDPEDLKKPDPASLRAASCGEGKKRKACKNCTCGLAEELEKEK----SREQMSSQPKSA 260
Query: 256 CGSCGLGDAFRCSTCPYKGLPAFKMGETVLLLSPSL 291
CG+C LGDAFRC++CPY G+PAFK GE VLL +L
Sbjct: 261 CGNCYLGDAFRCASCPYLGMPAFKPGEKVLLSDSNL 296
>UniRef100_O15692 Hypothetical protein rsc43 [Dictyostelium discoideum]
Length = 256
Score = 126 bits (316), Expect = 8e-28
Identities = 92/251 (36%), Positives = 129/251 (50%), Gaps = 23/251 (9%)
Query: 45 DEAVLPVSQVFDAIRELGNEGVEKLDPLVITSASSLSKFPVESSSVDLVVLIWKSLDFPI 104
++ VL +S V E + ++ L T +SLSK + + V+I S F
Sbjct: 11 EQEVLIISDV-----ENSESIINQVKELSKTVTTSLSKEQQQIQNNFDHVIIISSKPFN- 64
Query: 105 DQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQALQSS----V 160
L +LK GG I++++++ M L+ G + +A +S +
Sbjct: 65 SALISMYSNLLKKGGKLSIYQTNENETLVNSGM------DFLIGGLVDFKATSNSTYKTI 118
Query: 161 IKAKKPSWKIGSSFALKKFVKSS--PKVQIDFDSDLIDKNSLLSEED-LKKPELPSGDCE 217
+ A KPSW S + SS P I+ D I++N L+SE D KP DCE
Sbjct: 119 VHADKPSWDTNESSTINIPSTSSNNPWASIE-GGDRINENDLVSENDKTSKPATTLDDCE 177
Query: 218 IGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNPQ--SACGSCGLGDAFRCSTCPYKGL 275
+G T+KACKNC+CGRAEEE + K LT E I NP S+CG+C LGDAFRC CPY+GL
Sbjct: 178 VGKTKKACKNCTCGRAEEENQS-KPKLTKEMIENPGVGSSCGNCSLGDAFRCGGCPYRGL 236
Query: 276 PAFKMGETVLL 286
P FK+GE + L
Sbjct: 237 PTFKVGEKIQL 247
>UniRef100_Q5XID1 Similar to RIKEN cDNA 2810413N20 [Rattus norvegicus]
Length = 309
Score = 125 bits (315), Expect = 1e-27
Identities = 92/280 (32%), Positives = 136/280 (47%), Gaps = 58/280 (20%)
Query: 62 GNEGVEKLDPLVITSASSLSKFPVESSSVDLVV--LIWKSLDFPIDQLTQEVLRVLKAGG 119
G+EG + + + + L + + SS D+++ ++ S + ++ R+L+ GG
Sbjct: 37 GSEG-----QVFVENITQLLQSAHKESSFDVILSGIVPGSTSLHSPEALADMARILRPGG 91
Query: 120 TTLIHKSSQSAVGSGDKM--IPDLENKLLLAGFSEIQALQSSVIK--------------- 162
+ + ++ + DK+ L + L L+G EI+ LQ +
Sbjct: 92 CLFLKEPVETTGVNNDKIKTASKLCSALTLSGLVEIKELQREALSPEEAQSMQEHLGYHS 151
Query: 163 ---------AKKPSWKIGSSFALKKFVKSSPKVQI------------------DFDSDLI 195
KKP++++GSS LK K S V+ D DLI
Sbjct: 152 DSLLSVHVTGKKPNFEVGSSSQLKLLHKKSSSVKPVVDPATAKLWTLSANDMEDDSMDLI 211
Query: 196 DKNSLLSEEDLKKPE---LPSGDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNP 252
D + LL EDLKKP+ L + C G RKACKNC+CG AEE EK + Q + P
Sbjct: 212 DSDELLDPEDLKKPDPASLKAPSCGEGKKRKACKNCTCGLAEELEKEQ----SKAQSSQP 267
