

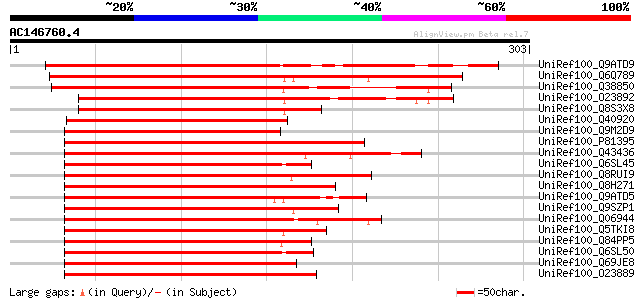
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146760.4 + phase: 0
(303 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ATD9 BNLGHi233 [Gossypium hirsutum] 298 2e-79
UniRef100_Q6Q789 Myb transcription factor [Vitis vinifera] 296 6e-79
UniRef100_Q38850 Myb-related protein Atmyb5 [Arabidopsis thaliana] 263 6e-69
UniRef100_O23892 OSMYB4 [Oryza sativa] 241 2e-62
UniRef100_Q8S3X8 Typical P-type R2R3 Myb protein [Oryza sativa] 221 2e-56
UniRef100_Q40920 MYB-like transcriptional factor MBF1 [Picea mar... 209 7e-53
UniRef100_Q9M2D9 Putative transcription factor [Arabidopsis thal... 208 1e-52
UniRef100_P81395 MYB-related protein 330 [Antirrhinum majus] 208 2e-52
UniRef100_Q43436 MYB1 [Gossypium hirsutum] 207 2e-52
UniRef100_Q6SL45 MYB6 [Tradescantia fluminensis] 207 3e-52
UniRef100_Q8RUI9 Putative myb-related protein [Oryza sativa] 206 5e-52
UniRef100_Q8H271 Myb-like transcription factor 1 [Gossypium hirs... 206 5e-52
UniRef100_Q9ATD5 GHMYB10 [Gossypium hirsutum] 205 1e-51
UniRef100_Q9SZP1 Transcription repressor MYB4 [Arabidopsis thali... 204 2e-51
UniRef100_Q06944 MybHv5 [Hordeum vulgare] 204 3e-51
UniRef100_Q5TKI8 Putative myb protein [Oryza sativa] 203 4e-51
UniRef100_Q84PP5 Transcription factor MYB101 [Lotus japonicus] 203 5e-51
UniRef100_Q6SL50 MYB1 [Tradescantia fluminensis] 203 5e-51
UniRef100_Q69JE8 Putative Myb-related protein Zm38 [Oryza sativa] 203 5e-51
UniRef100_O23889 OSMYB1 [Oryza sativa] 202 7e-51
>UniRef100_Q9ATD9 BNLGHi233 [Gossypium hirsutum]
Length = 247
Score = 298 bits (762), Expect = 2e-79
Identities = 165/266 (62%), Positives = 180/266 (67%), Gaps = 28/266 (10%)
Query: 22 NPNKKQKT-TTTTPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGK 80
NP KK T TTPCCSKVGLKRGPWT EEDE+LS YI KEGEGRWRTLPKRAGLLRCGK
Sbjct: 3 NPTKKDNVGTKTTPCCSKVGLKRGPWTPEEDELLSNYINKEGEGRWRTLPKRAGLLRCGK 62
Query: 81 SCRLRWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTH 140
SCRLRWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTH
Sbjct: 63 SCRLRWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTH 122
Query: 141 LSKKLINQGIDPRTHKPLNNPSSSSSIIPNTNHQTSTPFFAPSSSDHHHPITITNNEPDP 200
LSKKLI+QGIDPRTHKPL NP S P+ S SSS P NN P P
Sbjct: 123 LSKKLISQGIDPRTHKPL-NPQQLSPSPPSLKPSPS------SSSSMAKP----NNPPSP 171
Query: 201 SQTQQAQ-GCGNLVNDVSAMDNHHVLTNNNRGDYGDDNGNNNYGGDDVFTSFLDSLINDD 259
N +D S D+ ++ NN+ D+ DDVF+SFL+SLIN+D
Sbjct: 172 LPVHVVNANKQNYYDDGSNEDHQGMIMNNDHYQQQQDH-------DDVFSSFLNSLINED 224
Query: 260 AFAAHRSENDPLILSAAPPALWESPP 285
+D L+ + WES P
Sbjct: 225 --------DDALVSNLGLSQGWESTP 242
>UniRef100_Q6Q789 Myb transcription factor [Vitis vinifera]
Length = 320
Score = 296 bits (757), Expect = 6e-79
Identities = 153/252 (60%), Positives = 172/252 (67%), Gaps = 11/252 (4%)
Query: 24 NKKQKTTTTTPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCR 83
N +T+ TPCC+KVGLKRGPWT EEDE+L+ Y+K+EGEGRWRTLPKRAGLLRCGKSCR
