

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146758.11 + phase: 0 /pseudo
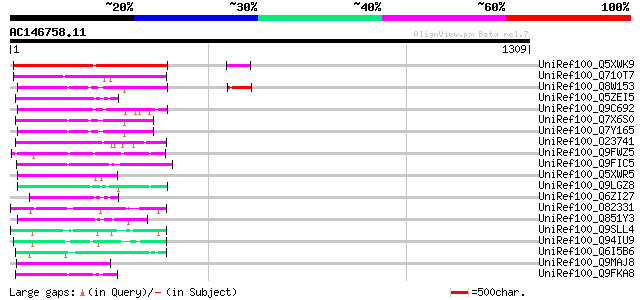
(1309 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q5XWK9 Gag-pol polyprotein-like [Solanum tuberosum] 408 e-112
UniRef100_Q710T7 Gag-pol polyprotein [Populus deltoides] 171 1e-40
UniRef100_Q8W153 Polyprotein [Oryza sativa] 109 5e-22
UniRef100_Q5ZEI5 Hypothetical protein P0009G03.10 [Oryza sativa] 107 2e-21
UniRef100_Q9C692 Polyprotein, putative [Arabidopsis thaliana] 106 4e-21
UniRef100_Q7X6S0 OSJNBb0011N17.2 protein [Oryza sativa] 105 9e-21
UniRef100_Q7Y165 Putative retrotransposon protein [Oryza sativa] 104 2e-20
UniRef100_O23741 SLG-Sc and SLA-Sc genes and Melmoth retrotransp... 101 1e-19
UniRef100_Q9FWZ5 Putative retroelement polyprotein [Arabidopsis ... 100 4e-19
UniRef100_Q9FIC5 Retroelement pol polyprotein-like [Arabidopsis ... 99 9e-19
UniRef100_Q5XWR5 Putative retroelement pol polyprotein-like [Sol... 96 6e-18
UniRef100_Q9LGZ8 Retroelement pol polyprotein-like [Arabidopsis ... 96 6e-18
UniRef100_Q6ZI27 Hypothetical protein OJ1004_H01.32 [Oryza sativa] 92 1e-16
UniRef100_O82331 Putative retroelement pol polyprotein [Arabidop... 91 2e-16
UniRef100_Q851Y3 Putative polyprotein [Oryza sativa] 89 7e-16
UniRef100_Q9SLL4 Putative retroelement pol polyprotein [Arabidop... 89 1e-15
UniRef100_Q94IU9 Copia-like polyprotein [Arabidopsis thaliana] 87 3e-15
UniRef100_Q6I5B6 Putative polyprotein [Oryza sativa] 86 6e-15
UniRef100_Q9MAJ8 F27F5.19 [Arabidopsis thaliana] 86 8e-15
UniRef100_Q9FKA8 Retroelement pol polyprotein-like [Arabidopsis ... 86 8e-15
>UniRef100_Q5XWK9 Gag-pol polyprotein-like [Solanum tuberosum]
Length = 1212
Score = 408 bits (1048), Expect = e-112
Identities = 197/388 (50%), Positives = 276/388 (70%), Gaps = 6/388 (1%)
Query: 10 DNMCVRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALEEWETKDAQIITWIL 69
++ VRF+GKNYS+WEFQF+++V G+ LW +++G APT+ L EW+ KDA+++TWIL
Sbjct: 7 ESFSVRFTGKNYSSWEFQFQLFVTGKELWGYIDGSDPAPTDATKLGEWKIKDARVMTWIL 66
Query: 70 STINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGF 129
+I+P ++ NLR + + MW+YL+++YNQDN A+RFQLE EIANY QG VQ+Y+SGF
Sbjct: 67 GSIDPLIVLNLRPYKTVKAMWDYLQKVYNQDNSARRFQLEYEIANYSQGGLFVQDYFSGF 126
Query: 130 LNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLD 189
NLW E + I++A +P L+ +Q V+ SK+DQFLM LR +FE +R +L+NR+ PSLD
Sbjct: 127 QNLWAEFTDIVYAKIPTESLSVIQAVHEQSKRDQFLMKLRSDFESIRSNLMNRDPSPSLD 186
Query: 190 TCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIAR 249
C ELLREEQRL+TQ ++ TVA+AAQ +GKG DM + QC+SCK++GHIA
Sbjct: 187 VCFRELLREEQRLVTQNVFKKE----NDVTVAFAAQGKGKGRDMSRTQCYSCKEYGHIAS 242
Query: 250 SCSKKFCNYCKQRGHIITECYVRPPPSTQSPM*A-LHAYSTTNATNGGVSQSEMIQQMVI 308
+CSKKF NYCKQ+GHII EC +RP + A ++ + N++ G V EM+QQM++
Sbjct: 243 NCSKKFYNYCKQQGHIIKECPMRPQNRRINAFQARINGSTDDNSSLGQVLTPEMVQQMIV 302