Query: 253 QSACGSCGLGDAFRCSTCPYKGLPAFKMGETVLLLSPSLK 292
+SACG+C LGDAFRC+ CPY G+PAFK GE VLL S +L+
Sbjct: 268 KSACGNCYLGDAFRCANCPYLGMPAFKPGEQVLLSSSNLQ 307
>UniRef100_Q8WTY4 Anamorsin [Mus musculus]
Length = 309
Score = 125 bits (313), Expect = 2e-27
Identities = 89/256 (34%), Positives = 131/256 (50%), Gaps = 53/256 (20%)
Query: 86 ESSSVDLVV--LIWKSLDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKM--IPDL 141
+ SS D+++ ++ S ++ E+ R+L+ GG + + ++A + DKM L
Sbjct: 56 KESSFDVILSGVVPGSTSLHSAEVLAEMARILRPGGCLFLKEPVETAEVNNDKMKTASKL 115
Query: 142 ENKLLLAGFSEIQALQSSVIK------------------------AKKPSWKIGSSFALK 177
+ L L+G EI+ LQ + KKP++++GSS LK
Sbjct: 116 CSALTLSGLVEIKELQREALSPEEVQSVQEHLGYHSDSLRSVRVTGKKPNFEVGSSSQLK 175
Query: 178 KFVKSSPKVQ-----------------IDFDS-DLIDKNSLLSEEDLKKPE---LPSGDC 216
K S V+ ++ DS DLID + LL EDLK+P+ L + C
Sbjct: 176 LPNKKSSSVKPVVDPAAAKLWTLSANDMEDDSVDLIDSDELLDPEDLKRPDPASLKAPSC 235
Query: 217 EIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNPQSACGSCGLGDAFRCSTCPYKGLP 276
G RKACKNC+CG AEE E+ + Q + P+SACG+C LGDAFRC+ CPY G+P
Sbjct: 236 GEGKKRKACKNCTCGLAEELEREQ----SKAQSSQPKSACGNCYLGDAFRCANCPYLGMP 291
Query: 277 AFKMGETVLLLSPSLK 292
AFK GE VLL + +L+
Sbjct: 292 AFKPGEQVLLSNSNLQ 307
>UniRef100_UPI000001AB38 UPI000001AB38 UniRef100 entry
Length = 312
Score = 120 bits (300), Expect = 6e-26
Identities = 88/276 (31%), Positives = 135/276 (48%), Gaps = 62/276 (22%)
Query: 62 GNEGVEKLDPLVITSASSLSKFPVESSSVDLVV--LIWKSLDFPIDQLTQEVLRVLKAGG 119
G VE ++ L+++S + S+ D VV ++ S + E+ R+LK GG
Sbjct: 40 GKVSVENVERLLLSSHLA--------STFDCVVSCVLADSSTVHSSETLAELARILKPGG 91
Query: 120 TTLIHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQALQSSV------------------- 160
++ ++ A + +L + L L+GF+ + + +
Sbjct: 92 KLILDEAVTGAEVQSVRTSANLISTLKLSGFTSVAEISKAELSPEALSAYRTATGYQGNT 151
Query: 161 -----IKAKKPSWKIGSSFALK-KFVKSSPKVQI-------------------DFDSDLI 195
I A KP++++GSS +K F +PK + D D DL+
Sbjct: 152 LCRIRISAFKPNFEVGSSSQIKLSFGSKTPKPEKPALDPNTVKMWMLSANDMNDDDLDLV 211
Query: 196 DKNSLLSEEDLKKPE-----LPSGDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQIN 250
D ++LL EED KKP+ +P+ G +KACKNC+CG A+E E+ K E+ N
Sbjct: 212 DSDALLDEEDFKKPDPSSLTVPTCGEGAGKKKKACKNCTCGLADELEEESK---GKEKTN 268
Query: 251 NPQSACGSCGLGDAFRCSTCPYKGLPAFKMGETVLL 286
