

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146759.6 - phase: 0
(1825 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
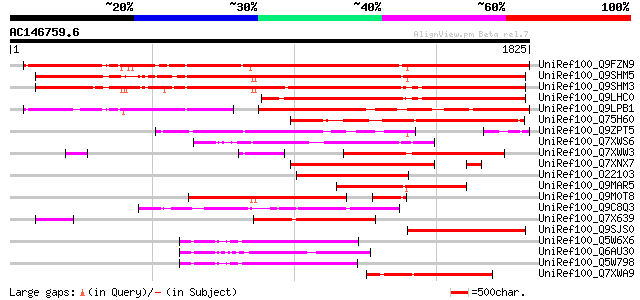
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FZN9 Retroelement pol polyprotein-like [Arabidopsis ... 1561 0.0
UniRef100_Q9SHM5 F7F22.15 [Arabidopsis thaliana] 1536 0.0
UniRef100_Q9SHM3 F7F22.17 [Arabidopsis thaliana] 1468 0.0
UniRef100_Q9LHC0 Retroelement pol polyprotein-like [Arabidopsis ... 1144 0.0
UniRef100_Q9LPB1 T32E20.9 [Arabidopsis thaliana] 1039 0.0
UniRef100_Q75H60 Putative reverse transcriptase [Oryza sativa] 803 0.0
UniRef100_Q9ZPT5 Hypothetical protein At2g14040 [Arabidopsis tha... 691 0.0
UniRef100_Q7XWS6 OSJNBa0091C12.6 protein [Oryza sativa] 682 0.0
UniRef100_Q7XWW3 OSJNBb0058J09.16 protein [Oryza sativa] 659 0.0
UniRef100_Q7XNX7 OSJNBb0026I12.3 protein [Oryza sativa] 639 0.0
UniRef100_O22103 Reverse transcriptase-like protein [Vicia faba] 605 e-171
UniRef100_Q9MAR5 F28H19.8 protein [Arabidopsis thaliana] 573 e-161
UniRef100_Q9M0T8 Putative athila transposon protein [Arabidopsis... 562 e-158
UniRef100_Q9C8Q3 Polyprotein, putative; 77260-80472 [Arabidopsis... 554 e-156
UniRef100_Q7X639 OSJNBb0067G11.5 protein [Oryza sativa] 533 e-149
UniRef100_Q9SJS0 Putative retroelement pol polyprotein [Arabidop... 526 e-147
UniRef100_Q5W6X6 Putative polyprotein [Oryza sativa] 494 e-138
UniRef100_Q6AU30 Putative reverse transcriptase [Oryza sativa] 493 e-137
UniRef100_Q5W798 Putative polyprotein [Oryza sativa] 491 e-137
UniRef100_Q7XWA9 OSJNBa0061A09.17 protein [Oryza sativa] 464 e-128
>UniRef100_Q9FZN9 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1864
Score = 1561 bits (4042), Expect = 0.0
Identities = 865/1879 (46%), Positives = 1167/1879 (62%), Gaps = 145/1879 (7%)
Query: 48 ETETTTPPPVITMEEGDPPPARPKLGDYGLANHRGRLTHVFRPANPVAFEIKASVQNGLK 107
+TET T V+ + P GD NH R V P FEIK+ + ++
Sbjct: 27 QTETDTMAAVV---DEQVQPNNIGAGD-APRNHNQRNGIVPPPVQNNNFEIKSGLIAMVQ 82
Query: 108 ERQFDGTGTISPHEHLSHFAETCEFCVPPANVNEDQKKLRLFPFTLIGKAKDWLLTLPNG 167
+F G P +HL F C V+ED KLRLFPF+L KA W +L G
Sbjct: 83 SNKFHGLPMEDPLDHLDEFDRLCSL-TKINGVSEDGFKLRLFPFSLGDKAHQWEKSLLQG 141
Query: 168 TIQTWEELELKFLEKYFPMSKYLDKKQEIASFRQGEGESLYDAWERFNLLLKRCPGHEFS 227
+I +W + + FL K+F S+ + +I+ F Q E+ +AWERF +CP H FS
Sbjct: 142 SITSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNNETFCEAWERFKGYQTQCPHHGFS 201
Query: 228 EKQYLQFFTEGLTHSNRMFLDASAGGSLRVKTDHEVQILIENMASNEYRAETKKKERGVF 287
+ L G+ RM LD ++ G+ K + L+EN+A ++ + +R V
Sbjct: 202 KASLLSTLYRGVLPKIRMLLDTASNGNFLNKDVEDGWELVENLAQSDGNYN-EDYDRSVR 260
Query: 288 GVSDTTSILANQA-AMNKQIE--ILTKEVHAYQLSNKQQAAAIRCDLCGEGHPNGECVPE 344
SD+ + AMN +++ +L ++ H + L + + +GE +
Sbjct: 261 TSSDSDEKHRREMKAMNDKLDKLLLVQQKHIHFLGDDETFQV----------QDGETMQ- 309
Query: 345 GASEEANYVNYQRQNPYSMN------KHPNLSYSNNNTLNP---LFPNPQQQQQPRK--- 392
SEE +YV Q Q Y+ HPNLSY + N NP ++P+ QQQ QPR
Sbjct: 310 --SEEVSYV--QNQGGYNKGFNNFKQNHPNLSYRSTNVANPQDQVYPS-QQQNQPRPFVP 364
Query: 393 ----------------------PPGIEETMMNFMKMT-QSNFEEMKK---------SQDL 420
PPG + T S+ + M + + DL
Sbjct: 365 YNQGQGYVPKQQYQGNYQPQLPPPGFTQQQQQPASTTPDSDLKNMLQQILQGQATGAMDL 424
Query: 421 ERKNNEASRKM-------------LETQLGQLAKQLAEQNKGGFSGNTQENPKN--ETCN 465
++ E K+ L +++ + Q F+G ++++ N E +
Sbjct: 425 SKRMAEIHNKVDCSYNDINIKVEALTSKIRYIEGQTGSTAAPKFTGPSRKSMSNSEEYAH 484
Query: 466 VIELRSKKVLAPLVPKVPNKVDESVVEVD--ENIEDEEVVEKESDQGVVENEKKKKTEGE 523
I LRS K L + PN+ E V+ D + ++ EK ++ +++
Sbjct: 485 AITLRSGKELP--TKESPNQNTEDSVDQDGEDFCQNGNSAEKAIEEPILD---------- 532
Query: 524 KSERLIDEDSILRKSKSQILKDGDEPQIIPSYVKLPYPHLAKKKKKEEGQFKKFMQL-FS 582
+ RL+ + K K D + P Y K P P + KK ++K ++
Sbjct: 533 QPTRLLAPAASPLVEKPAATKTKDNVFVPPPY-KPPLPFPGRFKKVMIQKYKALLEKQLK 591
Query: 583 QLQVNIPFGDALDQMPVYAKFMKEMLTGRRKPKDDENISLSENCSAILQRKL-PPKLKDP 641
L+V +P D L +P K++K+M+T R K + LS CSAI+Q+K+ P KL DP
Sbjct: 592 NLEVTMPLVDCLALIPDSNKYVKDMITERIKEVQGM-VVLSHECSAIIQQKIIPKKLGDP 650
Query: 642 GAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLGGGEPKPTRMTLTLADRSISYPFG 701
G+FT+PC++GP+ + LCDLGAS++LMPLS+ KKLG + KP ++L LADRS+ P G
Sbjct: 651 GSFTLPCALGPLAFNKCLCDLGASVSLMPLSVAKKLGFNKYKPCNISLILADRSVRIPHG 710
Query: 702 VLEDVLVKVNDLIFPADFVILDMAEDEDMPLILGRPFLATGRALIDVEMGQLMLRFHNE- 760
+LED+ V + + P DFV+L+M E+ PLILGRPFL T A+IDV+ G++ L +
Sbjct: 711 LLEDLPVMIGMVEVPTDFVVLEMDEEPKDPLILGRPFLVTVGAIIDVKKGKIDLNLGRDL 770
Query: 761 QVVFNIFEAMKHRAENPKCYRIDVIDEIVEDNSREHKPTHPLEETIVNSIENCELVEDLE 820
++ F+I MK + I+ +D + ++ E T L+ + + +L +LE
Sbjct: 771 KMTFDITNTMKKPTIERNIFWIEEMDMLADEMLEELGETDHLQSALTKDSKEGDL--NLE 828
Query: 821 VQECVKQLEANEQLLKPTKLEEMSN--------------EGKLGAKSLSNEEEKIPELKE 866
+ K L+ ++ + P + E++++ E L + S + +LK
Sbjct: 829 ILGYQKLLDEHKAVENPGEYEDLAHSVYSTELLDHNNPSEANLVSDDWSELKAPKVDLKP 888
Query: 867 LPSHLKYVFLSKDVSKPAIISSTLTPLEEEKLMRVLRENEGALGWKISDLKGISPAYCMH 926
LP L+YVFL + + P I++ LT + L+ L++ A+G+ + D+KGISP C H
Sbjct: 889 LPKGLRYVFLGLNSTYPVIVNDGLTADQVNLLITELKKYRKAIGYSLDDIKGISPTLCTH 948
Query: 927 RIHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGM 986
RIH+E E S ++PQRRLNP +KEVVKKE+LKLLDAG+IYPISDS+WVSPVH VPKKGGM
Sbjct: 949 RIHLENESYSSIEPQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHYVPKKGGM 1008
Query: 987 TVVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDG 1046
TVV N K+ELIPTRT TG RMCIDYR+LN A+RKDHFPLPF+DQM+ERLA +YCFLDG
Sbjct: 1009 TVVKNSKDELIPTRTTTGHRMCIDYRKLNAASRKDHFPLPFIDQMLERLANHPYYCFLDG 1068
Query: 1047 YSGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEV 1106
YSG+ QI + P DQEKT FTCP+G FAY+RMPFGLC APATFQRCM SIFSD+IE+ +EV
Sbjct: 1069 YSGFFQIPIHPNDQEKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEV 1128
Query: 1107 FMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVD 1166
FMDDFSV+G SF CL +L VLKRC ETNL+LNWEKCHFMV EGIVLGHKIS +GIEVD
Sbjct: 1129 FMDDFSVYGSSFSSCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGHKISEEGIEVD 1188
Query: 1167 QAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECL 1226
+AK++V+ +L PP VK +RSFLGHAGFYRRFIKDFSK+A+PL LL KETEF FD ECL
Sbjct: 1189 KAKVDVMMQLQPPKTVKDIRSFLGHAGFYRRFIKDFSKLARPLTRLLCKETEFAFDDECL 1248
Query: 1227 NAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNE 1286
AF LIK L+TAPI+ APNWD FE+MC+ASDYAVGAVLGQR +K H IYYAS+ +++
Sbjct: 1249 TAFKLIKEALITAPIVQAPNWDFPFEIMCNASDYAVGAVLGQRIDKKLHVIYYASRTMDD 1308
Query: 1287 SQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLL 1346
+QV Y+TTEKELLAV+FA EKFRSYL+GSKV V+TDHAAL+++ K D+KPRLLRW+LLL
Sbjct: 1309 AQVRYATTEKELLAVVFAFEKFRSYLVGSKVTVYTDHAALRHIYAKKDTKPRLLRWILLL 1368
Query: 1347 QEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAIRE---------- 1396
QEFD+EI DKKG+EN VADHLSR+ + E I P+EQL AI++
Sbjct: 1369 QEFDMEIVDKKGIENGVADHLSRMR----IEDEVPIDDSMPEEQLMAIQQLNESAQIRKS 1424
Query: 1397 ----------RPWFADMANFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGL 1446
PW+AD N+ P ++ +++KKFFKD NH+ WD+PYL+ + D +
Sbjct: 1425 LDQVCTIEEKLPWYADHVNYLVSGEEPPNLSSYEKKKFFKDINHFYWDEPYLYTLCKDKI 1484
Query: 1447 IRRCVAGEEIKNIVWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQ 1506
RRCV+ +EI+ I+ HCH SAYGGH + +T +K+LQ+GFWWP++FKD +F+ +CD+CQ
Sbjct: 1485 YRRCVSEDEIEGILLHCHGSAYGGHFATFKTVSKILQAGFWWPSMFKDAQEFISKCDSCQ 1544
Query: 1507 RTGSISKRNEMPLTGIIEVEPFDCWGIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCVA 1566
R G+IS+RNEMP I+EVE FD WGIDFMGPFP S +ILV VDYV+KWVEAI
Sbjct: 1545 RRGNISRRNEMPQNPILEVEIFDVWGIDFMGPFPSSYGNKYILVAVDYVSKWVEAIASPT 1604
Query: 1567 NDSKTVVNFLRKNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSG 1626
ND++ V+ + IF RFG PR++ISDGGKHF N E +LKK+ +KHKVATPYHPQTSG
Sbjct: 1605 NDARVVLKLFKTIIFPRFGVPRIMISDGGKHFINKVFENLLKKHGVKHKVATPYHPQTSG 1664
Query: 1627 QVEVSNRQLKQILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPV 1686
QVE+SNR++K ILEK V S+RKDWS KLDDALWAYR AFKT +G +P+ L++GK+CHLPV
Sbjct: 1665 QVEISNREIKAILEKIVGSTRKDWSAKLDDALWAYRTAFKTPIGTTPFNLLYGKSCHLPV 1724
Query: 1687 ELEHKAYWAIKALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRR 1746
ELE+KA WA+K LNFD A +KRL++LN+L E+RL AYE++ IYKERTK +HDK +V R
Sbjct: 1725 ELEYKAMWAVKLLNFDIKTAEEKRLIQLNDLNEIRLEAYESSKIYKERTKSFHDKKIVSR 1784
Query: 1747 EFYVGQLVLLFNSRLKLFPGKLKSKWSGPFMIESISPYGAVELSKPGEPGTFKVNAQRIK 1806
+F VG VLLFNSRL+LFPGKLKS+WSGPF + ++ PYGA+ L+ G+ G F VN QR+K
Sbjct: 1785 DFKVGDQVLLFNSRLRLFPGKLKSRWSGPFSVTAVRPYGAITLA--GKNGDFTVNGQRLK 1842
Query: 1807 PYLGGELPTNKGGVVLNDP 1825
Y+ + V L +P
Sbjct: 1843 KYMIDQFIPEGTSVPLEEP 1861
>UniRef100_Q9SHM5 F7F22.15 [Arabidopsis thaliana]
Length = 1862
Score = 1536 bits (3977), Expect = 0.0
Identities = 843/1790 (47%), Positives = 1120/1790 (62%), Gaps = 163/1790 (9%)
Query: 92 NPVAFEIKASVQNGLKERQFDGTGTISPHEHLSHFAETCEFCVPPANVNEDQKKLRLFPF 151
N A EI+ +F G P +HL F C V+ED KLRLFPF
Sbjct: 153 NRTAREIRKRRTTVNPGNKFHGLPMEDPLDHLDEFDRLCNL-TKINGVSEDGFKLRLFPF 211
Query: 152 TLIGKAKDWLLTLPNGTIQTWEELELKFLEKYFPMSKYLDKKQEIASFRQGEGESLYDAW 211
+L KA W LP+ +I TW++ + FL K+F ++ + EI+ F Q GES +AW