Query: 309 SALPSIGIQGKSSNASHPWFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITD 368
SA ++G+QG + S+ W +DSGAS HMT S L N+ Y G +IQIA+G+ L IT
Sbjct: 303 SAFSALGLQG-NDVTSNFWIVDSGASNHMTNSTSILKNVRKYQGPSQIQIANGSNLPITK 361
Query: 369 VGDINSDFRDVLVSPGLASNLLSVGQLV 396
VGDI F++V VSP L+++L+SVGQLV
Sbjct: 362 VGDITPTFKNVFVSPKLSTSLISVGQLV 389
Score = 50.8 bits (120), Expect = 3e-04
Identities = 26/62 (41%), Positives = 37/62 (58%), Gaps = 1/62 (1%)
Query: 547 FFDPSLKCFLYLRNF*HMLKLNFKQVLKFF-TDSGGEYMSHEFQEYLQHKGILSQ*SCPN 605
F + F + F ++ F +K +DSGGEYMS+EF+++L KGI+SQ SCP
Sbjct: 542 FLRSKSEVFSMFKTFLAYIETQFSTCIKLLRSDSGGEYMSYEFKKFLLDKGIVSQHSCPY 601
Query: 606 TP 607
TP
Sbjct: 602 TP 603
>UniRef100_Q710T7 Gag-pol polyprotein [Populus deltoides]
Length = 1382
Score = 171 bits (433), Expect = 1e-40
Identities = 120/410 (29%), Positives = 199/410 (48%), Gaps = 29/410 (7%)
Query: 9 IDNMCVRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPT-----EKAALEEWETKDAQ 63
+ ++ VR GKNYS W + R ++KG+++W +++G P + +++ WE +A+
Sbjct: 9 LQSVSVRLDGKNYSYWSYVMRNFLKGKKMWGYVSGTYVVPKNTEEGDTVSIDTWEANNAK 68
Query: 64 IITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQ 123
IITWI + + + L + A+E+W++L+R++ Q N AK++QLE +I Q N S+Q
Sbjct: 69 IITWINNYVEHSIGTQLAKYETAKEVWDHLQRLFTQSNFAKQYQLENDIRALHQKNMSIQ 128
Query: 124 EYYSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRN 183
E+YS +LW + + V A E + QFL LR +FE +RGS+L+R+
Sbjct: 129 EFYSAMTDLWDQ--LALTESVELKACGAYIERREQQRLVQFLTALRSDFEGLRGSILHRS 186
Query: 184 VVPSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQV------Q 237
+PS+D+ VSELL EE RL Q + L + +V + H + +
Sbjct: 187 PLPSVDSVVSELLAEEIRL--QSYSEKGILSASNPSVLAVPSKPFSNHQNKPYTRVGFDE 244
Query: 238 CFSCKQFGHIARSCSK--------KFCNYCKQRGHIITECYVRPPPSTQSPM*ALHAYST 289
C CKQ GH C K K + + H + Y +PP + + + + +
Sbjct: 245 CSFCKQKGHWKAQCPKLRQQNQAWKSGSQSQSNAHRSPQGY-KPPHHNTAAVASPGSITD 303
Query: 290 TNATNGGVSQSEMIQQMVISALPSIGIQGKSSNASH-PWFLDSGASYHMTGSLEYLHNLH 348
N + +Q +SA + SS SH W LDSGAS+HM+ ++
Sbjct: 304 PNTLAEQFQKFLSLQPQAMSASSIGQLPHSSSGISHSEWVLDSGASHHMSPDSSSFTSV- 362
Query: 349 SYDGNKKIQIADGNTLSITDVGDI---NSDFRDVLVSPGLASNLLSVGQL 395
S + + ADG + + VG + + +V + P L NL S+GQ+
Sbjct: 363 SPLSSIPVMTADGTPMPLAGVGSVVTLHLSLPNVYLIPKLKLNLASIGQI 412
>UniRef100_Q8W153 Polyprotein [Oryza sativa]
Length = 1472
Score = 109 bits (273), Expect = 5e-22
Identities = 100/401 (24%), Positives = 182/401 (44%), Gaps = 37/401 (9%)
Query: 19 KNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALE--EWETKDAQIITWILSTINPQM 76
KNY +W + + +K + L ++ G K P +++E W T ++ ++ W+L+++ P +
Sbjct: 55 KNYLSWSRRALLILKTKGLEGYVTGEVKEPENTSSVEWKTWSTTNSLVVAWLLTSLIPAI 114
Query: 77 INNLRSFSFAQEMWNYLKRIYN-QDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTE 135
+ + S A EMW L ++Y+ + N + + +I+ +QG SV EY + +LW++
Sbjct: 115 ATTVETISSASEMWKTLTKLYSGEGNVMLMVEAQEKISALRQGERSVAEYVAELKSLWSD 174