P+SACGSC LGDAFRC++CPY G+PAFK GE +LL
Sbjct: 269 LPKSACGSCYLGDAFRCASCPYLGMPAFKPGEKILL 304
>UniRef100_Q6PB17 LOC398841 protein [Xenopus laevis]
Length = 320
Score = 119 bits (297), Expect = 1e-25
Identities = 94/284 (33%), Positives = 139/284 (48%), Gaps = 59/284 (20%)
Query: 51 VSQVFDAIRELGNEGVEKLDPLVITSASSLSKFPVESSSVDLVVL--IWKSLDFPIDQLT 108
VS++ + + G VE ++ L++++ + SS D ++L + + ++
Sbjct: 40 VSKLQEVVAPRGKVSVENIERLLLSAHAD--------SSFDAILLGMVQGTQCIHSSEVL 91
Query: 109 QEVLRVLKAGGTTLIHKSSQSAVGSGDKMIPDLENKLLLAGFSEI-QALQSSV------- 160
EV R+LK GG +I + + G+ + L + L L+G +E+ Q LQ +
Sbjct: 92 AEVARILKPGGALIIQEPVAAGAGARLRTPEHLSSVLKLSGLTEVTQLLQEPLNPEQKQG 151
Query: 161 ----------------IKAKKPSWKIGSSFAL---KKFVKSSPKVQI------------- 188
I+AKKP++++GSS L K+ P V
Sbjct: 152 VVELLGYNGNDVSTIRIRAKKPNYEVGSSRQLSLPKRKTAEKPSVDPAAAKLWTLSASDM 211
Query: 189 -DFDSDLIDKNSLLSEEDLKKP---ELPSGDCEIGPT--RKACKNCSCGRAEEEEKVLKL 242
D D D++D + LL +EDLKKP L + C G RKACKNC+CG AEE E K
Sbjct: 212 NDDDVDILDSDELLDQEDLKKPAPSSLLASGCGEGSEKKRKACKNCTCGLAEELEAE-KT 270
Query: 243 GLTAEQINNPQSACGSCGLGDAFRCSTCPYKGLPAFKMGETVLL 286
T + SACG+C LGDAFRC++CPY G+PAFK GE VLL
Sbjct: 271 PSTVPKA--APSACGNCYLGDAFRCASCPYLGMPAFKPGEKVLL 312
>UniRef100_Q6INI9 LOC398841 protein [Xenopus laevis]
Length = 328
Score = 119 bits (297), Expect = 1e-25
Identities = 94/284 (33%), Positives = 139/284 (48%), Gaps = 59/284 (20%)
Query: 51 VSQVFDAIRELGNEGVEKLDPLVITSASSLSKFPVESSSVDLVVL--IWKSLDFPIDQLT 108
VS++ + + G VE ++ L++++ + SS D ++L + + ++
Sbjct: 48 VSKLQEVVAPRGKVSVENIERLLLSAHAD--------SSFDAILLGMVQGTQCIHSSEVL 99
Query: 109 QEVLRVLKAGGTTLIHKSSQSAVGSGDKMIPDLENKLLLAGFSEI-QALQSSV------- 160
EV R+LK GG +I + + G+ + L + L L+G +E+ Q LQ +
Sbjct: 100 AEVARILKPGGALIIQEPVAAGAGAQLRTPEHLSSVLKLSGLTEVTQLLQEPLNPEQKQG 159
Query: 161 ----------------IKAKKPSWKIGSSFAL---KKFVKSSPKVQI------------- 188
I+AKKP++++GSS L K+ P V
Sbjct: 160 VVELLGYNGNDVSTIRIRAKKPNYEVGSSRQLSLPKRKTAEKPSVDPAAAKLWTLSASDM 219
Query: 189 -DFDSDLIDKNSLLSEEDLKKP---ELPSGDCEIGPT--RKACKNCSCGRAEEEEKVLKL 242
D D D++D + LL +EDLKKP L + C G RKACKNC+CG AEE E K
Sbjct: 220 NDDDVDILDSDELLDQEDLKKPAPSSLLASGCGEGSEKKRKACKNCTCGLAEELEAE-KT 278
Query: 243 GLTAEQINNPQSACGSCGLGDAFRCSTCPYKGLPAFKMGETVLL 286
T + SACG+C LGDAFRC++CPY G+PAFK GE VLL
