

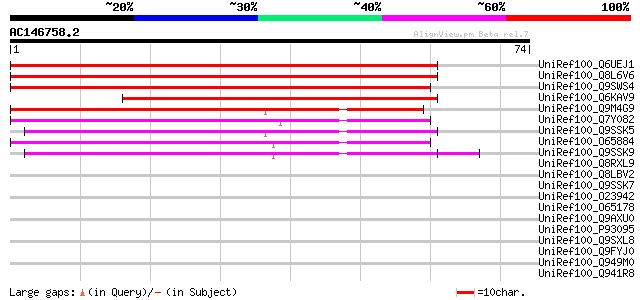
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146758.2 + phase: 0
(74 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6UEJ1 Ripening-related protein [Pisum sativum] 105 3e-22
UniRef100_Q8L6V6 Putative ripening related protein [Cicer arieti... 101 5e-21
UniRef100_Q9SWS4 Ripening related protein [Glycine max] 86 3e-16
UniRef100_Q6KAV9 Putative Bet v I family protein [Cicer arietinum] 74 1e-12
UniRef100_Q9M4G9 Putative ripening-related protein [Vitis vinifera] 67 9e-11
UniRef100_Q7Y082 Major latex protein homologue [Datisca glomerata] 54 1e-06
UniRef100_Q9SSK5 MLP-like protein 43 [Arabidopsis thaliana] 46 3e-04
UniRef100_O65884 Major latex-like protein [Rubus idaeus] 45 4e-04
UniRef100_Q9SSK9 MLP-like protein 28 [Arabidopsis thaliana] 45 5e-04
UniRef100_Q8RXL9 MLP-like protein 31 [Arabidopsis thaliana] 44 0.001
UniRef100_Q8LBV2 Major latex protein (MLP149), putative [Arabido... 43 0.002
UniRef100_Q9SSK7 MLP-like protein 34 [Arabidopsis thaliana] 43 0.002
UniRef100_O23942 Ripening-induced protein [Fragaria vesca] 42 0.005
UniRef100_O65178 Major latex protein homolog [Mesembryanthemum c... 41 0.007
UniRef100_Q9AXU0 Major latex-like protein [Prunus persica] 40 0.020
UniRef100_P93095 Hypothetical protein [Cucumis melo] 40 0.020
UniRef100_Q9SXL8 Csf-2 protein [Cucumis sativus] 39 0.033
UniRef100_Q9FYJ0 F17F8.9 [Arabidopsis thaliana] 39 0.044
UniRef100_Q949M0 Major latex like protein homolog [Beta vulgaris] 34 0.82
UniRef100_Q941R8 Major latex-like protein [Arabidopsis thaliana] 33 1.8
>UniRef100_Q6UEJ1 Ripening-related protein [Pisum sativum]
Length = 159
Score = 105 bits (262), Expect = 3e-22
Identities = 43/61 (70%), Positives = 55/61 (89%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTH 60
MVLAGKLSTE+GIKA +DKFFK++ S +HE+QN CE++H KLH+GD+WHHSD+VKHWT+
Sbjct: 1 MVLAGKLSTELGIKAPADKFFKVYTSELHELQNHCENIHAAKLHEGDDWHHSDTVKHWTY 60
Query: 61 V 61
V
Sbjct: 61 V 61
>UniRef100_Q8L6V6 Putative ripening related protein [Cicer arietinum]
Length = 152
Score = 101 bits (251), Expect = 5e-21
Identities = 44/61 (72%), Positives = 52/61 (85%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTH 60
MVLAGKLSTE+GIKA + KF+ LFA+ +HEVQN CE VH TKL QG++WHHSDSVKHW +
Sbjct: 1 MVLAGKLSTELGIKAPASKFYNLFATQLHEVQNHCERVHHTKLQQGEDWHHSDSVKHWKY 60
Query: 61 V 61
V
Sbjct: 61 V 61
>UniRef100_Q9SWS4 Ripening related protein [Glycine max]
Length = 152
Score = 85.5 bits (210), Expect = 3e-16
Identities = 33/60 (55%), Positives = 50/60 (83%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTH 60
M LAGK+S+EIG++A++ K+F LF + IH VQN+ + +H T+LH G++WHH++SVKHWT+
Sbjct: 1 MSLAGKISSEIGVQATAAKWFNLFTTQIHHVQNLTDRIHGTELHHGEDWHHNESVKHWTY 60
>UniRef100_Q6KAV9 Putative Bet v I family protein [Cicer arietinum]
Length = 136
Score = 73.6 bits (179), Expect = 1e-12
Identities = 30/45 (66%), Positives = 37/45 (81%)
Query: 17 SDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTHV 61
+DKFF LF+ +H+VQN CE VHET LH+GD+WH SDSVK WT+V
Sbjct: 1 ADKFFHLFSKKLHDVQNHCERVHETILHEGDDWHVSDSVKQWTYV 45
>UniRef100_Q9M4G9 Putative ripening-related protein [Vitis vinifera]
Length = 151
Score = 67.4 bits (163), Expect = 9e-11
Identities = 31/60 (51%), Positives = 41/60 (67%), Gaps = 2/60 (3%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNIC-ESVHETKLHQGDEWHHSDSVKHWT 59
M L GKL TE IKA +DKFFK+F S H + NIC + +H+ ++H+GD W SVKHW+
Sbjct: 1 MALTGKLETETEIKAPADKFFKIFRSQAHHLPNICSDKIHKIEVHEGD-WETQGSVKHWS 59