Sbjct: 3 NPASASTSKTPCCTKVGLKRGPWTPEEDELLANYVKREGEGRWRTLPKRAGLLRCGKSCR 62
Query: 84 LRWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSK 143
LRWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSK
Sbjct: 63 LRWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSK 122
Query: 144 KLINQGIDPRTHKPLN---NPSSS-----SSIIPNTNHQTSTPFFAPSSSDHH-HPITIT 194
KLI+QGIDPRTHKPLN NPS S IPN N S+ S+H I
Sbjct: 123 KLISQGIDPRTHKPLNPKPNPSPDVNAPVSKSIPNANPNPSSSRVGEIGSNHEVKEIESN 182
Query: 195 NNEPDPSQTQQAQG--CGNLVNDVSAMDNHHVLTNNNRGDYGDDNGNNNYGGDDVFTSFL 252
N +P Q + + + D + G DD + + DD F SFL
Sbjct: 183 ENHKEPPNLDQYHSPLAADSNENWQSADGLVTGLQSTHGTSNDDEDDIGFCNDDTFPSFL 242
Query: 253 DSLINDDAFAAH 264
+SLIN+D F H
Sbjct: 243 NSLINEDVFGNH 254
>UniRef100_Q38850 Myb-related protein Atmyb5 [Arabidopsis thaliana]
Length = 249
Score = 263 bits (671), Expect = 6e-69
Identities = 137/238 (57%), Positives = 158/238 (65%), Gaps = 34/238 (14%)
Query: 25 KKQKTTTTTPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRL 84
KK + TTPCC+K+G+KRGPWTVEEDE+L +IKKEGEGRWR+LPKRAGLLRCGKSCRL
Sbjct: 7 KKPVSKKTTPCCTKMGMKRGPWTVEEDEILVSFIKKEGEGRWRSLPKRAGLLRCGKSCRL 66
Query: 85 RWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKK 144
RWMNYLRPSVKRG I DEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHL KK
Sbjct: 67 RWMNYLRPSVKRGGITSDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLRKK 126
Query: 145 LINQGIDPRTHKPL--NNPSSSSSIIPNTNHQTSTPFFAPSSSDHHHPITITNNEPDPSQ 202
L+ QGIDP+THKPL NN + P SS H T+ + D
Sbjct: 127 LLRQGIDPQTHKPLDANNIHKPEEEVSGGQKYP----LEPISSSHTDDTTVNGGDGD--- 179
Query: 203 TQQAQGCGNLVNDVSAMDNHHVLTNNNRGDYGDDNGNNNYG--GDDVFTSFLDSLIND 258
+ N+ +G ++G ++G DD F+SFL+SLIND
Sbjct: 180 -----------------------SKNSINVFGGEHGYEDFGFCYDDKFSSFLNSLIND 214
>UniRef100_O23892 OSMYB4 [Oryza sativa]
Length = 275
Score = 241 bits (615), Expect = 2e-62
Identities = 130/235 (55%), Positives = 158/235 (66%), Gaps = 25/235 (10%)
Query: 41 LKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPSVKRGQIA 100
LKRGPWT EEDEVL+ ++ +EG RWRTLP+RAGLLRCGKSCRLRWMNYLRP +KR IA
Sbjct: 20 LKRGPWTPEEDEVLARFVAREGCDRWRTLPRRAGLLRCGKSCRLRWMNYLRPDIKRCPIA 79
Query: 101 PDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDPRTHKPLN- 159
DEEDLILRLHRLLGNRWSLIAGR+PGRTDNEIKNYWN+HLSKKLI QGIDPRTHKPL
Sbjct: 80 DDEEDLILRLHRLLGNRWSLIAGRLPGRTDNEIKNYWNSHLSKKLIAQGIDPRTHKPLTA 139
Query: 160 --NPSSSSSIIPNTNHQTSTPFFAPSSSDHHHPITITNNEPDPSQTQQAQGCGNLVNDVS 217
+ S++++ + T+++ + P P ++ E + + A G G+ VS
Sbjct: 140 AADHSNAAAAVAATSYKKAVPAKPPRTASS----PAAGIECSDDRARPADGGGDFAAMVS 195
Query: 218 AMDNHHVLTNNNRGDYGDD-------NGNNNYG------GDDVFTSFLDSLINDD 259
A D G +GD +G + G GDD F+SFLDSLIND+
Sbjct: 196 AAD-----AEGFEGGFGDQFCAEDAVHGGFDMGSASAMVGDDDFSSFLDSLINDE 245
>UniRef100_Q8S3X8 Typical P-type R2R3 Myb protein [Oryza sativa]
Length = 166
Score = 221 bits (563), Expect = 2e-56