Sbjct: 212 SLGDKAHIWEKNLPHDSITTWDDCKKAFLSKFFSNARTARLRNEISGFSQKTGESFCEAW 271
Query: 212 ERFNLLLKRCPGHEFSEKQYLQFFTEGLTHSNRMFLDASAGGSLRVKTDHEVQILIENMA 271
ERF +C H F++ L G+ RM LD ++ G+ + K E L+EN+A
Sbjct: 272 ERFKGYTNQCSHHGFTKASLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVENLA 331
Query: 272 SNEYRAETKKKERGVFGVSDTTSILANQA-AMNKQIE--ILTKEVHAYQLSNKQQAAAIR 328
+ + +R V G +D+ + A+N +++ +L++ H + L + +Q
Sbjct: 332 QSNGNYN-ENCDRTVRGTADSDDKHRKEIKALNDKLDRILLSQHKHVHFLVDDEQYEVQD 390
Query: 329 CDLCGEGHPNGECVPEGASEEANYVN-----YQRQNPYSMNKHPNLSYSNNNTLNP---L 380
GEG+ EE +Y+N Y+ N + N +PNLSY + N NP +
Sbjct: 391 ----GEGNQ---------LEEVSYINNNQGGYKGYNNFKTN-NPNLSYRSTNVANPQDQV 436
Query: 381 FPNPQQQQQPRKPPGIEETMMNFMKMTQSNFEEMKKSQDLERKNNEASRKMLETQLGQLA 440
+P PQQQQ KP F+ Q+
Sbjct: 437 YP-PQQQQSQNKP---------FVPYNQAT------------------------------ 456
Query: 441 KQLAEQNKGGFSGNTQENPKNETCNVIELRSKKVLAPLVPKVPNKVDESVVEVDENIEDE 500
KQ ++ G +NPK E + I L S K L P + + V D +D
Sbjct: 457 KQTSQ-----LPGKAVQNPK-EYAHAITLHSGKAL-------PTREEPKTVTEDSEDQDG 503
Query: 501 EVVEKESDQGVVENEKKKKTEGEKSERLIDEDSILRKSKSQILKDGDEPQIIPSYVKLPY 560
E + + DQ +K E+ L + S L + P I + P
Sbjct: 504 EDLSLKKDQA----DKPLDLSLEQPLDLSLQQS-LDPPLDSFTRPTTRPVIPAASPTAPK 558
Query: 561 PHLAKKKKKEEGQFKKFMQLFSQLQVNIPFGDALDQMPVYAKFMKEMLTGRRKPKDDENI 620
P K K+K E + IP DAL +P KF+K+++ R + +
Sbjct: 559 PVAVKNKEKVE--------------LRIPLVDALALIPDSHKFLKDLIVERIQEVQGM-V 603
Query: 621 SLSENCSAILQRKL-PPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLGG 679
LS CSAI+Q+K+ P KL DPG+FT+PCS+GP+ R LCDLGAS++LMPLS+ K+LG
Sbjct: 604 VLSHECSAIIQKKIIPKKLSDPGSFTLPCSLGPLAFNRCLCDLGASVSLMPLSVAKRLGF 663
Query: 680 GEPKPTRMTLTLADRSISYPFGVLEDVLVKVNDLIFPADFVILDMAEDEDMPLILGRPFL 739
+ K ++L LADRS+ P G+LE++ +++ + P DFV+L+M E+ PLILGRPFL
Sbjct: 664 TQYKSCNISLILADRSVRIPHGLLENLPIRIGAVEIPTDFVVLEMDEEPKDPLILGRPFL 723
Query: 740 ATGRALIDVEMGQLMLRFHNE-QVVFNIFEAMKHRAENPKCYRIDVIDEIVEDNSREHKP 798
AT A+IDV+ G++ L + ++ F++ +AMK + + I+ +D++ ++ E
Sbjct: 724 ATAGAMIDVKKGKIDLNLGKDFRMTFDVKDAMKKPTIEGQLFWIEEMDQLADELLEELAE 783
Query: 799 THPLEETIVNS-------IENCELVEDLEVQECVKQLEANEQLLKP-TKLEEMSNEGKLG 850
L + S +E + L+ + +++ E E+L P T++ MS EG
Sbjct: 784 EDHLNSALTKSGEDGFLHLETLGYQKLLDSHKAMEESEPFEELNGPATEVMVMSEEGSTR 843
Query: 851 AK------------SLSNEEEKIP----------------ELKELPSHLKYVFLSKDVSK 882
+ +LS +E + P +LK LP L+Y FL + +
Sbjct: 844 VQPALSRTYSSNHSTLSTDEPREPIISTSDDWSELKAPKVDLKPLPKGLRYAFLGPNSTY 903
Query: 883 PAIISSTLTPLEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEYKSVVQPQR 942
P II++ L E L+ LR+ A+G+ +SD+KGISP+ C HRIH+E E S ++P R
Sbjct: 904 PVIINAELNSDEVNLLLSELRKYRRAIGYSLSDIKGISPSLCNHRIHLENESYSSIEPHR 963
Query: 943 RLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPTRTV 1002
RLNP +KEVVKKE+LKLLDAG+IYPISDS+WVSPVH VPKK GM VV NEK+ELIPTRT+
Sbjct: 964 RLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKDGMIVVKNEKDELIPTRTI 1023
Query: 1003 TGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEK 1062
TG RMCIDYR+LN A+RKDHFPLPF+DQM+ERLA +YCFLDGYSG+ QI + P DQEK
Sbjct: 1024 TGHRMCIDYRKLNAASRKDHFPLPFIDQMLERLANHPYYCFLDGYSGFFQIPIHPNDQEK 1083
Query: 1063 TAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCL 1122
T FTCP+G FAY+RMPFGLC APATFQRCM SIFSD+I++ +EVFMDDFSV+G SF CL
Sbjct: 1084 TTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIKEMVEVFMDDFSVYGPSFSSCL 1143
Query: 1123 FHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNV 1182
+L VL RC ETNL+LNWEKCHFMV EGIVLGHKIS KGIEVD+ K+EV+ +L PP V
Sbjct: 1144 LNLGRVLTRCEETNLVLNWEKCHFMVKEGIVLGHKISEKGIEVDKGKVEVMMQLQPPKTV 1203
Query: 1183 KGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLVTAPII 1242
K +RSFLGHAGFYRRFIKDFSKIA+PL LL KETEF FD +CL +F IK+ LV+AP++
Sbjct: 1204 KDIRSFLGHAGFYRRFIKDFSKIARPLTRLLCKETEFKFDDDCLKSFQTIKDALVSAPVV 1263
Query: 1243 IAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKELLAVI 1302
APNWD FE+MCDASDYAVGAVLGQ+ +K H IYYAS+ L+++Q Y+TTEKELLAV+
Sbjct: 1264 RAPNWDYPFEIMCDASDYAVGAVLGQKIDKKLHVIYYASRTLDDAQGRYATTEKELLAVV 1323
Query: 1303 FALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENV 1362
FA EKFRSYL+GSKV V+TDHAAL++L K D+KPRLLRW+LLLQEFD+EI DKKG+EN
Sbjct: 1324 FAFEKFRSYLVGSKVTVYTDHAALRHLYAKKDTKPRLLRWILLLQEFDMEIVDKKGIENG 1383
Query: 1363 VADHLSRLENNEVTKKEGAIMAEFPDEQLFAIR-------------------ERPWFADM 1403
ADHLSR+ E I P+EQL + E PW+AD
Sbjct: 1384 AADHLSRMRIEEPL----LIDDSMPEEQLMVVEFFGKSYSGKEFHQLNAVEGESPWYADH 1439
Query: 1404 ANFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKNIVWHC 1463
N+ A + P ++ ++RKKFF+D +HY WD+PYL+ + D + RRCV+ +E++ I+ HC
Sbjct: 1440 VNYLACGVEPPNLTSYERKKFFRDIHHYYWDEPYLYTLCKDKIYRRCVSEDEVEGILLHC 1499
Query: 1464 HSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEMPLTGII 1523
H SAYGGH + +T +K+LQ+GFWWPT+FKD +FV +CD+CQR G+IS+RNEMP I+
Sbjct: 1500 HGSAYGGHFATFKTVSKILQAGFWWPTMFKDAQEFVSKCDSCQRKGNISRRNEMPQNPIL 1559
Query: 1524 EVEPFDCWGIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVVNFLRKNIFTR 1583
EVE FD WGIDFMGPFP S +ILV VDYV+KWVEAI ND+K V+ + IF R
Sbjct: 1560 EVEIFDVWGIDFMGPFPSSYGNKYILVAVDYVSKWVEAIASPTNDAKVVLKLFKTIIFPR 1619
Query: 1584 FGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTV 1643
FG PRV+ISDGGKHF N E +LKK+ +KHKVATPY+PQTSGQVE+SNR++K ILEKTV
Sbjct: 1620 FGVPRVVISDGGKHFINKVFENLLKKHGVKHKVATPYNPQTSGQVEISNREIKTILEKTV 1679
Query: 1644 ASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAYWAIKALNFDQ 1703
+RKDWS KLDDALWAYR FKT +G +P+ L++GK+CHLPVELE+KA WA+K LNFD
Sbjct: 1680 GITRKDWSAKLDDALWAYRTTFKTPIGTTPFNLLYGKSCHLPVELEYKAMWAVKLLNFDI 1739
Query: 1704 TLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQLVLLFNSRLKL 1763
A +KRL++L++L+E+RL AYE++ IYKERTK +HDK ++ ++F VG VLLFNSRLKL
Sbjct: 1740 KTAEEKRLIQLSDLDEIRLEAYESSKIYKERTKLFHDKKIITKDFQVGDQVLLFNSRLKL 1799
Query: 1764 FPGKLKSKWSGPFMIESISPYGAVELSKPGEPGTFKVNAQRIKPYLGGEL 1813
FPGKLKS+WSGPF I + PYGAV L+ G+ G F VN QR+K YL ++
Sbjct: 1800 FPGKLKSRWSGPFCITEVRPYGAVTLT--GKSGDFTVNGQRLKKYLADQI 1847
>UniRef100_Q9SHM3 F7F22.17 [Arabidopsis thaliana]
Length = 1799
Score = 1468 bits (3800), Expect = 0.0
Identities = 832/1836 (45%), Positives = 1122/1836 (60%), Gaps = 183/1836 (9%)
Query: 92 NPVAFEIKASVQNGLKERQFDGTGTISPHEHLSHFAETCEFCVPPANVNEDQKKLRLFPF 151
N A EI+ +F G P +HL F C V+ED KLRLFPF
Sbjct: 18 NRTAREIRKRRTTVNPGNKFHGLPMEDPLDHLDEFDRLCNL-TKINGVSEDGFKLRLFPF 76
Query: 152 TLIGKAKDWLLTLPNGTIQTWEELELKFLEKYFPMSKYLDKKQEIASFRQGEGESLYDAW 211
+L KA W LP+ +I TW++ + FL K+F ++ + EI+ F Q GES +AW
Sbjct: 77 SLGDKAHIWEKNLPHDSITTWDDCKKVFLSKFFSNARTARLRNEISGFSQKTGESFCEAW 136
Query: 212 ERFNLLLKRCPGHEFSEKQYLQFFTEGLTHSNRMFLDASAGGSLRVKTDHEVQILIENMA 271
ERF +CP H F++ L G+ RM LD ++ G+ + K E L+EN+A
Sbjct: 137 ERFKGYTNQCPHHGFTKASLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVENLA 196
Query: 272 SNE--YRAETKKKERGVFGVSDTTSILANQAAMNKQIE--ILTKEVHAYQLSNKQQAAAI 327
++ Y + + RG D A+N +++ +L++ H + L + +Q
Sbjct: 197 QSDGNYNEDCDRTVRGTADSDDKHR--KEIKALNDKLDRILLSQHKHVHFLVDDEQYEVQ 254
Query: 328 RCDLCGEGHPNGECVPEGASEEANYVN-----YQRQNPYSMNKHPNLSYSNNNTLNP--- 379
GEG+ EE +Y+N Y+ N + N +PNLSY + N NP
Sbjct: 255 D----GEGNQ---------LEEVSYINNNQGGYKGYNYFKTN-NPNLSYRSTNVANPQDQ 300
Query: 380 LFPNPQQQQQPR---------------------KPPGIEETM--------MNFMKMTQS- 409
++P QQQ Q + +PPG + +M Q
Sbjct: 301 VYPPQQQQSQNKPFVPYNQGFVPKQQFQGNYQQQPPGFAPPQHQGPAAPDADMKQMLQQL 360
Query: 410 -------NFEEMKKSQDLERK----NNEASRKM--LETQL----GQLAKQLAEQNKGGFS 452
+ E KK +L K N+ + KM L+T++ G A +
Sbjct: 361 LHGQASCSMEMAKKISELHNKLDCSYNDLNVKMETLDTKVRYLEGHSTSSSATKQTSQLP 420
Query: 453 GNTQENPKNETCNVIELRSKKVLAPLVPKVPNKVDESVVEVDENIEDEEVVEKESDQGVV 512
G +NPK E + I LRS K L P + + V D +D E + E DQ
Sbjct: 421 GKAVQNPK-EYAHAITLRSGKAL-------PTREEPKTVTEDSEDQDGEDLSLEKDQADK 472
Query: 513 ENEKKKKTEGEKS-ERLIDE--DSILRKSKSQIL-------------KDGDEPQIIPSYV 556
+ + + S ++ +D DS R + ++ K+ ++ + P Y
Sbjct: 473 PLDLSLEQPLDLSLQQSLDPPLDSFTRPTTRPVIPAASPTAPKPVAVKNKEKVFVPPPYK 532
Query: 557 -KLPYPHLAKKKKKEEGQFKKFMQLFSQLQVNIPFGDALDQMPVYAKFMKEMLTGRRKPK 615
+LP+P KK ++ + F + ++++ IP DAL +P KF+K+++ R +
Sbjct: 533 SQLPFPGRHKKALADKYR-AMFAKNIKEVELRIPLVDALALIPDSHKFLKDLIVERIQEV 591
Query: 616 DDENISLSENCSAILQRKL-PPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMM 674
+ LS CSAI+Q+K+ P KL DPG+FT+PCS+GP+ R LCDLGAS++LMPLS+
Sbjct: 592 QGM-VVLSHECSAIIQKKIIPKKLSDPGSFTLPCSLGPLAFNRCLCDLGASVSLMPLSVA 650
Query: 675 KKLGGGEPKPTRMTLTLADRSISYPFGVLEDVLVKVNDLIFPADFVILDMAEDEDMPLIL 734
K+LG + K ++L LADRS+ P G+LE++ +++ + P DFV+L+M E+ PLIL
Sbjct: 651 KRLGFTQYKSCNISLILADRSVRIPHGLLENLPIRIGAVEIPTDFVVLEMDEEPKDPLIL 710
Query: 735 GRPFLATGRALIDVEMGQLMLRFHNE-QVVFNIFEAMKHRAENPKCYRIDVIDEIVEDNS 793
GRPFLAT A+IDV+ G++ L + ++ F++ +AMK + + I+ +D++ ++
Sbjct: 711 GRPFLATAGAMIDVKKGKIDLNLGKDFRMTFDVKDAMKKPTIEGQLFWIEEMDQLADELL 770
Query: 794 REHKPTHPLEETIVNS-------IENCELVEDLEVQECVKQLEANEQLLKP-TKLEEMSN 845
E L + S +E + L+ + +++ E E+L P T++ MS
Sbjct: 771 EELAEEDHLNSALTKSGEDGFLHLETLGYQKLLDSHKAMEESEPFEELNGPATEVMVMSE 830