>UniRef100_Q7Y082 Major latex protein homologue [Datisca glomerata]
Length = 141
Score = 53.5 bits (127), Expect = 1e-06
Identities = 26/61 (42%), Positives = 36/61 (58%), Gaps = 2/61 (3%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICES-VHETKLHQGDEWHHSDSVKHWT 59
MVLAGKL EI I A +DK++++F H V NI + E +H+GD W S+K W
Sbjct: 1 MVLAGKLEAEIDINAPADKYYRIFKGQAHHVPNISPGIIQEVNVHEGD-WDTHGSIKIWN 59
Query: 60 H 60
+
Sbjct: 60 Y 60
>UniRef100_Q9SSK5 MLP-like protein 43 [Arabidopsis thaliana]
Length = 158
Score = 45.8 bits (107), Expect = 3e-04
Identities = 24/60 (40%), Positives = 33/60 (55%), Gaps = 2/60 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNIC-ESVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL TE+ IKAS+ KF +F H V + +H +LH+GD W S+ W +V
Sbjct: 7 LVGKLETEVEIKASAKKFHHMFTERPHHVSKATPDKIHGCELHEGD-WGKVGSIVIWKYV 65
>UniRef100_O65884 Major latex-like protein [Rubus idaeus]
Length = 152
Score = 45.4 bits (106), Expect = 4e-04
Identities = 23/62 (37%), Positives = 34/62 (54%), Gaps = 3/62 (4%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICE--SVHETKLHQGDEWHHSDSVKHW 58
MVL GK+ +I I A +DKF+ LF S H V + ++ +H+GD W S+K W
Sbjct: 1 MVLQGKVEADIEISAPADKFYNLFKSEAHHVPKTSQTGTITGVAVHEGD-WETDGSIKIW 59
Query: 59 TH 60
+
Sbjct: 60 NY 61
>UniRef100_Q9SSK9 MLP-like protein 28 [Arabidopsis thaliana]
Length = 335
Score = 45.1 bits (105), Expect = 5e-04
Identities = 24/60 (40%), Positives = 33/60 (55%), Gaps = 2/60 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL T++ IKAS+DKF +FA H V ++ LH+GD W S+ W +V
Sbjct: 22 LVGKLETDVEIKASADKFHHMFAGKPHHVSKASPGNIQGCDLHEGD-WGTVGSIVFWNYV 80
Score = 44.7 bits (104), Expect = 6e-04
Identities = 24/66 (36%), Positives = 35/66 (52%), Gaps = 2/66 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL T++ IKAS++KF +FA H V ++ LH+GD W S+ W +V
Sbjct: 184 LVGKLETDVEIKASAEKFHHMFAGKPHHVSKASPGNIQGCDLHEGD-WGQVGSIVFWNYV 242
Query: 62 RVLSSK 67
+K
Sbjct: 243 HDREAK 248
>UniRef100_Q8RXL9 MLP-like protein 31 [Arabidopsis thaliana]
Length = 171
Score = 43.5 bits (101), Expect = 0.001
Identities = 24/60 (40%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL T+I IKAS+ KF +FA H V + +LH+GD W S+ W +V
Sbjct: 20 LCGKLETDIEIKASAGKFHHMFAGRPHHVSKATPGKIQGCELHEGD-WGKVGSIVFWNYV 78
>UniRef100_Q8LBV2 Major latex protein (MLP149), putative [Arabidopsis thaliana]
Length = 180
Score = 42.7 bits (99), Expect = 0.002
Identities = 23/60 (38%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL TE+ IKAS+ +F +FA H V ++ LH+GD W S+ W +V
Sbjct: 9 LVGKLETEVEIKASAGQFHHMFAGKPHHVSKASPGNIQSCDLHEGD-WGTVGSIVFWNYV 67
>UniRef100_Q9SSK7 MLP-like protein 34 [Arabidopsis thaliana]
Length = 316
Score = 42.7 bits (99), Expect = 0.002
Identities = 23/60 (38%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL TE+ IKAS+ +F +FA H V ++ LH+GD W S+ W +V
Sbjct: 9 LVGKLETEVEIKASAGQFHHMFAGKPHHVSKASPGNIQSCDLHEGD-WGTVGSIVFWNYV 67
Score = 39.7 bits (91), Expect = 0.020
Identities = 21/56 (37%), Positives = 30/56 (53%), Gaps = 2/56 (3%)
Query: 7 LSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
L TE+ IKAS++KF +FA H V ++ LH+GD W S+ W +V
Sbjct: 169 LETEVEIKASAEKFHHMFAGKPHHVSKATPGNIQSCDLHEGD-WGTVGSIVFWNYV 223
>UniRef100_O23942 Ripening-induced protein [Fragaria vesca]
Length = 152
Score = 41.6 bits (96), Expect = 0.005
Identities = 22/62 (35%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICE--SVHETKLHQGDEWHHSDSVKHW 58
M L GK+ E I A +DKF+ +F H V N + S+ +H+GD W S+K W