Identities = 104/145 (71%), Positives = 122/145 (83%), Gaps = 3/145 (2%)
Query: 41 LKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPSVKRGQIA 100
LKRGPWT EEDEVL+ ++ +EG RWRTLP+RAGLLRCGKSCRLRWMNYLRP +KR IA
Sbjct: 20 LKRGPWTPEEDEVLARFVAREGCDRWRTLPRRAGLLRCGKSCRLRWMNYLRPDIKRCPIA 79
Query: 101 PDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDPRTHKPLN- 159
DEEDLILRLHRLLGNRWSLIAGR+PGRTDNEIKNYWN+HLSKKLI QGIDPRTHKPL
Sbjct: 80 DDEEDLILRLHRLLGNRWSLIAGRLPGRTDNEIKNYWNSHLSKKLIAQGIDPRTHKPLTA 139
Query: 160 --NPSSSSSIIPNTNHQTSTPFFAP 182
+ S++++ + T+++ + P P
Sbjct: 140 AADHSNAAAAVAATSYKKAVPAKPP 164
>UniRef100_Q40920 MYB-like transcriptional factor MBF1 [Picea mariana]
Length = 388
Score = 209 bits (532), Expect = 7e-53
Identities = 90/129 (69%), Positives = 109/129 (83%)
Query: 34 PCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPS 93
PCC+KVGL +G W+ EED +L +YI+ GEG WR+LPK+AGL RCGKSCRLRW+NYLRP
Sbjct: 5 PCCAKVGLNKGAWSAEEDSLLGKYIQTHGEGNWRSLPKKAGLRRCGKSCRLRWLNYLRPC 64
Query: 94 VKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDPR 153
+KRG I DEE+LI+R+H LLGNRWS+IAGR+PGRTDNEIKNYWNT+LSKKL +GIDP+
Sbjct: 65 IKRGNITADEEELIIRMHALLGNRWSIIAGRVPGRTDNEIKNYWNTNLSKKLAVRGIDPK 124
Query: 154 THKPLNNPS 162
THK + S
Sbjct: 125 THKKVTTDS 133
>UniRef100_Q9M2D9 Putative transcription factor [Arabidopsis thaliana]
Length = 299
Score = 208 bits (530), Expect = 1e-52
Identities = 89/126 (70%), Positives = 105/126 (82%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
TPCC K+GLK+GPWT EEDEVL +IKK G G WRTLPK AGLLRCGKSCRLRW NYLRP
Sbjct: 4 TPCCDKIGLKKGPWTPEEDEVLVAHIKKNGHGSWRTLPKLAGLLRCGKSCRLRWTNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG DEE L+++LH +LGNRW+ IA ++PGRTDNEIKN WNTHL K+L++ G+DP
Sbjct: 64 DIKRGPFTADEEKLVIQLHAILGNRWAAIAAQLPGRTDNEIKNLWNTHLKKRLLSMGLDP 123
Query: 153 RTHKPL 158
RTH+PL
Sbjct: 124 RTHEPL 129
>UniRef100_P81395 MYB-related protein 330 [Antirrhinum majus]
Length = 274
Score = 208 bits (529), Expect = 2e-52
Identities = 92/175 (52%), Positives = 125/175 (70%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L YI+ GEG WR+LPK AGLLRCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDQRLINYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +E+++I++LH LLGN+WSLIAG +PGRTDNEIKNYWNTH+ +KL+++GIDP
Sbjct: 64 DLKRGNFTEEEDEIIIKLHSLLGNKWSLIAGALPGRTDNEIKNYWNTHIKRKLVSRGIDP 123
Query: 153 RTHKPLNNPSSSSSIIPNTNHQTSTPFFAPSSSDHHHPITITNNEPDPSQTQQAQ 207
+TH+ LN+ +++++ P N+ +PS+S + T NN S T +
Sbjct: 124 QTHRSLNSATTTATATPTVNNSCLDFRTSPSNSKNICMPTTDNNNNSSSSTDDTK 178
>UniRef100_Q43436 MYB1 [Gossypium hirsutum]
Length = 294
Score = 207 bits (528), Expect = 2e-52
Identities = 100/216 (46%), Positives = 136/216 (62%), Gaps = 13/216 (6%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L YI+ GEG WR+LPK AGLLRCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDQRLINYIRVHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +E++LI++LH LLGN+WSLIAGR+PGRTDNEIKNYWNTH+ +KLI++GIDP