Query: 846 EGKLGAK------------SLSNEEEKIP----------------ELKELPSHLKYVFLS 877
EG + +LS +E + P +LK LP L+Y FL
Sbjct: 831 EGSTRVQPALSRTYSSNHSTLSTDEPREPIIPTSDDWSELKAPKVDLKPLPKGLRYAFLG 890
Query: 878 KDVSKPAIISSTLTPLEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEYKSV 937
+ + P II++ L E L+ L++ A+G+ +SD+KGISP+ C HRIH+E E S
Sbjct: 891 PNSTYPVIINAELNSDEVNLLLSELKKYRRAIGYSLSDIKGISPSLCNHRIHLENESYSS 950
Query: 938 VQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELI 997
++PQRRLNP +KEVVKKE+LKLLDAG+IYPISD +H VPKK GMTVV NEK+ELI
Sbjct: 951 IEPQRRLNPNLKEVVKKEILKLLDAGVIYPISD------MHCVPKKDGMTVVKNEKDELI 1004
Query: 998 PTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAP 1057
PTRT+TG RMCIDYR+LN A+RKDHFPLPF+DQM+ERLA +YCFLDGY+G+ QI + P
Sbjct: 1005 PTRTITGHRMCIDYRKLNAASRKDHFPLPFIDQMLERLANHPYYCFLDGYNGFFQIPIHP 1064
Query: 1058 EDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKS 1117
DQEKT FTCP+G FAY+RMPFGLC APATFQRCM SIFSD+IE+ +EVFMDDFSV+G S
Sbjct: 1065 NDQEKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGPS 1124
Query: 1118 FDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLP 1177
F CL +L VL RC ETNL+LNWEKCHFMV EGIVLGHKIS KGIEVD+ K+EV+ +L
Sbjct: 1125 FSSCLLNLGRVLTRCEETNLVLNWEKCHFMVKEGIVLGHKISEKGIEVDKGKVEVMMQLQ 1184
Query: 1178 PPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLV 1237
PP VK +RSFLGHAGFYRRFIKDFSKIA+PL LL KETEF FD +CL +F IK+ LV
Sbjct: 1185 PPKTVKDIRSFLGHAGFYRRFIKDFSKIARPLTRLLCKETEFKFDDDCLKSFQTIKDALV 1244
Query: 1238 TAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKE 1297
+AP++ APNWD FE+MCDASDYAVGAVLGQ+ +K H IYYAS+ L+++Q Y+TTEKE
Sbjct: 1245 SAPVVRAPNWDYPFEIMCDASDYAVGAVLGQKIDKKLHVIYYASRTLDDAQGRYATTEKE 1304
Query: 1298 LLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKK 1357
LL V+FA EKFRSYL+GSKV V+TDHAAL++L K D+KPRLLRW+LLLQEFD+EI DKK
Sbjct: 1305 LLVVVFAFEKFRSYLVGSKVTVYTDHAALRHLYAKKDTKPRLLRWILLLQEFDMEIVDKK 1364
Query: 1358 GVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAIRERPWFADMANFKAGNIIPDDME 1417
G+EN ADHLSR+ E I P+EQL + +
Sbjct: 1365 GIENGAADHLSRMRIEEPL----LIDDSMPEEQLMVV-------------------EFFG 1401
Query: 1418 QHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKNIVWHCHSSAYGGHHSGERT 1477
+ K F N + P+ RCV+ +E++ I+ HCH SAYGGH + +T
Sbjct: 1402 KSYSGKEFHQLNAVEGESPW-----------RCVSEDEVEGILLHCHGSAYGGHFATFKT 1450
Query: 1478 AAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEMPLTGIIEVEPFDCWGIDFMG 1537
+K+LQ+GFWWPT+FKD +FV +CD+CQR G+IS+RNEMP I+EVE FD WGIDFMG
Sbjct: 1451 VSKILQAGFWWPTMFKDAQEFVSKCDSCQRKGNISRRNEMPQNPILEVEIFDVWGIDFMG 1510
Query: 1538 PFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVVNFLRKNIFTRFGTPRVLISDGGKH 1597
PFP S +ILV VDYV+KWVEAI ND+K V+ + IF RFG PRV+ISDGGKH
Sbjct: 1511 PFPSSYGNKYILVAVDYVSKWVEAIASPTNDAKVVLKLFKTIIFPRFGVPRVVISDGGKH 1570
Query: 1598 FCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSRKLDDA 1657
F N E +LKK+ +KHKVATPY+PQTSGQVE+SNR++K ILEKTV +RKDWS KLDDA
Sbjct: 1571 FINKVFENLLKKHGVKHKVATPYNPQTSGQVEISNREIKTILEKTVGITRKDWSAKLDDA 1630
Query: 1658 LWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAYWAIKALNFDQTLAGKKRLLKLNEL 1717
LWAYR AFKT +G +P+ L++GK+CHLPVELE+KA WA+K LNFD A +KRL++L++L
Sbjct: 1631 LWAYRTAFKTPIGTTPFNLLYGKSCHLPVELEYKAMWAVKLLNFDIKTAEEKRLIQLSDL 1690
Query: 1718 EEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQLVLLFNSRLKLFPGKLKSKWSGPFM 1777
+E+RL AYE++ IYKERTK +HDK ++ ++F G VLLFNSRLKLFPGKLKS+WSGPF
Sbjct: 1691 DEIRLEAYESSKIYKERTKLFHDKKIITKDFQFGVQVLLFNSRLKLFPGKLKSRWSGPFC 1750
Query: 1778 IESISPYGAVELSKPGEPGTFKVNAQRIKPYLGGEL 1813
I + PYGAV L+ G+ G F VN QR+K YL ++
Sbjct: 1751 ITEVRPYGAVTLA--GKSGDFTVNGQRLKKYLADQI 1784
>UniRef100_Q9LHC0 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 897
Score = 1144 bits (2960), Expect = 0.0
Identities = 558/929 (60%), Positives = 692/929 (74%), Gaps = 48/929 (5%)
Query: 885 IISSTLTPLEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEYKSVVQPQRRL 944
II++ L E + LR+ A+G+ +SD+KGISP+ C HRIH+E E S ++PQRRL
Sbjct: 2 IINAELNSDEVNLQLSELRKYRRAIGYSLSDIKGISPSLCNHRIHLENESYSSIEPQRRL 61
Query: 945 NPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPTRTVTG 1004
NP DAG+IYPISDS+WVS V+ VPKKGGMTVV NEK+ELIPTRT+TG
Sbjct: 62 NPNF------------DAGVIYPISDSTWVSLVYCVPKKGGMTVVKNEKDELIPTRTITG 109
Query: 1005 RRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTA 1064
RMCIDYR+LN A+RKDHFPLPF+DQM+ERLA +YCFLDGYSG+ QI + P DQEKT
Sbjct: 110 HRMCIDYRKLNAASRKDHFPLPFIDQMLERLANHPYYCFLDGYSGFFQIPIHPNDQEKTT 169
Query: 1065 FTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFH 1124
FTCP+G FAY+RMPFGLC APATFQRCM SIFSD+IE+ +EVFMDDFS +G SF CL +
Sbjct: 170 FTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSGYGPSFSSCLLN 229
Query: 1125 LNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKG 1184
L VL RC ETNL+LNWEKCHFMV EGIVLGHKIS KGIEVD+ K+EV+ +L PP VK
Sbjct: 230 LGRVLTRCEETNLVLNWEKCHFMVKEGIVLGHKISEKGIEVDKGKVEVMMQLQPPKTVKE 289
Query: 1185 VRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIA 1244
+RSFLGHAGFYRRFIKDFSKI +PL LL KETEF+FD +CL +F IK+ LV+AP++ A
Sbjct: 290 IRSFLGHAGFYRRFIKDFSKIVRPLTRLLCKETEFEFDEDCLKSFQTIKDALVSAPVVRA 349
Query: 1245 PNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFA 1304
PNWD FE+MCDASDY VGAVLGQ+ +K H IYYAS+ L+++Q Y+TTEKELLAV+FA
Sbjct: 350 PNWDYPFEIMCDASDYVVGAVLGQKIDKKLHVIYYASRTLDDAQGRYATTEKELLAVVFA 409
Query: 1305 LEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENVVA 1364
EKFRSYL+GSKV V+TDHAAL++L K D+KPRLLRW+LLLQEFD+EI DKKG+EN A
Sbjct: 410 FEKFRSYLVGSKVTVYTDHAALRHLYAKKDTKPRLLRWILLLQEFDMEIVDKKGIENGAA 469
Query: 1365 DHLSRLENNEVTKKEGAIMAEFPDEQLFAIRERPWFADMANFKAGNIIPDDMEQHQRKKF 1424
DHL R+ E + ++ P+EQL + + + K
Sbjct: 470 DHLLRMRIEEPLPIDDSM----PEEQLMVV-------------------EFFGKSYSGKE 506
Query: 1425 FKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKNIVWHCHSSAYGGHHSGERTAAKVLQS 1484
F N + P+ RCV+ +E++ I+ HCH SAYGGH + +T +K+LQ+
Sbjct: 507 FHQLNDVEGESPW-----------RCVSEDEVEGILLHCHCSAYGGHFATFKTVSKILQA 555
Query: 1485 GFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEMPLTGIIEVEPFDCWGIDFMGPFPPSSS 1544
GFWWPT+FKD +FV +CD+CQR +IS+ NEMP I+EVE FD WGIDFMG FP S
Sbjct: 556 GFWWPTMFKDAQEFVSKCDSCQRKDNISRINEMPQNPIVEVEIFDVWGIDFMGLFPSSYG 615
Query: 1545 YLHILVCVDYVTKWVEAIPCVANDSKTVVNFLRKNIFTRFGTPRVLISDGGKHFCNNFLE 1604
+ILV +DYV+KWVEAI ND+K V+ + IF RFG PRV+ISDGGKHF N E
Sbjct: 616 NKYILVAIDYVSKWVEAIAIPTNDAKVVLKLFKTIIFPRFGVPRVVISDGGKHFINKVFE 675
Query: 1605 TVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSRKLDDALWAYRIA 1664
+LKK+ +KHKVATPYHPQTSGQVE+S+R++K ILEKTV +RKDWS KLDDALWAYR A
Sbjct: 676 NLLKKHGVKHKVATPYHPQTSGQVEISDREIKTILEKTVGITRKDWSAKLDDALWAYRTA 735
Query: 1665 FKTHLGLSPYQLVFGKACHLPVELEHKAYWAIKALNFDQTLAGKKRLLKLNELEEMRLGA 1724
FKT +G +P+ +++GK+CHLPVELE+KA WA+K LNFD A +KRL++L++L+E+RL A
Sbjct: 736 FKTPIGTTPFNILYGKSCHLPVELEYKAMWAVKLLNFDIKTAEEKRLIQLSDLDEIRLEA 795
Query: 1725 YENAVIYKERTKRYHDKGLVRREFYVGQLVLLFNSRLKLFPGKLKSKWSGPFMIESISPY 1784
YE++ IYKERTK +HDK ++ ++F VG VLLFNSRLKLFPGKLKS+WSGPF I + PY
Sbjct: 796 YESSKIYKERTKLFHDKKIITKDFQVGDQVLLFNSRLKLFPGKLKSRWSGPFCITKVRPY 855
Query: 1785 GAVELSKPGEPGTFKVNAQRIKPYLGGEL 1813
GAV L+ G+ G F VN QR+K YL ++
Sbjct: 856 GAVTLA--GKSGDFTVNGQRLKKYLADQI 882
>UniRef100_Q9LPB1 T32E20.9 [Arabidopsis thaliana]
Length = 1586
Score = 1039 bits (2686), Expect = 0.0
Identities = 517/951 (54%), Positives = 653/951 (68%), Gaps = 115/951 (12%)
Query: 876 LSKDVSKPAIISSTLT-PLEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEY 934
L +D+ I++T+ P E + + + A+G+ + D+KGISP C HRIH+E E
Sbjct: 747 LGRDLKMTFDITNTMKKPTIEGNIFWIEEMDMKAIGYSLDDIKGISPTLCTHRIHLENES 806
Query: 935 KSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKN 994
S ++PQRRLNP +KEVVKKE+LKLLDAG+IYPISDS+WVSPVH VPKKGGMTVV N K+
Sbjct: 807 YSSIEPQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKD 866
Query: 995 ELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIA 1054
ELIPTRT+TG RMCI+YR+LN A+RK+HFPLPF+D M+ERLA +YCFLD YSG+ QI
Sbjct: 867 ELIPTRTITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIP 926
Query: 1055 VAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVF 1114
+ P DQ KT FTCP+G FAY+RMPFGLC APATFQRCM SIFSD+IE+ +EVFMDDFSV+
Sbjct: 927 IHPNDQGKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVY 986
Query: 1115 GKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIE 1174
G SF CL +L VLKRC ETNL+LNWEKCHFMV EGIVLG KIS +GIEVD+AKI+V+
Sbjct: 987 GSSFSSCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEGIEVDKAKIDVMM 1046
Query: 1175 KLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKN 1234
+L PP VK +RSFLGHAGFYR FIKDFSK+A+PL LL KETEF FD ECL AF LIK
Sbjct: 1047 QLQPPKTVKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEFAFDDECLTAFKLIKE 1106
Query: 1235 KLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTT 1294
L+TAPI+ APNWD FE++ ++++QV Y+TT
Sbjct: 1107 ALITAPIVQAPNWDFPFEII----------------------------TMDDAQVRYATT 1138