Sbjct: 1 MSLQGKVEAEFEITAPADKFYNIFKIEAHLVPNTSQTGSITGVAVHEGD-WETDGSIKIW 59
Query: 59 TH 60
+
Sbjct: 60 NY 61
>UniRef100_O65178 Major latex protein homolog [Mesembryanthemum crystallinum]
Length = 152
Score = 41.2 bits (95), Expect = 0.007
Identities = 22/61 (36%), Positives = 33/61 (54%), Gaps = 2/61 (3%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICES-VHETKLHQGDEWHHSDSVKHWT 59
M + GKL E+ IK+ D F +LF H V NI S +H +H+G E+ S+ +W
Sbjct: 1 MGITGKLEVEVDIKSCGDMFHELFGKKPHHVSNITPSKIHGCDVHEG-EFGKPGSIINWD 59
Query: 60 H 60
+
Sbjct: 60 Y 60
>UniRef100_Q9AXU0 Major latex-like protein [Prunus persica]
Length = 156
Score = 39.7 bits (91), Expect = 0.020
Identities = 19/57 (33%), Positives = 32/57 (55%), Gaps = 2/57 (3%)
Query: 5 GKLSTEIGIKASSDKFFKLFASNIHEVQNI-CESVHETKLHQGDEWHHSDSVKHWTH 60
GK+ T++ IKA + KF ++F H + N+ ++ LH+G EW SV +W +
Sbjct: 6 GKVETDVEIKAPATKFHEVFTHRPHHISNVSSNNIQGCDLHEG-EWGTVGSVVYWNY 61
>UniRef100_P93095 Hypothetical protein [Cucumis melo]
Length = 151
Score = 39.7 bits (91), Expect = 0.020
Identities = 21/61 (34%), Positives = 35/61 (56%), Gaps = 4/61 (6%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNIC-ESVHETKLHQGDEW--HHSDSVKH 57
M L GKL +E+ I A+++KF+++F +V NI + + ++H G W H S+K
Sbjct: 1 MSLIGKLVSELEINAAAEKFYEIFKDQCFQVPNITPRCIQQVEIH-GTNWDGHGHGSIKS 59
Query: 58 W 58
W
Sbjct: 60 W 60
>UniRef100_Q9SXL8 Csf-2 protein [Cucumis sativus]
Length = 151
Score = 38.9 bits (89), Expect = 0.033
Identities = 20/61 (32%), Positives = 33/61 (53%), Gaps = 2/61 (3%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNI-CESVHETKLHQGDEWHHSDSVKHWT 59
M L GKL ++ I+AS+ KF ++F H + N + +H +L +G EW S+ W
Sbjct: 1 MSLCGKLEKDVPIRASASKFHEMFHKKPHHICNCSTDKIHGVELQEG-EWGQVGSIICWK 59
Query: 60 H 60
+
Sbjct: 60 Y 60
>UniRef100_Q9FYJ0 F17F8.9 [Arabidopsis thaliana]
Length = 152
Score = 38.5 bits (88), Expect = 0.044
Identities = 18/61 (29%), Positives = 34/61 (55%), Gaps = 2/61 (3%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQN-ICESVHETKLHQGDEWHHSDSVKHWT 59
M ++G T++ + S++K +KL++S H + + I + LH+GD W S+K W
Sbjct: 1 MAMSGTYMTDVPLNGSAEKHYKLWSSETHRIPDTIGHLIQGVILHEGD-WDSHGSIKTWK 59
Query: 60 H 60
+
Sbjct: 60 Y 60
>UniRef100_Q949M0 Major latex like protein homolog [Beta vulgaris]
Length = 150
Score = 34.3 bits (77), Expect = 0.82
Identities = 15/48 (31%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNIC-ESVHETKLHQGD 47
M + GKL E+ I D F ++F++ H+V + E++H +H G+
Sbjct: 1 MHITGKLEVEVDINCHGDIFHEIFSTRPHDVSTMSPENIHGCDVHDGE 48
>UniRef100_Q941R8 Major latex-like protein [Arabidopsis thaliana]
Length = 75
Score = 33.1 bits (74), Expect = 1.8
Identities = 17/60 (28%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHE-TKLHQGDEWHHSDSVKHWTHV 61
L G L T + +K+S++KF +F H + ++ S + +LH+G E ++ W +V
Sbjct: 8 LVGILETTVDLKSSTEKFHDMFVGRPHNISDVSPSNFQGCELHEG-EMGQVGAILFWNYV 66
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.129 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 112,060,546
Number of Sequences: 2790947
Number of extensions: 3416024
Number of successful extensions: 9182
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 9164
Number of HSP's gapped (non-prelim): 27
length of query: 74
length of database: 848,049,833
effective HSP length: 50
effective length of query: 24
effective length of database: 708,502,483
effective search space: 17004059592
effective search space used: 17004059592
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146758.2