Sbjct: 279 PSTVPKA--APSACGNCYLGDAFRCASCPYLGMPAFKPGEKVLL 320
>UniRef100_Q7PWQ6 ENSANGP00000013898 [Anopheles gambiae str. PEST]
Length = 254
Score = 119 bits (297), Expect = 1e-25
Identities = 74/187 (39%), Positives = 104/187 (55%), Gaps = 13/187 (6%)
Query: 111 VLRVLKAGGTTLIHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQALQSSVIKAKKPSWKI 170
++++LK G + S ++ I + LLLAGF I A S+V A+KP +++
Sbjct: 73 LVKLLKPKGKCVFRDDSAAS-------IEQARSNLLLAGFINIVASDSNVYVAEKPDYEV 125
Query: 171 GS----SFALKKFVKSSPKVQIDFDSDLIDKNSLLSEEDLKKPELPS-GDCEIGPTRKAC 225
GS SFA K V + K+ + + + ID LL E+D KP S C RKAC
Sbjct: 126 GSKSKLSFAKKSNVAAVWKLDDNEEEERIDDEELLDEDDKAKPTEESLRVCGTTGKRKAC 185
Query: 226 KNCSCGRAEEEEKVLKLGLTAEQINNPQSACGSCGLGDAFRCSTCPYKGLPAFKMGETVL 285
K+CSCG AEE + K G + +S+CGSC LGDAFRC+TCPY G+PAFK GE ++
Sbjct: 186 KDCSCGLAEELDAEAK-GKALTDTSAAKSSCGSCYLGDAFRCATCPYLGMPAFKPGEKIV 244
Query: 286 LLSPSLK 292
L ++
Sbjct: 245 LTDTQMQ 251
>UniRef100_Q9V3Y2 CG4180-PA (L(2)35Bg protein) [Drosophila melanogaster]
Length = 248
Score = 118 bits (295), Expect = 2e-25
Identities = 85/230 (36%), Positives = 120/230 (51%), Gaps = 28/230 (12%)
Query: 72 LVITSASSLSKFPVESSSVDLVVLIWKSLDFPIDQLTQ---EVLRVLKAGGTTLIHKSSQ 128
+ + + LS +SS DL+V+ QLT ++L +LK G +
Sbjct: 35 VALENVHRLSFSSYANSSFDLIVI-------ECAQLTDSYVKLLHMLKPSGKLHL----V 83
Query: 129 SAVGSGDKMIPDLENKLLLAGFSEIQALQSSVIKAKKPSWKIGSSFALKKFVKSSPKVQI 188
S +G ++ +++ L+GF + + A+KP ++ GSS L K++ V +
Sbjct: 84 SYIGPAASLLQEIK----LSGFINCREDSPDALTAEKPGYETGSSARLSFAKKNASAVNV 139
Query: 189 ----DFDSDLIDKNSLLSEEDLKKPELPSG--DCEIGPTRKACKNCSCGRAEEEEKVLKL 242
D +LID+ LL EED +KP+ P+G C RKACKNCSCG AEE E +
Sbjct: 140 WKISGDDEELIDEEELLDEEDKQKPD-PAGLRVCSTTGKRKACKNCSCGLAEELETEKQS 198
Query: 243 GLTAEQINNPQSACGSCGLGDAFRCSTCPYKGLPAFKMGETVLLLSPSLK 292
E N +S+CG+C LGDAFRCSTCPY G+PAFK GE V L LK
Sbjct: 199 QKATE---NAKSSCGNCYLGDAFRCSTCPYLGMPAFKPGEKVQLADNLLK 245
>UniRef100_Q6AZB3 Zgc:92009 [Brachydanio rerio]
Length = 311
Score = 117 bits (293), Expect = 4e-25
Identities = 82/255 (32%), Positives = 123/255 (48%), Gaps = 53/255 (20%)
Query: 87 SSSVDLVV--LIWKSLDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKMIPDLENK 144
+SS D V+ L+ S ++ E+ RV+K GGT ++ + + +G + L +
Sbjct: 57 ASSYDCVLSGLLTDSSLIHSSEMLVEMARVMKPGGTLVLEEPVTGSEDTGVRTAEKLMSA 116