Sbjct: 64 DLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIKRKLISRGIDP 123
Query: 153 RTHKPLNNPSSSSSIIPNT--NHQTSTPFFAPSSSDHHHPITITNNE------PDPSQTQ 204
+TH+PLN ++++++ T + + S + SSS + + NE D +
Sbjct: 124 QTHRPLNQTANTNTVTAPTELDFRNSPTSVSKSSSIKNPSLDFNYNEFQFKSNTDSLEEP 183
Query: 205 QAQGCGNLVNDVSAMDNHHVLTNNNRGDYGDDNGNN 240
+ D + H + YG NG +
Sbjct: 184 NCTASSGMTTDEEQQEQLH-----KKQQYGPSNGQD 214
>UniRef100_Q6SL45 MYB6 [Tradescantia fluminensis]
Length = 268
Score = 207 bits (527), Expect = 3e-52
Identities = 92/144 (63%), Positives = 114/144 (78%), Gaps = 2/144 (1%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L YIK GEG WR+LPK AGLLRCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDQRLIAYIKVHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +E++LI++LH LLGN+WSLIAGR+PGRTDNEIKNYWNTH+ +KLI++GIDP
Sbjct: 64 DLKRGNFTEEEDELIIKLHALLGNKWSLIAGRLPGRTDNEIKNYWNTHIKRKLISRGIDP 123
Query: 153 RTHKPLNNPSSSSSIIPNTNHQTS 176
+TH+P+N + S I + NHQ S
Sbjct: 124 QTHRPIN--AGSQFTISSANHQGS 145
>UniRef100_Q8RUI9 Putative myb-related protein [Oryza sativa]
Length = 287
Score = 206 bits (525), Expect = 5e-52
Identities = 95/181 (52%), Positives = 126/181 (69%), Gaps = 2/181 (1%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L YIK GEG WR+LPK AGLLRCGKSCRLRWMNYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDQRLIAYIKAHGEGCWRSLPKAAGLLRCGKSCRLRWMNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG D+++LI++LH LLGN+WSLIAG++PGRTDNEIKNYWNTH+ +KL+++GIDP
Sbjct: 64 DLKRGNFTDDDDELIIKLHALLGNKWSLIAGQLPGRTDNEIKNYWNTHIKRKLLSRGIDP 123
Query: 153 RTHKPLNNPSS--SSSIIPNTNHQTSTPFFAPSSSDHHHPITITNNEPDPSQTQQAQGCG 210
+TH+P++ SS ++S + T + P AP+ H + + PS A+
Sbjct: 124 QTHRPVSAGSSAAAASGLTTTASTAAFPSLAPAPPPQQHRLHNPVHAAAPSNASFARSAA 183
Query: 211 N 211
+
Sbjct: 184 S 184
>UniRef100_Q8H271 Myb-like transcription factor 1 [Gossypium hirsutum]
Length = 294
Score = 206 bits (525), Expect = 5e-52
Identities = 92/159 (57%), Positives = 120/159 (74%), Gaps = 1/159 (0%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L YI+ GEG WR+LPK AGLLRCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDQRLINYIRVHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +E++LI++LH LLGN+WSLIAGR+PGRTDNEIKNYWNTH+ +KLI++GIDP
Sbjct: 64 DLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIKRKLISRGIDP 123
Query: 153 RTHKPLNNPSSSSSI-IPNTNHQTSTPFFAPSSSDHHHP 190
+TH+PLN ++++++ P +TP SS +P
Sbjct: 124 QTHRPLNQTANTNTVTAPTELDFRNTPTSVSKSSSIKNP 162
>UniRef100_Q9ATD5 GHMYB10 [Gossypium hirsutum]
Length = 302
Score = 205 bits (522), Expect = 1e-51
Identities = 104/192 (54%), Positives = 126/192 (65%), Gaps = 22/192 (11%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCCSK GL RG WT ED++L +YIK GEGRWR LPKRAGL RCGKSCRLRW+NYLRP
Sbjct: 4 SPCCSKEGLNRGAWTALEDKILKDYIKVHGEGRWRNLPKRAGLKRCGKSCRLRWLNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG I+PDEE+LI++LH+LLGNRWSLIAGR+PGRTDNEIKNYWNT+LSK++ ++ P