Query: 1295 EKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIR 1354
EKELLAV+FA EKFRSYL+GSKV ++TDHAAL+++ K D+KPRLLRW+LLLQEFD+EI
Sbjct: 1139 EKELLAVVFAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKPRLLRWILLLQEFDMEIV 1198
Query: 1355 DKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAIRERPWFADMANFKAGNIIPD 1414
DKKG+EN + PW+AD N+ P
Sbjct: 1199 DKKGIEN---------------------------------EKLPWYADHVNYLVSGEEPP 1225
Query: 1415 DMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKNIVWHCHSSAYGGHHSG 1474
++ +++KKFFKD NH+ WD+PYL+ + D + R CV+ +EI+ I+ HCH AYGGH +
Sbjct: 1226 NLSSYEKKKFFKDINHFYWDEPYLYTLCKDKIYRTCVSEDEIEGILLHCHGFAYGGHFAT 1285
Query: 1475 ERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEMPLTGIIEVEPFDCWGID 1534
+T +K+LQ+ EVE FD WGID
Sbjct: 1286 FKTMSKILQA---------------------------------------EVENFDVWGID 1306
Query: 1535 FMGPFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVVNFLRKNIFTRFGTPRVLISDG 1594
FMGPFP S +ILV +DYV+KWVEAI ND++ V+ + IF RFG PR++ISDG
Sbjct: 1307 FMGPFPSSYGNKYILVAIDYVSKWVEAIASHTNDARVVLKLFKTIIFPRFGVPRIVISDG 1366
Query: 1595 GKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSRKL 1654
GKHF N E +LKK+ +KHK VE+SNR++K ILEKTV S+RKDWS KL
Sbjct: 1367 GKHFINKGFENLLKKHGVKHK------------VEISNREIKAILEKTVGSTRKDWSAKL 1414
Query: 1655 DDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAYWAIKALNFDQTLAGKKRLLKL 1714
+D LWAYR AFKT +G +P+ L++GK+CHLPVELE+KA WA+K LNFD A +KRL++L
Sbjct: 1415 NDTLWAYRTAFKTPIGTTPFNLLYGKSCHLPVELEYKAMWAVKLLNFDIKTAEEKRLIQL 1474
Query: 1715 NELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQLVLLFNSRLKLFPGKLKSKWSG 1774
N+L ++RL AYE++ IYKERTK +HDK +V R+F VG VLLFNSRL+LFPGKLKS+WSG
Sbjct: 1475 NDLNKIRLEAYESSKIYKERTKSFHDKKIVSRDFKVGDQVLLFNSRLRLFPGKLKSRWSG 1534
Query: 1775 PFMIESISPYGAVELSKPGEPGTFKVNAQRIKPYLGGELPTNKGGVVLNDP 1825
PF + ++ PYGA+ L+ G+ G F VN QR+K Y+ + V L +P
Sbjct: 1535 PFSVTAVRPYGAITLA--GKNGDFTVNGQRLKKYMIDQFIPEGTSVPLEEP 1583
Score = 264 bits (674), Expect = 2e-68
Identities = 232/795 (29%), Positives = 369/795 (46%), Gaps = 100/795 (12%)
Query: 48 ETETTTPPPVITMEEGDPPPARPKLGDYGLANHRGRLTHVFRPANPVAFEIKASVQNGLK 107
+TET T V+ + P GD NH R V P FEIK+ + ++
Sbjct: 27 QTETDTMAAVV---DEQVQPNNIGAGD-APRNHNQRNGIVPPPVQNNNFEIKSGLIAMVQ 82
Query: 108 ERQFDGTGTISPHEHLSHFAETCEFCVPPANVNEDQKKLRLFPFTLIGKAKDWLLTLPNG 167
+F G P +HL F C V+ED KLRLFPF+L KA W +LP G
Sbjct: 83 SNKFHGLPMEDPLDHLDEFDRLCSL-TKINRVSEDGFKLRLFPFSLGDKAHQWEKSLPQG 141
Query: 168 TIQTWEELELKFLEKYFPMSKYLDKKQEIASFRQGEGESLYDAWERFNLLLKRCPGHEFS 227
+I +W + + FL K+F S+ + +I+ F Q E+ Y+AWERF +CP HE
Sbjct: 142 SITSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNNETFYEAWERFKGYQTQCPHHE-- 199
Query: 228 EKQYLQFFTEGLTHSNRMFLDASAGGSLRVKTDHEVQILIENMASNEYRAETKKKERGVF 287
M LD ++ G+ K + ++EN+A ++ + +R +
Sbjct: 200 -----------------MLLDTASNGNFLNKDVEDGWEVVENLAQSDGNYN-EDYDRSIR 241
Query: 288 GVSDTTSILANQA-AMNKQIE--ILTKEVHAYQLSNKQQAAAIRCDLCGEGHPNGECVPE 344
SD+ + AMN +++ +L ++ H + L + + +GE +
Sbjct: 242 TSSDSDEKHRREMKAMNDKLDKLLLMQQKHIHFLGDDETLQV----------QDGETLQ- 290
Query: 345 GASEEANYVNYQRQNPYSMN------KHPNLSYSNNNTLN---PLFPNPQQQQQPRKPPG 395
EE +YV Q Q Y+ HPNLSY + N N ++P+ QQQ QP+ P
Sbjct: 291 --LEEVSYV--QNQGGYNKGFNNFKQNHPNLSYRSTNVANRQDQVYPS-QQQNQPK--PF 343
Query: 396 I-----------EETMMNFMK-MTQSNFEEMKKSQDLERKNNEASRKMLETQLGQ----- 438
+ ++ N+ + + F + ++ L +++ + + GQ
Sbjct: 344 VPYNQGQGYVPKQQYQGNYQQQLPPPGFTQQQQQPALTTPDSDLKNMLQQILQGQATGAM 403
Query: 439 -LAKQLAE-QNKGGFSGNTQENPKNETCNVIELRSKKVLAPLVPKVPNKVDESVVEVDEN 496
L+K++AE NK S N + I + + PK +S+ E
Sbjct: 404 DLSKRMAEIHNKVDCSYNDINIKVEALTSKIRYIEGQTGSTAAPKFTGPSGKSMSNSKEY 463
Query: 497 IE------DEEVVEKESDQGVVENEKKKKTE-----GEKSERLIDEDSI------LRKSK 539
+E+ KES E+ + E G +E+ I+E + L +
Sbjct: 464 AHAITLRSGKELPTKESPNQNTEDSLDQDGEDFCQNGNSAEKAIEEPILHQPTRPLAPAA 523
Query: 540 SQILKDG-----DEPQIIPSYVKLPYPHLAKKKKKEEGQFKKFMQL-FSQLQVNIPFGDA 593
S +++ E IP K P P + KK ++K ++ L+V +P D
Sbjct: 524 SPLVEKPAAAKTKENVFIPPPYKPPLPFPGRFKKVMIQKYKALLEKQLKNLEVTMPLVDC 583
Query: 594 LDQMPVYAKFMKEMLTGRRKPKDDENISLSENCSAILQRKL-PPKLKDPGAFTIPCSIGP 652
L +P K++K+M+T R K + LS CSAI+Q+K+ P KL DPG+FT+PC++GP
Sbjct: 584 LALIPDSNKYVKDMITERIKEVQGM-VVLSHECSAIIQQKIIPKKLGDPGSFTLPCALGP 642
Query: 653 VDIGRALCDLGASINLMPLSMMKKLGGGEPKPTRMTLTLADRSISYPFGVLEDVLVKVND 712
+ + LCDLGAS++LMPL + KKLG + KP ++L LADRS+ G+LED+ V +
Sbjct: 643 LAFSKCLCDLGASVSLMPLPVAKKLGFNKYKPCNISLILADRSVRISHGLLEDLPVMIGV 702
Query: 713 LIFPADFVILDMAEDEDMPLILGRPFLATGRALIDVEMGQLMLRFHNE-QVVFNIFEAMK 771
+ P DFV+L+M E+ PLILGRPFLA RA+IDV+ G++ L + ++ F+I MK
Sbjct: 703 VEVPTDFVVLEMDEEPKDPLILGRPFLARARAIIDVKKGKIDLNLGRDLKMTFDITNTMK 762
Query: 772 HRAENPKCYRIDVID 786
+ I+ +D
Sbjct: 763 KPTIEGNIFWIEEMD 777
>UniRef100_Q75H60 Putative reverse transcriptase [Oryza sativa]
Length = 743
Score = 803 bits (2073), Expect = 0.0
Identities = 420/826 (50%), Positives = 534/826 (63%), Gaps = 106/826 (12%)
Query: 986 MTVVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLD 1045
MTVV N +NELIP RTVTG RMCIDYR+LN AT++DHFPLPF+D+M+ER+A +F+CFLD
Sbjct: 1 MTVVANAQNELIPQRTVTGWRMCIDYRKLNKATKEDHFPLPFIDEMLERMANHSFFCFLD 60
Query: 1046 GYSGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIE 1105
GYSGY+QI + PEDQ KT FTCP+G +AYRRM FGLC APA+FQRCM+SIFSDMIE
Sbjct: 61 GYSGYHQIPIHPEDQSKTTFTCPYGTYAYRRMSFGLCNAPASFQRCMMSIFSDMIE---- 116
Query: 1106 VFMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEV 1165
D VF F V G
Sbjct: 117 ---DIMEVFMDDFS---------------------------------VYG---------- 130
Query: 1166 DQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVEC 1225
+ +++ PP+N+KG+RSFLGHAGFYRRFIKDFS IA+PL NLL K+ F+ D C
Sbjct: 131 -----KTFDQIRPPVNIKGIRSFLGHAGFYRRFIKDFSTIARPLTNLLAKDAPFESDDAC 185
Query: 1226 LNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLN 1285
L +F ++K LV+APII P+W L FE+MCDASD+AVGA
Sbjct: 186 LKSFEVLKKALVSAPIIQPPDWTLSFEIMCDASDFAVGA--------------------- 224
Query: 1286 ESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLL 1345
FRSYL+ +KVI++TDHAALKY LTK D+KPRLLRW+LL
Sbjct: 225 ----------------------FRSYLVRAKVIIYTDHAALKYPLTKKDAKPRLLRWILL 262
Query: 1346 LQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAIRE-RPWFADMA 1404
LQEFDLEI+DKKGVEN VADHLSRL+ + E I D+ L +R+ PW+A++
Sbjct: 263 LQEFDLEIKDKKGVENSVADHLSRLQITNMP--EQPINDFLRDDMLMTVRDSNPWYANIV 320
Query: 1405 NFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKNIVWHCH 1464
N+ IP Q K ++ ++WD+PYL++V +DGL+RRCV EE I+ CH
Sbjct: 321 NYMVFKYIPPGKNQRNLKY---ESRRHIWDEPYLYRVCSDGLLRRCVPTEEGLKIIERCH 377
Query: 1465 SSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEMPLTGIIE 1524
+S YGGH+ RT AK+ QSGF+WPT+++D +FVRRC +CQR G I+ R+ MPLT ++
Sbjct: 378 ASPYGGHYGVFRTQAKIWQSGFFWPTMYEDSKEFVRRCTSCQRQGGITARDAMPLTYNLQ 437
Query: 1525 VEPFDCWGIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVVNFLRKNIFTRF 1584
VE FD WGIDFMGPFP S + +ILV VDYV+KWV+A+PC A D++ + IF +F
Sbjct: 438 VEIFDVWGIDFMGPFPKSRNCEYILVAVDYVSKWVKAMPCSATDARHAKKMFIEIIFPKF 497
Query: 1585 GTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVA 1644
TPR++ISDGG HF + +L++ KH VATPYHPQTSGQ E SN+Q+K IL+KTV
Sbjct: 498 DTPRMVISDGGSHFIDKTFRDLLREMGAKHNVATPYHPQTSGQAETSNKQIKNILQKTVN 557
Query: 1645 SSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAYWAIKALNFDQT 1704
W +L DAL AY IA+KT + +SPYQ+V+ K C LPVELEH+AYWAI+ N D
Sbjct: 558 KMGTGWKDRLPDALCAYWIAYKTPIRMSPYQIVYRKPCRLPVELEHRAYWAIRNWNMDFE 617
Query: 1705 LAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQLVLLFNSRLKLF 1764
AG+ R +++ ELEE R AY NA IYKERTKR+HDK ++F G VL+FNSR+KLF
Sbjct: 618 GAGEWRKMQIAELEEWREKAYHNAKIYKERTKRWHDKRKKIKKFKPGDKVLMFNSRVKLF 677
Query: 1765 -PGKLKSKWSGPFMIESISPYGAVELSKPGEPGTFKVNAQRIKPYL 1809
GKL+SKW GPF + S +GA+ L + FKVN QR+K +L
Sbjct: 678 GHGKLRSKWEGPFDVIDTSSHGAITL-RDELGNIFKVNGQRLKIFL 722
>UniRef100_Q9ZPT5 Hypothetical protein At2g14040 [Arabidopsis thaliana]
Length = 1048
Score = 691 bits (1784), Expect = 0.0
Identities = 400/948 (42%), Positives = 560/948 (58%), Gaps = 129/948 (13%)
Query: 511 VVENEKKKKTEGEKSERLIDEDSILRKSKSQILKDGDEPQIIPSYV-KLPYPHLAKKKKK 569
+++ EK K+ S+ L ++++ +S L++ + ++P YV KLP+P +K ++
Sbjct: 53 ILKKEKAKEKSKAASDDLEEDNT---GKESDTLEEKAKKTVLPPYVPKLPFPGRQRKIQR 109
Query: 570 EEGQFKKFMQLFSQLQVNIPFGDALDQMPVYAKFMKEMLTGRRKPKDDENISLSENCSAI 629
E+ ++ F ++ QLQV +PF D + + +Y K +K++LT +R ++ ++ +S CSAI
Sbjct: 110 EK-EYSLFDEIMRQLQVKLPFLDLVQNVSIYRKHLKDILTNKRT-LEEGHVLISHECSAI 167
Query: 630 LQRKLPPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLGGGEPKPTRMTL 689
LQ IG R LCDLGA ++LMP S+ K+LG PT+M+L
Sbjct: 168 LQN----------VALFSRMIGEYTFDRCLCDLGAGVSLMPFSVAKRLGDTNFTPTKMSL 217
Query: 690 TLADRSISYPFGVLEDVLVKVNDLIFPADFVILDMAEDEDMPLILGRPFLATGRALIDVE 749
L DRSIS+P GV EDV V+V + P DFVI+++ E+ LILGRPFL ALIDV
Sbjct: 218 VLGDRSISFPVGVAEDVQVRVGNFYIPTDFVIIELDEEPRHRLILGRPFLNIVAALIDVR 277
Query: 750 MGQLMLRFHNEQVVFNIFEAMKHRAENPKCYRIDVIDEIVEDNSREHKPTHPLEETIVNS 809
++ LR + FN+ M + + +D++DE+V + E PL+ +