Query: 145 LLLAGFSEIQALQSSVIKAK------------------------KPSWKIGSS------F 174
+ LAG + +++ + K KP +++GSS F
Sbjct: 117 IKLAGLVSVTEVKTEPLSPKAAAALTERTGFSGNTLSRVRMTASKPDFEVGSSTQLKLSF 176
Query: 175 ALKKFVKSSPKVQI--------------DFDSDLIDKNSLLSEEDLKKPELPSGDCEIGP 220
KK P + D D DL+D ++LL +DLKKP+ S G
Sbjct: 177 GKKKTTTVKPALDPGTAQKWTLSANDMDDDDVDLVDSDALLDADDLKKPDPSSLKASCGD 236
Query: 221 T----RKACKNCSCGRAEEEEKVLKLGLTAEQINNPQSACGSCGLGDAFRCSTCPYKGLP 276
+KACKNC+CG AEE E+ K A++ + P+SACG+C LGDAFRC +CPY G+P
Sbjct: 237 DSKKKKKACKNCTCGLAEELEQESK---AAQKASQPKSACGNCYLGDAFRCGSCPYLGMP 293
Query: 277 AFKMGETVLLLSPSL 291
AFK GE VLL + +
Sbjct: 294 AFKPGEKVLLANTDI 308
>UniRef100_UPI00003AA558 UPI00003AA558 UniRef100 entry
Length = 308
Score = 117 bits (292), Expect = 5e-25
Identities = 80/230 (34%), Positives = 113/230 (48%), Gaps = 56/230 (24%)
Query: 106 QLTQEVLRVLKAGGTTLIHKS--SQSAVGSGDKMIPDLENKLLLAGFSEIQALQSSVIKA 163
++ E+ R+LK GG L+ + ++S S K L L L+G E++ LQ + A
Sbjct: 78 EVLAEIARILKPGGRVLLKEPVVTESENNSQIKTAAKLPAALTLSGLVEVKELQKEPLTA 137
Query: 164 ------------------------KKPSWKIGSS------FALKKFVKSSPKVQI----- 188
+KP++++GSS FA K P V
Sbjct: 138 EEAQSVREHLGYQGNDLLIVQIEGRKPNFEVGSSSQLKLSFAKKTSPSGKPSVDPATAKL 197
Query: 189 ---------DFDSDLIDKNSLLSEEDLKKPE---LPSGDCEIGPTRKACKNCSCGRAEEE 236
D + DL+D + LL EDLKKP+ L + C+ +KACKNC+CG AEE
Sbjct: 198 WTLSASDMNDEEMDLLDSDELLDSEDLKKPDPASLRAPSCKEKGKKKACKNCTCGLAEEL 257
Query: 237 EKVLKLGLTAEQINNPQSACGSCGLGDAFRCSTCPYKGLPAFKMGETVLL 286
E+ K + P+SACG+C LGDAFRC++CPY G+PAFK GE +LL
Sbjct: 258 EQEKKS-------SQPKSACGNCYLGDAFRCASCPYLGMPAFKPGEKILL 300
>UniRef100_UPI000021E8FD UPI000021E8FD UniRef100 entry
Length = 354
Score = 111 bits (277), Expect = 3e-23
Identities = 58/103 (56%), Positives = 70/103 (67%), Gaps = 7/103 (6%)
Query: 193 DLIDKNSLLSEEDLKKPE---LPSGDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQI 249
DLID + LL EDLKKP+ L + C G RKACKNC+CG AEE EK + Q
Sbjct: 254 DLIDSDELLDPEDLKKPDPASLKAPSCGEGKKRKACKNCTCGLAEELEKEQ----SKAQS 309
Query: 250 NNPQSACGSCGLGDAFRCSTCPYKGLPAFKMGETVLLLSPSLK 292
+ P+SACG+C LGDAFRC+ CPY G+PAFK GE VLL S +L+
Sbjct: 310 SQPKSACGNCYLGDAFRCANCPYLGMPAFKPGEQVLLSSSNLQ 352
>UniRef100_Q6FI81-2 Splice isoform 2 of Q6FI81 [Homo sapiens]
Length = 108