Sbjct: 64 DIKRGNISPDEEELIIKLHKLLGNRWSLIAGRLPGRTDNEIKNYWNTNLSKRVSDRQKSP 123
Query: 153 R--THKPL--------------NNPSSSSSIIPNTNHQTSTPFFAPSSSDHHHPITITNN 196
+ KP N SSS+ + T + F + HHH N
Sbjct: 124 AAPSKKPEAARRGTAGNGNTNGNGSGSSSTHVVRTRATRCSKVFI---NPHHH---TQNR 177
Query: 197 EPDPSQTQQAQG 208
P PS T G
Sbjct: 178 HPKPSSTCSNHG 189
>UniRef100_Q9SZP1 Transcription repressor MYB4 [Arabidopsis thaliana]
Length = 282
Score = 204 bits (520), Expect = 2e-51
Identities = 95/166 (57%), Positives = 120/166 (72%), Gaps = 6/166 (3%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EEDE L YIK GEG WR+LPK AGLLRCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDERLVAYIKAHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +E++LI++LH LLGN+WSLIAGR+PGRTDNEIKNYWNTH+ +KLIN+GIDP
Sbjct: 64 DLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKLINRGIDP 123
Query: 153 RTHKPLNNPSSS-----SSIIPNTNHQTSTPF-FAPSSSDHHHPIT 192
+H+P+ S+S + + P T++ + F AP H I+
Sbjct: 124 TSHRPIQESSASQDSKPTQLEPVTSNTINISFTSAPKVETFHESIS 169
>UniRef100_Q06944 MybHv5 [Hordeum vulgare]
Length = 251
Score = 204 bits (518), Expect = 3e-51
Identities = 98/195 (50%), Positives = 130/195 (66%), Gaps = 12/195 (6%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L YI+ GEG WR+LPK AGLLRCGKSCRLRWMNYLRP
Sbjct: 4 SPCCEKEHTNKGAWTKEEDQRLIAYIRANGEGCWRSLPKAAGLLRCGKSCRLRWMNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG DE++LI+RLH LLGN+WSLIAG++PGRTDNEIKNYWNTH+ +KL+++G+DP
Sbjct: 64 DLKRGNFTDDEDELIIRLHSLLGNKWSLIAGQLPGRTDNEIKNYWNTHIKRKLLSRGMDP 123
Query: 153 RTHKPLNNPSSSSSIIPNTNHQTSTP-------FFAPSSSDHHHPITITNNEPDPSQTQQ 205
TH+PL +++ P Q + P FA + P+ + +++ D S
Sbjct: 124 HTHRPLTAVIDAAA--PTRPAQIAVPARAAPTTMFALPTKQQQPPVPVESSDDDGSSGAT 181
Query: 206 AQG---CGNLVNDVS 217
+ G C +L D+S
Sbjct: 182 STGEPRCPDLNLDLS 196
>UniRef100_Q5TKI8 Putative myb protein [Oryza sativa]
Length = 260
Score = 203 bits (517), Expect = 4e-51
Identities = 92/157 (58%), Positives = 117/157 (73%), Gaps = 4/157 (2%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L YI+ GEG WR+LPK AGLLRCGKSCRLRWMNYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDQRLIAYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWMNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG DE++LI+RLH LLGN+WSLIAG++PGRTDNEIKNYWNTH+ +KL+ +GIDP
Sbjct: 64 DLKRGNFTDDEDELIIRLHSLLGNKWSLIAGQLPGRTDNEIKNYWNTHIKRKLLARGIDP 123
Query: 153 RTHKPL----NNPSSSSSIIPNTNHQTSTPFFAPSSS 185
+TH+PL + ++S+ P H S P A +++
Sbjct: 124 QTHRPLLSGGDGIAASNKAAPPPPHPISVPAKAAAAA 160
>UniRef100_Q84PP5 Transcription factor MYB101 [Lotus japonicus]
Length = 348
Score = 203 bits (516), Expect = 5e-51
Identities = 91/148 (61%), Positives = 110/148 (73%), Gaps = 4/148 (2%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
TPCC ++GLK+GPWT EED +L +YI+K G G WR LPK AGL RCGKSCRLRW NYLRP