Sbjct: 278 KSKINLRIGDIVQEFNMERIMSKPTTECQTFWVDIMDELVNELLAELNTEDPLQTVLTKE 337
Query: 810 IENCELVEDLEVQECVKQLEANEQLLKPTKLEEMSNEGKLGAKSLSNEEE-------KIP 862
+ + + + L+++ + K E+ + A +S+ ++ P
Sbjct: 338 ESEFGYLGEATTR-FARILDSSSPMTKVVAFAELGDNEVEKALVVSSPKDCDDWSELNAP 396
Query: 863 --ELKELPSHLKYVFLSKDVSKPAIISSTLTPLEEEKLMRVLRENEGALGWKISDLKGIS 920
ELK LP+ L+ FL + + II+ L +E L+ LR+ ALG+ + D+ GIS
Sbjct: 397 KMELKPLPARLRVAFLGPNSTYLVIINPELNNVESALLLCELRKYRKALGYSLDDITGIS 456
Query: 921 PAYCMHRIHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVV 980
P CMH IH+E E + V+ QRRLN +++VVKKE++KLLDAG+IYPISDS+WVSPVHVV
Sbjct: 457 PTLCMHMIHLEGESITSVEHQRRLNSNLRDVVKKEIMKLLDAGIIYPISDSTWVSPVHVV 516
Query: 981 PKKGGMTVVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAF 1040
PKKGG+ V+ NEKNELIPTRTVTG RMCIDYR+LN+ATRKD+FPL F+DQM+ERL+ Q +
Sbjct: 517 PKKGGVIVIKNEKNELIPTRTVTGHRMCIDYRKLNSATRKDNFPLSFIDQMLERLSNQPY 576
Query: 1041 YCFLDGYSGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMI 1100
YCFLDGY G+ QI + P+DQEKT FTCP+G FAYRRMPFGLC APATFQ CM IFSDMI
Sbjct: 577 YCFLDGYLGFFQILIHPDDQEKTTFTCPYGTFAYRRMPFGLCNAPATFQHCMKYIFSDMI 636
Query: 1101 EKNIEVFMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISS 1160
E DF ++RC + +L+LNWEK HFMV +GIVLGHKIS
Sbjct: 637 E--------DF-----------------MERCEDKHLVLNWEKSHFMVRDGIVLGHKISE 671
Query: 1161 KGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFD 1220
KG+EVD+AKIEV+ + PP +VK +E + +
Sbjct: 672 KGVEVDRAKIEVMMSMQPPNSVKDH-----------------------------EEQKSE 702
Query: 1221 FDVECLNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYA 1280
FD CL AF+++K LV+A I+ P W+L FE++CDASDY VGAVLGQRK+K HAIYYA
Sbjct: 703 FDDTCLQAFNIVKESLVSALIVQPPEWELPFEVICDASDYVVGAVLGQRKDKKLHAIYYA 762
Query: 1281 SKVLNESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLL 1340
S+ L+ G++VIV T HAAL+YLL+K D+KPRLL
Sbjct: 763 SRTLD----------------------------GAQVIVHTVHAALRYLLSKKDAKPRLL 794
Query: 1341 RWVLLLQEFDLEIRDKKGVENVVADHLSRLE-NNEVTKKE---GAIMAEFPD---EQLFA 1393
RW+LLLQ+FDLEI+DKKG+EN VAD+LSRL EV ++ G +A D +++
Sbjct: 795 RWILLLQDFDLEIKDKKGIENEVADNLSRLRVQEEVLMRDILPGENLASIEDCYMDEVGR 854
Query: 1394 IR--------------ERPWFADMANFKAGNIIPDDMEQHQRKKFFKD 1427
+R PW+AD AN+ + + P D + +KK K+
Sbjct: 855 LRVSTLELMTLHTGESNLPWYADFANYLSCEVPPPDFTGYFKKKLLKE 902
Score = 125 bits (314), Expect = 1e-26
Identities = 71/163 (43%), Positives = 95/163 (57%), Gaps = 33/163 (20%)
Query: 1665 FKTHLGLSPYQLVFGKACHLPVELEHKAYWAIKALNFDQTLAG-KKRLLKLNELEEMRLG 1723
+KT +G +P+ L++GK+CHLPVELE+KA WA K +N+D AG ++RLL+LNEL+E++
Sbjct: 914 YKTPVGTTPFNLIYGKSCHLPVELEYKALWATKLMNYDIKPAGERRRLLQLNELDEIQ-- 971
Query: 1724 AYENAVIYKERTKRYHDKGLVRREFYVGQLVLLFNSRLKLFPGKLKSKWSGPFMIESISP 1783
G VLLFNS+LK+FPGKL+SKWSGPF+I + P
Sbjct: 972 ---------------------------GDKVLLFNSKLKIFPGKLRSKWSGPFVIMEMRP 1004
Query: 1784 YG-AVELSKPGEPGTFKVNAQRIKPYLGGELPTNKGGVVLNDP 1825
G A+ G+P F N QR+KPYL V L DP
Sbjct: 1005 NGSAILWDNNGKP--FSTNGQRLKPYLAHSTLEEGTAVPLTDP 1045
>UniRef100_Q7XWS6 OSJNBa0091C12.6 protein [Oryza sativa]
Length = 781
Score = 682 bits (1761), Expect = 0.0
Identities = 384/859 (44%), Positives = 519/859 (59%), Gaps = 112/859 (13%)
Query: 645 TIPCSIGPVDIGRALCDLGASINLMPLSM-MKKLGGGEPKPTRMTLTLADRSISYPFGVL 703
TI C IG + RAL + N+M + LG +PT T ++ S + G++
Sbjct: 24 TIMCHIGGNSV-RALYNPSVGANIMSATFAFDCLGDRSLEPTVRTFRISTNSTTEGLGII 82
Query: 704 EDVLVKVNDLIFPADFVILDMAEDEDMPLILGRPFLATGRALIDV-EMGQLMLRFHNEQV 762
V + N + DF + ++ D +++G P L+DV E G L
Sbjct: 83 SGVPTRHNSVEVILDFHVFEIY---DFDILIGHPI----EKLLDVPETGVL--------- 126
Query: 763 VFNIFEAMKHRAENPKCYRIDVIDEIVEDNSREHKPTHPLEETIVNSIENCELVEDLEVQ 822
N K RI + P+ ++ + +E+ + E +E
Sbjct: 127 -------------NLKIGRIAT--------------SVPMLQSTNSLMESLPVPEPIEGV 159
Query: 823 ECVKQLEANEQLLKPTKLEEMSNEGKLGAKSLSNEEEKIP-----ELKELPSHLKYVFLS 877
+ E E + + E E + G +++ + +P ELK LPS L Y FL+
Sbjct: 160 MAISPFETLESIFDESIEEFNEGEDETG-ETMDLPKTDLPSRAPIELKPLPSSLSYAFLN 218
Query: 878 KDVSKPAIISSTLTPLEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEYKSV 937
D IIS L+ E +L+ +L ++ A G+ + DLKGISP C HRI +E
Sbjct: 219 SDAESLVIISDKLSEREMARLIAILEKHSAAFGYSLQDLKGISPTLCTHRILLEPSSTPS 278
Query: 938 VQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELI 997
+PQRRLN M+EV+KKEVLKLL G+IYP+ S WVSPV VVPKKGGMTVV N NELI
Sbjct: 279 REPQRRLNNAMREVIKKEVLKLLHTGIIYPVPYSEWVSPVQVVPKKGGMTVVENSNNELI 338
Query: 998 PTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAP 1057
P RTVTG RMCIDYR+LN AT+KDHF LPF+D+M+ERLA +F+CFLDGYS Y+Q
Sbjct: 339 PQRTVTGWRMCIDYRKLNKATKKDHFSLPFIDEMLERLANHSFFCFLDGYSSYHQ----- 393
Query: 1058 EDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKS 1117
I ++ DD ++GK+
Sbjct: 394 ----------------------------------------------IPIYPDD-QIYGKT 406
Query: 1118 FDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLP 1177
F+ CL +L+ VL+RC E +L+LNWEK HFMV EGIVLGH+IS +G+EVD+AKIEVI +LP
Sbjct: 407 FNHCLENLDKVLQRCQEKDLVLNWEKYHFMVREGIVLGHQISERGVEVDRAKIEVIRQLP 466
Query: 1178 PPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLV 1237
PP+N GVRSFLGHAGFYRRFIKDFSKIA+PL LL + DFD EC+ +F ++K L+
Sbjct: 467 PPVNDMGVRSFLGHAGFYRRFIKDFSKIARPLTALLANDVPVDFDDECMKSFKILKESLI 526
Query: 1238 TAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKE 1297
+APII +W L FE+MCDASD+AVGAVLGQ K++ HAI YASK L +Q+NY+TTEKE
Sbjct: 527 SAPIIQPSDWSLLFEIMCDASDFAVGAVLGQTKDRKHHAITYASKTLTGAQLNYATTEKE 586
Query: 1298 LLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKK 1357
LLAV+FA+EKFRSYL+G+KV+V+TDHAALKYLLTK D+KPR++RW+LLLQEFD+EI DKK
Sbjct: 587 LLAVVFAIEKFRSYLVGAKVVVYTDHAALKYLLTKKDAKPRVIRWILLLQEFDIEIMDKK 646
Query: 1358 GVENVVADHLSRLENNEVTKKEGAIMAEF--PDEQLFAIRERPWFADMANFKAGNIIPDD 1415
GVEN VADHLSR+ ++T + ++ ++ D +L I PW+A + NF A +P
Sbjct: 647 GVENYVADHLSRM---QITNMQKLLINDYLRDDMRLKVIDSDPWYATIVNFMATGHVP-- 701
Query: 1416 MEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKNIVWHCHSSAYGGHHSGE 1475
+K+ ++ +LWD PYLF+V +DGL+R CV+ +E I+ CH++ YGGH+
Sbjct: 702 -PVENKKRLIYESRRHLWDAPYLFRVCSDGLLRICVSTDEGIMIIEKCHAAPYGGHYGAF 760
Query: 1476 RTAAKVLQSGFWWPTLFKD 1494
RT AK+ QSGF+WP+++ D
Sbjct: 761 RTHAKIWQSGFFWPSMYDD 779
>UniRef100_Q7XWW3 OSJNBb0058J09.16 protein [Oryza sativa]
Length = 1045
Score = 659 bits (1699), Expect = 0.0
Identities = 324/566 (57%), Positives = 414/566 (72%), Gaps = 22/566 (3%)
Query: 1175 KLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKN 1234
+LPPP+N+KG+ SFLGHAGFYRRFIKDFS IA+PL NLL K+ F+F+ CL +F ++K
Sbjct: 500 RLPPPVNIKGICSFLGHAGFYRRFIKDFSTIARPLTNLLAKDAPFEFNDACLRSFKILKK 559
Query: 1235 KLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTT 1294
LV+APII P+W L FE+MCDASD+AVGAVLGQ K+K HAI YASK L +Q+NY+TT
Sbjct: 560 ALVSAPIIQPPDWTLPFEIMCDASDFAVGAVLGQTKDKKHHAICYASKTLTGAQLNYATT 619
Query: 1295 EKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIR 1354
EKELL V+FA++KFRSYL+G+KVI++TDHAALKYLLTK D+KPRLLRW+LLLQEFD+EI+
Sbjct: 620 EKELLTVVFAIDKFRSYLVGAKVIIYTDHAALKYLLTKKDAKPRLLRWILLLQEFDIEIK 679
Query: 1355 DKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAIRERPWFADMANFKAGNIIPD 1414
DKKGVEN VADHLSRL + +E I D+ L IP
Sbjct: 680 DKKGVENYVADHLSRLHITNM--QEQPINDLLRDDMLMTC-----------------IPP 720
Query: 1415 DMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKNIVWHCHSSAYGGHHSG 1474
++QRK ++ H +WD+PYL++V +DGL+RRCV +E I+ CH+S YGGH+
Sbjct: 721 G--ENQRKLKYESHRH-IWDEPYLYRVCSDGLLRRCVPTDEGLKIIERCHASPYGGHYGA 777
Query: 1475 ERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEMPLTGIIEVEPFDCWGID 1534
RT AK+ QSGF+WPT+++D +F+RRC +CQR G I+ R+ MPLT ++VE FD WGID
Sbjct: 778 FRTQAKIWQSGFFWPTMYEDSKEFIRRCTSCQRQGGITARDAMPLTYNLQVEIFDVWGID 837
Query: 1535 FMGPFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVVNFLRKNIFTRFGTPRVLISDG 1594
FMGPFP S + +ILV VDYV+KWVEA+PC A D++ + IF RFGT R++ISD
Sbjct: 838 FMGPFPKSRNCEYILVAVDYVSKWVEAMPCSAADARHAKKMFTETIFPRFGTRRMVISDR 897
Query: 1595 GKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSRKL 1654
G HF + +L++ KH VATPYHPQTSGQ E SN+Q+K IL+KTV +W +L
Sbjct: 898 GFHFIDKTFRDLLREMEAKHNVATPYHPQTSGQAETSNKQIKNILQKTVNKMGTEWKDRL 957
Query: 1655 DDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAYWAIKALNFDQTLAGKKRLLKL 1714
DALWAYR A+KT +G+SPYQ+V+GK+C LPVELEH+AYWAI+ N D AG+ R +++
Sbjct: 958 QDALWAYRTAYKTPIGMSPYQIVYGKSCRLPVELEHRAYWAIRNSNMDFEGAGEWRKMQI 1017
Query: 1715 NELEEMRLGAYENAVIYKERTKRYHD 1740
ELEE R AY NA IYKERTKR+HD
Sbjct: 1018 AELEEWREMAYHNAKIYKERTKRWHD 1043
Score = 98.6 bits (244), Expect = 2e-18
Identities = 64/164 (39%), Positives = 94/164 (57%), Gaps = 7/164 (4%)
Query: 805 TIVNSIENCELVEDLEVQECVKQLEANEQLLK---PTKLEEMSNEGKLGAKSLSNEEEKI 861
T+ + + ELVE++ V E+ E LL+ P ++E ++ G+ + +
Sbjct: 307 TLTDPLPEIELVEEVTA---VPPHESPEALLEDEVPDFIKEEADPGETLDLPIMEPPPQP 363
Query: 862 P-ELKELPSHLKYVFLSKDVSKPAIISSTLTPLEEEKLMRVLRENEGALGWKISDLKGIS 920
P ELK LP L+Y FL D P IIS L+ E ++L VL ++ LG+ + DL+GI+
Sbjct: 364 PLELKPLPLGLRYAFLHNDREAPVIISDKLSKDETQRLPTVLEKHRSVLGYSLQDLRGIN 423
Query: 921 PAYCMHRIHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGM 964