Score = 110 bits (274), Expect = 6e-23
Identities = 59/107 (55%), Positives = 73/107 (68%), Gaps = 8/107 (7%)
Query: 189 DFDSDLIDKNSLLSEEDLKKPE---LPSGDCEIGPTRKACKNCSCGRAEEEEKVLKLGLT 245
D DLID + LL EDLKKP+ L + C G RKACKNC+CG AEE EK +
Sbjct: 3 DDSMDLIDSDELLDPEDLKKPDPASLRAASCGEGKKRKACKNCTCGLAEELEKEK----S 58
Query: 246 AEQINN-PQSACGSCGLGDAFRCSTCPYKGLPAFKMGETVLLLSPSL 291
EQ+++ P+SACG+C LGDAFRC++CPY G+PAFK GE VLL +L
Sbjct: 59 REQMSSQPKSACGNCYLGDAFRCASCPYLGMPAFKPGEKVLLSDSNL 105
>UniRef100_UPI000036A7F3 UPI000036A7F3 UniRef100 entry
Length = 367
Score = 109 bits (272), Expect = 1e-22
Identities = 58/103 (56%), Positives = 72/103 (69%), Gaps = 8/103 (7%)
Query: 193 DLIDKNSLLSEEDLKKPE---LPSGDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQI 249
DLID + LL EDLKKP+ L + C G RKACKNC+CG AEE EK + EQ+
Sbjct: 266 DLIDSDELLDPEDLKKPDPASLRAASCGEGKKRKACKNCTCGLAEELEKEK----SREQM 321
Query: 250 NN-PQSACGSCGLGDAFRCSTCPYKGLPAFKMGETVLLLSPSL 291
++ P+SACG+C LGDAFRC++CPY G+PAFK GE VLL +L
Sbjct: 322 SSQPKSACGNCYLGDAFRCASCPYLGMPAFKPGEKVLLSDSNL 364
Score = 35.4 bits (80), Expect = 1.9
Identities = 38/150 (25%), Positives = 65/150 (43%), Gaps = 32/150 (21%)
Query: 66 VEKLDPLVITSASSLSKFPVESSSVDLVVLIWKSLDFPIDQLTQEVLRVLKAGGTTLIHK 125
V+KL L + + VE+ + L L+ S ++ E+ R+L+ GG + +
Sbjct: 29 VDKLQALT----GNEGRVSVENINQLLQCLVPGSTTLHSAEILAEIARILRPGGCLFLKE 84
Query: 126 SSQSAVGSGDKM--IPDLENKLLLAGFSEIQALQSS------------------------ 159
++AV + K+ L + L L+G E++ LQ
Sbjct: 85 PVETAVDNNSKVKTASKLCSALTLSGLVEVKELQREPLTPEEVQSVREHLGHESDNLLFV 144
Query: 160 VIKAKKPSWKIGSSFALKKFV--KSSPKVQ 187
I KKP++++GSS LK + KSSP V+
Sbjct: 145 QITGKKPNFEVGSSRQLKFSITKKSSPSVK 174
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.135 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 474,816,329
Number of Sequences: 2790947
Number of extensions: 18717301
Number of successful extensions: 52914
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 52735
Number of HSP's gapped (non-prelim): 93
length of query: 304
length of database: 848,049,833
effective HSP length: 127
effective length of query: 177
effective length of database: 493,599,564
effective search space: 87367122828
effective search space used: 87367122828
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146776.17