Sbjct: 4 TPCCDEIGLKKGPWTPEEDAILVDYIQKHGHGSWRALPKLAGLNRCGKSCRLRWTNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG+ +EE LI+ LH +LGN+WS IAG +PGRTDNEIKN+WNTHL KKL+ G+DP
Sbjct: 64 DIKRGKFTEEEEQLIINLHAVLGNKWSAIAGHLPGRTDNEIKNFWNTHLKKKLLQMGLDP 123
Query: 153 RTHKP----LNNPSSSSSIIPNTNHQTS 176
TH+P LN S+ + TN TS
Sbjct: 124 VTHRPRLDHLNLLSNLQQFLAATNMVTS 151
>UniRef100_Q6SL50 MYB1 [Tradescantia fluminensis]
Length = 270
Score = 203 bits (516), Expect = 5e-51
Identities = 89/145 (61%), Positives = 115/145 (78%), Gaps = 2/145 (1%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L YIK GEG WR+LPK AGLLRCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDQRLIAYIKVHGEGCWRSLPKSAGLLRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +E+++I++LH LLGN+WSLIAGR+PGRTDNEIKNYWNTH+ +KLI++GIDP
Sbjct: 64 DLKRGNFTEEEDEVIIKLHALLGNKWSLIAGRLPGRTDNEIKNYWNTHIKRKLISRGIDP 123
Query: 153 RTHKPLNNPSSSSSIIPNTNHQTST 177
+TH+P+N S + I + N+Q ++
Sbjct: 124 QTHRPVN--SGAQFTISSANNQANS 146
>UniRef100_Q69JE8 Putative Myb-related protein Zm38 [Oryza sativa]
Length = 251
Score = 203 bits (516), Expect = 5e-51
Identities = 87/135 (64%), Positives = 111/135 (81%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L YIK GEG WR+LPK AGLLRCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDDRLIAYIKAHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +E++LI++LH LLGN+WSLIAGR+PGRTDNEIKNYWNTH+ +KL+++GIDP
Sbjct: 64 DLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKLLSRGIDP 123
Query: 153 RTHKPLNNPSSSSSI 167
TH+P+N+ +S+ +I
Sbjct: 124 VTHRPINDSASNITI 138
>UniRef100_O23889 OSMYB1 [Oryza sativa]
Length = 239
Score = 202 bits (515), Expect = 7e-51
Identities = 88/147 (59%), Positives = 111/147 (74%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L YI+ GEG WR+LPK AGLLRCGKSCRLRWMNYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDQRLIAYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWMNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG DE++LI+RLH LLGN+WSLIAG++PGRTDNEIKNYWNTH+ +KL+ +GIDP
Sbjct: 64 DLKRGNFTDDEDELIIRLHSLLGNKWSLIAGQLPGRTDNEIKNYWNTHIKRKLLARGIDP 123
Query: 153 RTHKPLNNPSSSSSIIPNTNHQTSTPF 179
+TH+PL + + +H+ P+
Sbjct: 124 QTHRPLLSGGDGIAASNKRHHRRRIPY 150
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.311 0.129 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 599,009,989
Number of Sequences: 2790947
Number of extensions: 28617977
Number of successful extensions: 142213
Number of sequences better than 10.0: 1753
Number of HSP's better than 10.0 without gapping: 1307
Number of HSP's successfully gapped in prelim test: 466
Number of HSP's that attempted gapping in prelim test: 122975
Number of HSP's gapped (non-prelim): 8752
length of query: 303
length of database: 848,049,833
effective HSP length: 127
effective length of query: 176
effective length of database: 493,599,564
effective search space: 86873523264
effective search space used: 86873523264
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146760.4