A C HRI ++ + +PQRRLN M+EVVKKEVLKLL AG+
Sbjct: 424 LALCTHRIPIDPKSTPSREPQRRLNNEMQEVVKKEVLKLLHAGI 467
Score = 50.8 bits (120), Expect = 4e-04
Identities = 27/79 (34%), Positives = 41/79 (51%)
Query: 195 EIASFRQGEGESLYDAWERFNLLLKRCPGHEFSEKQYLQFFTEGLTHSNRMFLDASAGGS 254
EI SF+Q + ES+ W RF L++ P E LQ+F GL + +LD + GGS
Sbjct: 47 EILSFKQIKNESIGATWSRFTNLVQSGPTLSLPEYVLLQYFHTGLDKESAFYLDITVGGS 106
Query: 255 LRVKTDHEVQILIENMASN 273
KT E + +++ + N
Sbjct: 107 FMHKTPSEGRTILDRILEN 125
>UniRef100_Q7XNX7 OSJNBb0026I12.3 protein [Oryza sativa]
Length = 589
Score = 639 bits (1647), Expect = 0.0
Identities = 312/508 (61%), Positives = 400/508 (78%), Gaps = 7/508 (1%)
Query: 986 MTVVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLD 1045
MTVV N NELIP RTVTG RMCIDY++LN AT+KD+FPLP +D+M+ERLA + +CFLD
Sbjct: 1 MTVVANVNNELIPQRTVTGWRMCIDYQKLNKATKKDNFPLPVIDEMLERLANHSLFCFLD 60
Query: 1046 GYSGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIE 1105
GYSGY+QI + P+DQ KT FTCP+G +AYRRM FGLC APA+FQRCM+SIFSDMIE +E
Sbjct: 61 GYSGYHQIPIHPKDQCKTTFTCPYGTYAYRRMSFGLCNAPASFQRCMMSIFSDMIEAIME 120
Query: 1106 VFMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEV 1165
VFMDDFS++GK+FD CL +L+ V +RC E +L+LNWEKCHFMV EGIVLGH++S +GIEV
Sbjct: 121 VFMDDFSLYGKTFDHCLQNLDKVFQRCQEKDLVLNWEKCHFMVHEGIVLGHRVSERGIEV 180
Query: 1166 DQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVEC 1225
D+AKIEVI++LPPP+N+KG+R+FLGHAGF+RRFIK+FS IA+PL NLL K+ F+FD C
Sbjct: 181 DRAKIEVIDQLPPPVNIKGIRNFLGHAGFHRRFIKEFSTIARPLTNLLAKDAPFEFDDVC 240
Query: 1226 LNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLN 1285
L +F +K LV+A II P+ L FE+MCDASD+ VGAVLGQ K+K HAI YAS+ L
Sbjct: 241 LKSFKTLKKALVSALIIQPPDGMLPFEIMCDASDFIVGAVLGQTKDKKLHAICYASETLA 300
Query: 1286 ESQVNYSTTEKELLAVIFALEKF-RSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVL 1344
+Q NY+TTEKELLAV+FA++KF RS L+G+KVIV+T+HAALKYLLTK D+KPRLLRW+
Sbjct: 301 GAQSNYATTEKELLAVVFAIDKFRRSDLVGAKVIVYTEHAALKYLLTKKDAKPRLLRWIP 360
Query: 1345 LLQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAIRER-PWFADM 1403
LLQEFDLEI+DKKGVEN VADHLSRL+ + +E I D+ L A+R PW+A++
Sbjct: 361 LLQEFDLEIKDKKGVENSVADHLSRLQLTNM--QEPPINYFLQDDMLMAVRNSDPWYANI 418
Query: 1404 ANFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKNIVWHC 1463
N+ +P ++QRK ++ H +WD+PYL++V ++GL+RRCV EE I+ C
Sbjct: 419 VNYMVSKYVPQG--ENQRKLKYESHCH-IWDEPYLYRVCSNGLLRRCVPKEEGFKIIERC 475
Query: 1464 HSSAYGGHHSGERTAAKVLQSGFWWPTL 1491
H++ Y GH+ RT AK+ QS F+WPT+
Sbjct: 476 HAAPYRGHYGAFRTQAKIWQSRFFWPTM 503
Score = 74.3 bits (181), Expect = 3e-11
Identities = 32/54 (59%), Positives = 40/54 (73%)
Query: 1606 VLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSRKLDDALW 1659
+L+K N KH +ATPYHPQT+GQ E+SN+Q+K IL+KTV W KL DALW
Sbjct: 508 LLRKMNAKHNIATPYHPQTNGQAEMSNKQIKNILQKTVHEMGTGWKDKLPDALW 561
>UniRef100_O22103 Reverse transcriptase-like protein [Vicia faba]
Length = 407
Score = 605 bits (1560), Expect = e-171
Identities = 288/394 (73%), Positives = 332/394 (84%)
Query: 1007 MCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFT 1066
+CIDYRRLN ATRKDHFPLPF+DQM+ERLA +YCFLDGYSGYNQIAV PEDQEKT FT
Sbjct: 1 VCIDYRRLNLATRKDHFPLPFIDQMLERLADHEYYCFLDGYSGYNQIAVVPEDQEKTTFT 60
Query: 1067 CPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLN 1126
CPFG+F+YRR+PFGLC APATFQRCM SIF+DM+EK +EVFMDDFSVFGKSFD CL +L
Sbjct: 61 CPFGIFSYRRIPFGLCNAPATFQRCMQSIFADMLEKYMEVFMDDFSVFGKSFDNCLSNLA 120
Query: 1127 AVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGVR 1186
VL+RC E+NLILNWEKCHFMV EGIVLGHKIS KGIEVDQAKIEVI KL PP N KG+R
Sbjct: 121 LVLERCQESNLILNWEKCHFMVREGIVLGHKISYKGIEVDQAKIEVISKLHPPTNEKGIR 180
Query: 1187 SFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIAPN 1246
SFLGH GFYRRFI+DFSKI KPL LLVK+ +F FD EC+ AF +K KLV+API++AP+
Sbjct: 181 SFLGHVGFYRRFIRDFSKIEKPLTTLLVKDKDFVFDKECVVAFETLKGKLVSAPIVVAPD 240
Query: 1247 WDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFALE 1306
W L FE+MCDASD AVGAVLGQR+ K H IYYAS VLN +Q+NY+TTEKELL +++A +
Sbjct: 241 WYLPFEIMCDASDIAVGAVLGQRREKRLHVIYYASHVLNPAQMNYATTEKELLVMVYAFD 300
Query: 1307 KFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENVVADH 1366
KFR Y++GSKVIV+TDHAALKYL +K DSKPRLLRW+LLLQEFD+EIRDK+G EN VADH
Sbjct: 301 KFRQYMLGSKVIVYTDHAALKYLFSKQDSKPRLLRWILLLQEFDVEIRDKRGCENTVADH 360
Query: 1367 LSRLENNEVTKKEGAIMAEFPDEQLFAIRERPWF 1400
LSR+ + E TK + I EF DE + + PWF
Sbjct: 361 LSRMSHIEETKVKQPIKDEFADEHILVVIGVPWF 394
>UniRef100_Q9MAR5 F28H19.8 protein [Arabidopsis thaliana]
Length = 640
Score = 573 bits (1478), Expect = e-161
Identities = 278/473 (58%), Positives = 343/473 (71%), Gaps = 22/473 (4%)
Query: 1150 EGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPL 1209
EGIVLGHKIS KGIEVD+AKI+V+ +L P VK +RSFLGHAGFYRRFIKDFSKIA+PL
Sbjct: 170 EGIVLGHKISGKGIEVDKAKIDVMVRLQPLKTVKDIRSFLGHAGFYRRFIKDFSKIARPL 229
Query: 1210 CNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQR 1269
LL KE EFDFD +CL AF IK LV API+ APNW+ FE+MCDASDYAVGAVLGQR
Sbjct: 230 TRLLCKEAEFDFDEDCLKAFHSIKEALVLAPIVQAPNWNHPFEIMCDASDYAVGAVLGQR 289
Query: 1270 KNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYL 1329
+K H IYYAS+ L+++Q Y+TTEKELL V+FA EKFRSYL+GSKV V+TDHAALK++
Sbjct: 290 IDKKLHVIYYASRTLDDTQSRYATTEKELLVVVFAFEKFRSYLVGSKVTVYTDHAALKHI 349
Query: 1330 LTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDE 1389
K D+KPRLLRW+L LQEFD+EI DK+G EN VADHLSR+ E + ++ P+E
Sbjct: 350 YAKKDTKPRLLRWILFLQEFDMEIVDKRGSENGVADHLSRMRMTEAVPIDDSM----PEE 405
Query: 1390 QLFAI-----------------RERPWFADMANFKAGNIIPDDMEQHQRKKFFKDANHYL 1432
QL A+ + PW+ D+ N+ I+P ++ +Q+KKFF+D NHY
Sbjct: 406 QLLAVGTSFDKFKIETMCATRSGKLPWYVDLVNYLTAGIVPSELSSYQKKKFFRDINHYY 465
Query: 1433 WDDPYLFKVSTDGLIRRCVAGEEIKNIVWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLF 1492
WD+P L+K DGL RRC+A EE++ ++ CH+S YGGH + +T KVLQ+G WWP++F
Sbjct: 466 WDEPVLYKKGVDGLFRRCIAEEEVQGVLELCHNSPYGGHFATFKTVQKVLQAGLWWPSMF 525
Query: 1493 KDCHDFVRRCDNCQRTGSISKRNEMPLTGIIEVEPFDCWGIDFMGPFPPSS-SYLHILVC 1551
KD H+ + RCD CQR G+IS+RNEMP I+EVE FD WGIDFMGPF P+S +ILV
Sbjct: 526 KDAHEHITRCDRCQRVGNISRRNEMPQNPILEVEVFDVWGIDFMGPFEPASYGNKYILVA 585
Query: 1552 VDYVTKWVEAIPCVANDSKTVVNFLRKNIFTRFGTPRVLISDGGKHFCNNFLE 1604
VDYV+KWVEAI NDSK V+ + IF RFG PRV+ISDGG HF N E
Sbjct: 586 VDYVSKWVEAIASPTNDSKVVLKLFKTIIFPRFGIPRVVISDGGSHFINKVFE 638
>UniRef100_Q9M0T8 Putative athila transposon protein [Arabidopsis thaliana]
Length = 724
Score = 562 bits (1449), Expect = e-158
Identities = 298/596 (50%), Positives = 395/596 (66%), Gaps = 38/596 (6%)
Query: 628 AILQRKL-PPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLGGGEPKPTR 686
AI Q+K+ P KL DPG+FT+PCS+GP+ R LCDLGA ++LMPLS+ K+LG + K
Sbjct: 3 AITQKKIVPKKLIDPGSFTLPCSLGPLAFKRCLCDLGALVSLMPLSVAKRLGFTQYKSCN 62
Query: 687 MTLTLADRSISYPFGVLEDVLVKVNDLIFPADFVILDMAEDEDMPLILGRPFLATGRALI 746
++L LADRS+ P + E++ +++ + P DFV+L+M E+ PLILGRPFLAT A+
Sbjct: 63 ISLILADRSVRIPHSLFENLPIRIGAVDIPTDFVVLEMDEEPKDPLILGRPFLATAGAMN 122
Query: 747 DVEMGQLMLRFHNE-QVVFNIFEAMKHRAENPKCYRIDVIDEIVEDNSREHKPTHPLEET 805
DV+ G++ L ++ F++ +AMK + + I+ ID++ ++ E L
Sbjct: 123 DVKKGKIDLNLGKYCRMTFDVKDAMKKPTIKGQLFWIEEIDQLADELLEERAEEDHLYSA 182
Query: 806 IVNS-------IENCELVEDLEVQECVKQLEANEQLLKP-TKLEEMSNEGKL-------- 849
+ +E + L+ + +++ E E+L P T++ MS EG
Sbjct: 183 LTKRGEDGFLHLETLGYQKLLDSHKAMEESEPFEELNGPETEVMVMSEEGSTQVQPAHSR 242
Query: 850 ------GAKSLSNEEEKI--------------PELKELPSHLKYVFLSKDVSKPAIISST 889
++SN E I +LK LP L+YVF + + P II+
Sbjct: 243 TYSTNNSTSTISNSGELIIPTSDDWSELKAPKVDLKSLPKGLRYVFFGPNSTYPVIINVE 302
Query: 890 LTPLEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEYKSVVQPQRRLNPTMK 949
L E L+ LR+ + A+G+ +SD+K ISP+ C HRIH+E E S ++PQRRLN +K
Sbjct: 303 LNDNEVNLLLSELRKYKRAIGYSLSDIKRISPSLCNHRIHLENESYSSIEPQRRLNLNLK 362
Query: 950 EVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPTRTVTGRRMCI 1009
EVVKKE+LKLLDAG+IYPISDS+WV PVH VPKKGGMTVV NEK+ELIPTRT+TG R+CI
Sbjct: 363 EVVKKEILKLLDAGVIYPISDSTWVFPVHCVPKKGGMTVVKNEKDELIPTRTITGHRVCI 422
Query: 1010 DYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFTCPF 1069
DYR+LN A+RKDHFPLPF +QM+E LA + CFLDGYSG+ QI + P DQEKT FTCP+
Sbjct: 423 DYRKLNAASRKDHFPLPFTNQMLEGLANHLYNCFLDGYSGFFQIPIHPNDQEKTTFTCPY 482
Query: 1070 GVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLNAVL 1129
G FAY+RMPFGLC AP TFQRCM SIFSD+IEK +EVFMDDFSV+G SF CL +L VL
Sbjct: 483 GTFAYKRMPFGLCNAPTTFQRCMTSIFSDLIEKMVEVFMDDFSVYGPSFSSCLLNLGRVL 542
Query: 1130 KRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGV 1185
+ ETNL+LNWEKC+FMV EGIVLGHKIS KGIEVD+ KI+V+ +L PP VK +
Sbjct: 543 TKWEETNLVLNWEKCYFMVKEGIVLGHKISEKGIEVDKEKIKVMMQLQPPKTVKDI 598
Score = 135 bits (340), Expect = 1e-29
Identities = 69/118 (58%), Positives = 88/118 (74%), Gaps = 4/118 (3%)
Query: 1277 IYYASKVLNESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSK 1336
IYYAS+ L+E+Q Y+TTEKELL V+FA +KFRSYL+ SKV V+TDHAAL+++ K D+K
Sbjct: 598 IYYASRTLDEAQGRYATTEKELLVVVFAFKKFRSYLVESKVTVYTDHAALRHMYAKKDTK 657
Query: 1337 PRLLRWVLLLQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAI 1394
PRLLR +LLLQEFD+EI +KKG+EN ADHLSR+ E I P+EQL +
Sbjct: 658 PRLLRGILLLQEFDMEIVEKKGIENGAADHLSRMRIEEPI----LIDDSMPEEQLMVV 711
>UniRef100_Q9C8Q3 Polyprotein, putative; 77260-80472 [Arabidopsis thaliana]
Length = 884
Score = 554 bits (1428), Expect = e-156
Identities = 352/924 (38%), Positives = 510/924 (55%), Gaps = 137/924 (14%)
Query: 454 NTQENPKNETCNVIELRSKKVLAPLVPKVPNKVDESVVEVDENIEDEEVVEKESDQG-VV 512
+T + P +T + I+ + A P K ES++E ++++ + + G ++
Sbjct: 70 STHQKPVQQTTSFIQNHASSTQAHPPPDT-TKTIESMLEQILKGQEKQTTDFDHKTGCII 128
Query: 513 ENEKKKKTEGEKSERLIDEDSILRKSKSQILKDGDEPQIIPSYVKLPYPHLAKKKKKEEG 572
+ + + E+ ++ ID R+S +++ K K+P+P + +K K+E
Sbjct: 129 RRCEWEDSNIEQLDQAIDYKQ--RRSAAKVTKP-----------KVPFPK-SPRKSKQEL 174
Query: 573 QFKKFMQLFSQLQVNIPFGDALDQMPVYAKFMKEMLTGRRKPKDDENISLSENCSAILQR 632
+ + +L V +P DA+ P+ +F+K ++T + + +++S S I+Q
Sbjct: 175 DDARCKVMMDKLIVEMPLIDAVKSSPMIRQFVKRIVT-KDMLTETVVMTMSTQVSDIIQN 233
Query: 633 KLPPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLGGGEPKPTRMTLTLA 692
K+P KL D + +PT++TL LA
Sbjct: 234 KIPQKLPDR---------------------------------------DLEPTQITLVLA 254
Query: 693 DRSISYPFGVLEDVLVKVNDLIFPADFVILDMAEDEDMPLILGRPFLATGRALIDVEMGQ 752
DRS+ G++ DV V+V P D V+L ++ PLILGRPFL T A+IDV G
Sbjct: 255 DRSVRRSDGIICDVPVQVGTSYIPTDLVVLSYEKEPKDPLILGRPFLVTTGAIIDVRKGI 314
Query: 753 LMLRFHNEQVVFNIFEAMKHRAENPKCYRIDVIDEIVEDNSREHKPTHPLEETIVNSIEN 812
+ L + + F+I + +K + K + +D I + ++ + PL+ ++ SI+
Sbjct: 315 IGLNVGDLTMQFDINKVVKKPTIDGKTFYLDTISFLADEFLMKMTLAVPLKHALIPSIDK 374
Query: 813 CE--LVEDLEVQECVKQLEANEQLLKPTKLEEMSNEGKLGAKSLSNEEEKIPELKELPSH 870
+ L+ E V QL A E+L+ + S +G SL EK P++
Sbjct: 375 VPKGYGKLLDRIEHVMQLVAQEELIGTS-----SQAATIGDWSL----EKAPKV------ 419
Query: 871 LKYVFLSKDVSKPAIISSTLTPLEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHM 930
DLK + P +
Sbjct: 420 --------------------------------------------DLKPLPPGLRYAFLGE 435
Query: 931 EAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSP---VHVVPKKGGMT 987
+ Y +V LN ++ ++ K A + Y + D + +SP +H + K
Sbjct: 436 NSTYHVIVNAS--LNKVELTLLLSKLRKYRKA-LGYSLDDITGISPDLCMHRIHLKDESK 492
Query: 988 VVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGY 1047
V + L P ++ + ++LN ATRK+HFPL F+DQ++ERL+ +YC LDGY
Sbjct: 493 PSVEHRRRLNPNLKDAVKKEIM--KKLNAATRKNHFPLQFIDQLLERLSNHKYYCVLDGY 550
Query: 1048 SGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVF 1107
SG+ QI + P+DQEKT FTCP+G FAY RMPFGLC APA F+RCM+SIF+DMIE IEVF
Sbjct: 551 SGFFQIPIHPDDQEKTMFTCPYGTFAYSRMPFGLCNAPAIFERCMMSIFTDMIENFIEVF 610
Query: 1108 MDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQ 1167
MDDFSV+G SF+ CL +L VL RC E NL+LNWEKCHFMV EGIVLGHK+S GIEV++
Sbjct: 611 MDDFSVYGSSFEACLENLRKVLARCEEKNLVLNWEKCHFMVQEGIVLGHKVSGAGIEVNK 670
Query: 1168 AKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLN 1227
AKIEV+ L +V RF+KDFSKIA+PL LL K+ +FDF+ EC +
Sbjct: 671 AKIEVMTSLQALDSVN------------LRFVKDFSKIARPLTALLCKDVKFDFNSECHD 718
Query: 1228 AFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNES 1287
AF+ IKN LV+API+ +W+L FE+MCDA DYAVGAVLGQRK HAI+YAS+ L+++
Sbjct: 719 AFNQIKNALVSAPIVQPLDWNLPFEIMCDAGDYAVGAVLGQRKGNKLHAIHYASRTLDDA 778
Query: 1288 QVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQ 1347
Q N +TTEKELLAV+FA +KFRSYL+GSKVIV TDH+ALKYL+ K D+KPRLLRW+LLLQ
Sbjct: 779 QRNNATTEKELLAVVFAFDKFRSYLVGSKVIVHTDHSALKYLMQKKDAKPRLLRWILLLQ 838
Query: 1348 EFDLEIRDKKGVENVVADHLSRLE 1371
EFD+E+RD+KG+EN VADHLSR++
Sbjct: 839 EFDIEVRDRKGIENGVADHLSRIK 862
>UniRef100_Q7X639 OSJNBb0067G11.5 protein [Oryza sativa]
Length = 791
Score = 533 bits (1372), Expect = e-149
Identities = 261/429 (60%), Positives = 323/429 (74%), Gaps = 15/429 (3%)
Query: 858 EEKIPEL--KELPSHLKYVFLSKDVSKPAIISSTLTPLEEEKLMRVLRENEGALGWKISD 915
E+++P+ +E L+Y FL D P IIS L+ E + L+ VL ++ LG+ + D
Sbjct: 373 EDEVPDFIKEEADPGLRYDFLHNDREAPVIISDKLSEDETQHLLTVLEKHRSILGYSLQD 432
Query: 916 LKGISPAYCMHRIHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVS 975
L+GI+PA C HRI ++ E +PQR LN M+EVVKKEVLKLL AG+IYPI S WVS
Sbjct: 433 LRGINPALCTHRILIDPESTPSREPQRWLNNAMREVVKKEVLKLLHAGIIYPIPYSEWVS 492
Query: 976 PVHVVPKKGGMTVVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERL 1035
PV VVPKKGGMTVV RMCIDYR+LN AT+KDHFPLPF+D+M+ERL
Sbjct: 493 PVQVVPKKGGMTVVYRW-------------RMCIDYRKLNKATKKDHFPLPFIDEMLERL 539
Query: 1036 AGQAFYCFLDGYSGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSI 1095
A +F+CFLDGYSGY+QI + PEDQ KT FTCP+G +AYRRM FGLC PA+FQRCM+SI
Sbjct: 540 ANHSFFCFLDGYSGYHQIPIHPEDQSKTTFTCPYGTYAYRRMSFGLCNTPASFQRCMMSI 599
Query: 1096 FSDMIEKNIEVFMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLG 1155
FSDMIE +EVFMDDFSV+GK+ CL +L+ VL+RC E +L+LN EKCHFMV EGIVLG
Sbjct: 600 FSDMIEDIMEVFMDDFSVYGKTLGHCLQNLDKVLQRCQEKDLVLNREKCHFMVREGIVLG 659
Query: 1156 HKISSKGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVK 1215
H++S +G+EVD+AKI+VI++LPPPMN+KG+ SF GHAGFYRRFIKDFS IA+PL NLL K
Sbjct: 660 HRVSERGVEVDRAKIDVIDQLPPPMNIKGIHSFFGHAGFYRRFIKDFSTIARPLTNLLAK 719
Query: 1216 ETEFDFDVECLNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFH 1275
+ +FD CL +F ++K LV+APII P+W L FE+MCDASD+AVGAVLGQ K+K H
Sbjct: 720 DAPIEFDDVCLKSFEILKKALVSAPIIQPPDWTLPFEIMCDASDFAVGAVLGQTKDKKHH 779
Query: 1276 AIYYASKVL 1284
AI YASK L
Sbjct: 780 AICYASKTL 788
Score = 73.9 bits (180), Expect = 4e-11
Identities = 39/133 (29%), Positives = 68/133 (50%), Gaps = 1/133 (0%)
Query: 90 PANPVAFEIKASVQNGLKERQFDGTGTISPHEHLSHFAETCEFCVPPANVNEDQKKLRLF 149
P +EI+ + + +E F G +P+ HL F + C C+ + ++ +LF
Sbjct: 24 PIYTTGYEIRPELISMFRENPFSGFDLENPYHHLRDFEQVCS-CLKIRGMRQETVWWKLF 82
Query: 150 PFTLIGKAKDWLLTLPNGTIQTWEELELKFLEKYFPMSKYLDKKQEIASFRQGEGESLYD 209
PF+L +AK W + +WE+L +F +FP+++ + EI +F+Q E ES+
Sbjct: 83 PFSLQERAKQWYTSTVGCVNGSWEKLRDRFCLTFFPVTRITALRVEILNFKQIENESIGA 142
Query: 210 AWERFNLLLKRCP 222
AW RF L++ P
Sbjct: 143 AWSRFTNLVQSGP 155
>UniRef100_Q9SJS0 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 466
Score = 526 bits (1356), Expect = e-147
Identities = 243/416 (58%), Positives = 315/416 (75%), Gaps = 2/416 (0%)
Query: 1398 PWFADMANFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIK 1457
PW+AD N+ A I P ++ ++RK FF+D +HY WD+PYL+ + D + R V+ +E++
Sbjct: 38 PWYADHVNYLACGIEPPNLTSYERKNFFRDIHHYYWDEPYLYTLCKDKIYRSYVSEDEVE 97
Query: 1458 NIVWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEM 1517
I+ HCH SAYGGH + +T +K+LQ+GFWWPT+FKD +FV +CD+CQR G+IS+RNEM
Sbjct: 98 GILLHCHDSAYGGHFATFKTVSKILQAGFWWPTMFKDAQEFVSKCDSCQRKGNISRRNEM 157
Query: 1518 PLTGIIEVEPFDCWGIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVVNFLR 1577
P I+EV+ FD WGIDFMGPFP S +ILV VDYV+KWVEAI ND+K V+ +
Sbjct: 158 PQNPILEVDIFDVWGIDFMGPFPSSYGNKYILVVVDYVSKWVEAIASPTNDAKVVLKLFK 217
Query: 1578 KNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQ 1637
IF RFG V+ISDGGKHF N E +LKK+ +KHKVATPYHPQTSGQVE+SNR++K
Sbjct: 218 TIIFPRFGVSWVVISDGGKHFINKVFENLLKKHGVKHKVATPYHPQTSGQVEISNREIKT 277
Query: 1638 ILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAYWAIK 1697
ILEKTV +RKDWS KLDDALWAY+ AFKT +G +P+ L+ K+CHL VELE+KA WA+K
Sbjct: 278 ILEKTVGITRKDWSTKLDDALWAYKTAFKTPIGTTPFNLLCVKSCHLHVELEYKAMWAVK 337
Query: 1698 ALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQLVLLF 1757
LNFD A +KRL++L+EL+E+RL AYE++ IYKERTK +HDK ++ ++F VG VLLF
Sbjct: 338 LLNFDIKTAEEKRLIQLSELDEIRLEAYESSKIYKERTKLFHDKKIITKDFQVGDQVLLF 397
Query: 1758 NSRLKLFPGKLKSKWSGPFMIESISPYGAVELSKPGEPGTFKVNAQRIKPYLGGEL 1813
NSRLK+FPGKLKS+WSGPF I + PYGAV L+ G+ G F VN QR+K YL ++
Sbjct: 398 NSRLKIFPGKLKSRWSGPFCITEVRPYGAVTLA--GKSGVFTVNGQRLKKYLANQI 451
>UniRef100_Q5W6X6 Putative polyprotein [Oryza sativa]
Length = 963
Score = 494 bits (1273), Expect = e-138
Identities = 287/634 (45%), Positives = 381/634 (59%), Gaps = 54/634 (8%)
Query: 598 PVYAKFMKEMLTGRRKPKDDENISLSENCSAILQRKLPPKLKDPGAFTIPCSIGPVDIGR 657
P+ +F++E + DE + +E S +++ P++ IPC + D+
Sbjct: 375 PMELRFLRETIRELTTIMSDEWLREAELSSEVIRINTAPRI-------IPCHLEGNDVS- 426
Query: 658 ALCDLGASINLMPLSM-MKKLGGGEPKPTRMTLTLADRSISYPFGVLEDVLVKVNDLIFP 716
L NL+ S L PT +I FG+++DV V D
Sbjct: 427 ILYSPTVGANLVSGSFAFAYLSDKAVTPTNKFFKHPSGNIIKRFGIMQDVPVCFEDKEAI 486
Query: 717 ADFVILDMAEDEDMPLILGRPFLATGRALIDVEMGQLMLRFHNEQVVFNIFEAMKHRAEN 776
DF + ++ +D +++G P EQ++ N R ++
Sbjct: 487 LDFHVFEI---QDFDILIGLPI---------------------EQLLINT-----PRLDS 517
Query: 777 PKCYRIDVIDEIVEDNSREHKPTHPLEETIVNSIENCELVEDLEVQECVKQLEANEQLLK 836
K + I +R T PL E +E E V E+ E LL+
Sbjct: 518 LKITLGETEFSIPFSRARIVL-TDPLPE-----------IESTEEVIAVPPHESPEALLE 565
Query: 837 ---PTKLEEMSNEGKLGAKSLSNEEEKIP-ELKELPSHLKYVFLSKDVSKPAIISSTLTP 892
P ++E ++ G+ + P ELK LP L Y FL D P IIS L
Sbjct: 566 DEVPDFIKEEADPGETLDLPIMEPPPWPPLELKPLPPSLHYAFLHHDREAPVIISDKLFE 625
Query: 893 LEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEYKSVVQPQRRLNPTMKEVV 952
E ++L+ VL ++ LG+ + DLKGI+P C+HRI ++ E +PQRRLN M+EVV
Sbjct: 626 DETQRLLAVLEKHCSVLGYSLQDLKGINPVLCIHRIPIDPESTPSREPQRRLNNAMREVV 685
Query: 953 KKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPTRTVTGRRMCIDYR 1012
KKEVLKLL AG+IYP+ S WVSPV VVPKKGG+ VV N +NELIP RTVT RMCIDYR
Sbjct: 686 KKEVLKLLHAGIIYPVPYSEWVSPVQVVPKKGGIMVVANAQNELIPQRTVTEWRMCIDYR 745
Query: 1013 RLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFTCPFGVF 1072
+LN AT+KDHFPLPF+D+M+ERLA +F+CFLDGYSGY+QI + PEDQ KT FTCP+G +
Sbjct: 746 KLNKATKKDHFPLPFIDEMLERLANHSFFCFLDGYSGYHQIPIHPEDQSKTTFTCPYGTY 805
Query: 1073 AYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLNAVLKRC 1132
AYRRM FGLC APA+FQRCM+SIFSDMIE +EVFMDDF V+GK+F L +L+ VL+RC
Sbjct: 806 AYRRMSFGLCNAPASFQRCMMSIFSDMIEYIMEVFMDDFLVYGKTFGHYLQNLDKVLQRC 865
Query: 1133 TETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHA 1192
E +L+LNWEKCHFMV EGIVLGH++S +GIEVD+AKI+VI++LPPP+N+KG+RSFLGHA
Sbjct: 866 QEKDLVLNWEKCHFMVREGIVLGHRVSERGIEVDRAKIDVIDQLPPPVNIKGIRSFLGHA 925
Query: 1193 GFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECL 1226
FYRRFI+D+S I +PL NLL K+ F+FD CL
Sbjct: 926 SFYRRFIEDYSTIVRPLTNLLAKDAPFEFDDACL 959
>UniRef100_Q6AU30 Putative reverse transcriptase [Oryza sativa]
Length = 1017
Score = 493 bits (1270), Expect = e-137
Identities = 296/672 (44%), Positives = 389/672 (57%), Gaps = 78/672 (11%)
Query: 598 PVYAKFMKEMLTGRRKPKDDENISLSENCSAILQRKLPPKLKDPGAFTIPCSIGPVDIGR 657
P+ F++E + DE +E S +++ P++ IPC + D+G
Sbjct: 422 PMELGFLRETVRELTSIMSDEWQREAELSSEVIRINTAPRI-------IPCHLEGNDVG- 473
Query: 658 ALCDLGASINLMPLSM-MKKLGGGEPKPTRMTLTLADRSISYPFGVLEDVLVKVNDLIFP 716
L NL+ +S L PT +R+I FG+++DV V D
Sbjct: 474 ILYSPTIGANLISVSFAFAYLSDKAVTPTNKFFKHPNRNIIEGFGIVQDVPVFFEDREAI 533
Query: 717 ADFVILDMAEDEDMPLILGRPFLATGRALIDVE-MGQLMLRFHNEQVVFNIFEAMKHRAE 775
DF + ++ +D +++G P + LI+ +G L + N F RA
Sbjct: 534 LDFHVFEI---QDFDILIGLPI---EQLLINTSRLGSLKITLGE-----NEFSIPFSRA- 581
Query: 776 NPKCYRIDVIDEIVEDNSREHKPTHPLEETIVNSIENCELVEDLEVQECVKQLEANEQLL 835
RI + D + E S E E T V E+ + + + EV + +K
Sbjct: 582 -----RITLTDPLPETESAE-------EVTAVPPHESTKALLEDEVPDFIK--------- 620
Query: 836 KPTKLEEMSNEGKLGAKSLSNEEEKIPELKELPSHLKYVFLSKDVSKPAIISSTLTPLEE 895
EE L + ELK LP L+Y FL D P IIS L+ E
Sbjct: 621 -----EEADPSETLDLPIMEPPPRPPLELKPLPPGLRYAFLHNDREAPVIISDKLSEDET 675
Query: 896 EKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEYKSVVQPQRRLNPTMKEVVKKE 955
++L+ VL ++ LG+ + DL+GI+PA C HRI ++ E +PQRRLN M+EVVKKE
Sbjct: 676 QRLLTVLEKHRSILGYSLQDLRGINPALCTHRIPIDLESTPSREPQRRLNNAMREVVKKE 735
Query: 956 VLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPTRTVTGRRMCIDYRRLN 1015
VLKLL AG+IYP+ S WVS V V+PKKGGMTVV N +NELIP RTVTG RMCIDYR+LN
Sbjct: 736 VLKLLHAGIIYPVPYSEWVSKVQVMPKKGGMTVVANSQNELIPQRTVTGWRMCIDYRKLN 795
Query: 1016 TATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFTCPFGVFAYR 1075
AT+KDHF LPF D+M+E LA +F+CFLDG
Sbjct: 796 KATKKDHFLLPFNDEMLEWLANHSFFCFLDG----------------------------- 826
Query: 1076 RMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLNAVLKRCTET 1135
M FGLC APA FQRCM+SIFSDMIE +EVFMDDFSV+GK+F CL +L+ VL+RC E
Sbjct: 827 -MSFGLCNAPAPFQRCMMSIFSDMIEDIMEVFMDDFSVYGKTFGHCLQNLDKVLQRCQEK 885
Query: 1136 NLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHAGFY 1195
+L+LNWEKCHFMV EGI+LGH++S +G EVD+AKI+VI++LPP +N+KG+ SFLGHAGFY
Sbjct: 886 DLVLNWEKCHFMVREGIILGHRVSERGSEVDRAKIDVIDQLPPLVNIKGIHSFLGHAGFY 945
Query: 1196 RRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIAPNWDLHFELMC 1255
RRFIKDFS IA+PL NLL K+ F+FD CL +F ++K LV APII P+W L FE+MC
Sbjct: 946 RRFIKDFSIIARPLTNLLAKDAPFEFDDACLKSFEILKKALVFAPIIQPPDWTLPFEIMC 1005
Query: 1256 DASDYAVGAVLG 1267
DASD+AV AVLG
Sbjct: 1006 DASDFAVAAVLG 1017
Score = 42.7 bits (99), Expect = 0.10
Identities = 24/71 (33%), Positives = 35/71 (48%)
Query: 203 EGESLYDAWERFNLLLKRCPGHEFSEKQYLQFFTEGLTHSNRMFLDASAGGSLRVKTDHE 262
E ES+ AW RF L++ P E LQ F GL + +LD +A GS KT E
Sbjct: 260 ENESIGAAWSRFTNLVQSGPTLSLPEYVLLQHFHTGLDKESAFYLDITARGSFMHKTPSE 319
Query: 263 VQILIENMASN 273
+ +++ + N
Sbjct: 320 GKTILDRILEN 330
>UniRef100_Q5W798 Putative polyprotein [Oryza sativa]
Length = 1005
Score = 491 bits (1264), Expect = e-137
Identities = 285/630 (45%), Positives = 379/630 (59%), Gaps = 54/630 (8%)
Query: 598 PVYAKFMKEMLTGRRKPKDDENISLSENCSAILQRKLPPKLKDPGAFTIPCSIGPVDIGR 657
P+ +F++E + DE + +E S +++ P++ IPC + D+
Sbjct: 380 PMELRFLRETIRELTTIMSDEWLREAELSSEVIRINTAPRI-------IPCHLEGNDVS- 431
Query: 658 ALCDLGASINLMPLSM-MKKLGGGEPKPTRMTLTLADRSISYPFGVLEDVLVKVNDLIFP 716
L NL+ S L PT +I FG+++DV V D
Sbjct: 432 ILYSPTVGANLVSGSFAFAYLSDKAVTPTNKFFKHPSGNIIKRFGIMQDVPVCFEDKEAI 491
Query: 717 ADFVILDMAEDEDMPLILGRPFLATGRALIDVEMGQLMLRFHNEQVVFNIFEAMKHRAEN 776
DF + ++ +D +++G P EQ++ N R ++
Sbjct: 492 LDFHVFEI---QDFDILIGLPI---------------------EQLLINT-----PRLDS 522
Query: 777 PKCYRIDVIDEIVEDNSREHKPTHPLEETIVNSIENCELVEDLEVQECVKQLEANEQLLK 836
K + I +R T PL E +E E V E+ E LL+
Sbjct: 523 LKITLGETEFSIPFSRARIVL-TDPLPE-----------IESTEEVIAVPPHESPEALLE 570
Query: 837 ---PTKLEEMSNEGKLGAKSLSNEEEKIP-ELKELPSHLKYVFLSKDVSKPAIISSTLTP 892
P ++E ++ G+ + P ELK LP L Y FL D P IIS L
Sbjct: 571 DEVPDFIKEEADPGETLDLPIMEPPPWPPLELKPLPPSLHYAFLHHDREAPVIISDKLFE 630
Query: 893 LEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEYKSVVQPQRRLNPTMKEVV 952
E ++L+ VL ++ LG+ + DLKGI+P C+HRI ++ E +PQRRLN M+EVV
Sbjct: 631 DETQRLLAVLEKHCSVLGYSLQDLKGINPVLCIHRIPIDPESTPSREPQRRLNNAMREVV 690
Query: 953 KKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPTRTVTGRRMCIDYR 1012
KKEVLKLL AG+IYP+ S WVSPV VVPKKGG+ VV N +NELIP RTVT RMCIDYR
Sbjct: 691 KKEVLKLLHAGIIYPVPYSEWVSPVQVVPKKGGIMVVANAQNELIPQRTVTEWRMCIDYR 750
Query: 1013 RLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFTCPFGVF 1072
+LN AT+KDHFPLPF+D+M+ERLA +F+CFLDGYSGY+QI + PEDQ KT FTCP+G +
Sbjct: 751 KLNKATKKDHFPLPFIDEMLERLANHSFFCFLDGYSGYHQIPIHPEDQSKTTFTCPYGTY 810
Query: 1073 AYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLNAVLKRC 1132
AYRRM FGLC APA+FQRCM+SIFSDMIE +EVFMDDF V+GK+F L +L+ VL+RC
Sbjct: 811 AYRRMSFGLCNAPASFQRCMMSIFSDMIEYIMEVFMDDFLVYGKTFGHYLQNLDKVLQRC 870
Query: 1133 TETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHA 1192
E +L+LNWEKCHFMV EGIVLGH++S +GIEVD+AKI+VI++LPPP+N+KG+RSFLGHA
Sbjct: 871 QEKDLVLNWEKCHFMVREGIVLGHRVSERGIEVDRAKIDVIDQLPPPVNIKGIRSFLGHA 930
Query: 1193 GFYRRFIKDFSKIAKPLCNLLVKETEFDFD 1222
FYRRFI+D+S I +PL NLL K+ F+FD
Sbjct: 931 SFYRRFIEDYSTIVRPLTNLLAKDAPFEFD 960
>UniRef100_Q7XWA9 OSJNBa0061A09.17 protein [Oryza sativa]
Length = 430
Score = 464 bits (1194), Expect = e-128
Identities = 227/445 (51%), Positives = 304/445 (68%), Gaps = 17/445 (3%)
Query: 1254 MCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFALEKFRSYLI 1313
MCDAS++AVGAVLGQ K++ HAI YASK L +Q+NYSTTEKELLAV+
Sbjct: 1 MCDASNFAVGAVLGQTKDRKQHAIAYASKTLTGAQLNYSTTEKELLAVV----------- 49
Query: 1314 GSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENVVADHLSRLENN 1373
G+KV+V+TDHAALKYLLTK D+KPRL+RW+L+LQEFD+EI +KKG+EN VADHLSR++
Sbjct: 50 GAKVVVYTDHAALKYLLTKKDAKPRLIRWILILQEFDIEINNKKGIENSVADHLSRMQIT 109
Query: 1374 EVTKKEGAIMAEFPDEQLFAIRER-PWFADMANFKAGNIIPDDMEQHQRKKFFKDANHYL 1432
+ +E +I D+ L + + PW+A + NF +P +K+ ++ L
Sbjct: 110 NM--QELSINDYLRDDMLLKVTDSDPWYAPIVNFMVAWHVPPG---ENKKRLSYESRKRL 164
Query: 1433 WDDPYLFKVSTDGLIRRCVAGEEIKNIVWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLF 1492
WD PYL++V +D L+RRCV +E I+ C + YG H+ RT AK+ QSGF+WPT++
Sbjct: 165 WDAPYLYRVCSDSLLRRCVPADEGMEIIEKCRVAPYGSHYGAFRTHAKIWQSGFFWPTMY 224
Query: 1493 KDCHDFVRRCDNCQRTGSISKRNEMPLTGIIEVEPFDCWGIDFMGPFPPSSSYLHILVCV 1552
D +F+RRC +CQ+ G I+ ++ MPLT ++VE FD WGIDFMGPFP +ILV V
Sbjct: 225 GDTKEFIRRCTSCQKHGGITAQDAMPLTYNLQVELFDVWGIDFMGPFPKFYDCEYILVAV 284
Query: 1553 DYVTKWVEAIPCVANDSKTVVNFLRKNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNI 1612
DYV+KWVEA+PC A ++K + IF FGTPR +ISDGG HF + + L++
Sbjct: 285 DYVSKWVEAMPCRAANTKHAHRMFDEIIFPCFGTPRTVISDGGSHFIDKTFQNFLQELGA 344
Query: 1613 KHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLS 1672
+H + TPYHP+T GQ E N+Q+ IL+KTV K W KL +ALWAYR + +G+S
Sbjct: 345 QHNIVTPYHPKTGGQAETRNKQIMNILQKTVNEMGKAWKNKLPNALWAYRTTYNMPIGMS 404
Query: 1673 PYQLVFGKACHLPVELEHKAYWAIK 1697
PYQLV+GK CHLPVELEHKA+WAI+
Sbjct: 405 PYQLVYGKTCHLPVELEHKAHWAIR 429
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,186,771,001
Number of Sequences: 2790947
Number of extensions: 145131137
Number of successful extensions: 520284
Number of sequences better than 10.0: 7725
Number of HSP's better than 10.0 without gapping: 4390
Number of HSP's successfully gapped in prelim test: 3424
Number of HSP's that attempted gapping in prelim test: 490260
Number of HSP's gapped (non-prelim): 19697
length of query: 1825
length of database: 848,049,833
effective HSP length: 142
effective length of query: 1683
effective length of database: 451,735,359
effective search space: 760270609197
effective search space used: 760270609197
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 82 (36.2 bits)
Medicago: description of AC146759.6