Query: 136 HS-----AIIHADVPNIYLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDT 190
+ H+D +A +++ + +FL L PEFE R ++ ++ +P+LD
Sbjct: 175 LDHYDPLGLEHSDC----IAKMKKWVERRRVIEFLKGLNPEFEGRRDAMFHQTTLPTLDE 230
Query: 191 CVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARS 250
++ + +EE L + + A S T A +GK + +CF+C + GH+ R
Sbjct: 231 AIAAMAQEE---LKKKVLPSAAPCSPSPTYAIV---QGK----ETRECFNCGEMGHLMRD 280
Query: 251 C-SKKFCNYCKQRGHIITECYVRPPPSTQSPM*ALHAY-------STTNATNGGVSQSEM 302
C + + Y + RG + +S + Y + T + +
Sbjct: 281 CHAPRKPTYGRGRGVDRGGTRGGRGYAGRSNRGRGYGYRGDYKANAVTLEEGSSGTTPDN 340
Query: 303 IQQMVISALPSIGIQGKSSNASH-PWFLDSGASYHMTG-SLEYL-HNLHSYDGNKKIQIA 359
+ S S S N SH W LDSGAS H+TG S E+ + +S+ + IQ A
Sbjct: 341 VANFAHSTSGSFNQAFMSMNTSHSSWILDSGASRHVTGMSGEFTSYKPYSFAHKETIQTA 400
Query: 360 DGNTLSITDVGDI----NSDFRDVLVSPGLASNLLSVGQLV 396
DG + + G + + VL NL+S+ LV
Sbjct: 401 DGTSCQVKGEGIVQCTPSITLSSVLYVHSFPVNLISISSLV 441
Score = 55.1 bits (131), Expect = 1e-05
Identities = 30/61 (49%), Positives = 40/61 (65%), Gaps = 4/61 (6%)
Query: 549 DPSLKCFLYLRNF*HMLKLNFKQVLKFF-TDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
D LKCF +NF +K +F ++F TD+GGEYM+ EF +L +GIL Q SCP+TP
Sbjct: 599 DEVLKCF---QNFYAYIKNHFNARVQFIRTDNGGEYMNSEFGHFLSLEGILHQTSCPDTP 655
Query: 608 P 608
P
Sbjct: 656 P 656
>UniRef100_Q5ZEI5 Hypothetical protein P0009G03.10 [Oryza sativa]
Length = 337
Score = 107 bits (268), Expect = 2e-21
Identities = 72/263 (27%), Positives = 127/263 (47%), Gaps = 14/263 (5%)
Query: 14 VRFSGKNYSAWEFQFRMYVKGERLWSHL-NGVSKAPTEKAALEEWETKDAQIITWILSTI 72
++ +G+NY WE RM ++ SHL + T+ ++ W+T D +++ +I ++
Sbjct: 18 IKLNGQNYQEWELSARMLLRSIGQASHLTDDPPDEKTDATKIKAWKTADDRVMGFIFMSV 77
Query: 73 NPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNL 132
+ + R S A+EMW+YLK+ Y Q++ A RF L + N +Q + S++E+Y+ F L
Sbjct: 78 EVPIRMSFRDHSTAKEMWDYLKQRYTQESGALRFSLLQNLHNLQQQDQSIEEFYNAFTRL 137
Query: 133 WTEHSAIIHADVPNIYLAAVQEVYNTSK-QDQFLMILRPEFEVVRGSLLNRNVVPSLDTC 191
+ A +E ++ QF+M L +FE +R LL R P++
Sbjct: 138 SGQLEAQTPKGASGCAQCKAREKHDQENLVYQFVMRLSSQFESIRVQLLGRPTRPTMAEA 197
Query: 192 VSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSC 251
+++L+ EE RL + + + V A QR G +F IA+
Sbjct: 198 LADLIAEETRLCSLDTTPTPVV---THNVMAAPQRVG-------APMGGIPEFSSIAK-- 245
Query: 252 SKKFCNYCKQRGHIITECYVRPP 274
S + C++CK+ GHI +C+ P
Sbjct: 246 SDRICSHCKKVGHISDDCFYLHP 268
>UniRef100_Q9C692 Polyprotein, putative [Arabidopsis thaliana]
Length = 1468
Score = 106 bits (265), Expect = 4e-21
Identities = 99/446 (22%), Positives = 192/446 (42%), Gaps = 86/446 (19%)
Query: 20 NYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAA-LEEWETKDAQIITWILSTINPQMIN 78
NY W F+ ++ + + L+G P + + LE+W T +A +++W+ TI+ +++
Sbjct: 42 NYEEWACGFKTALRSRKKFGFLDGTIPQPLDGSPDLEDWLTINALLVSWMKMTIDSELLT 101
Query: 79 NLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEHSA 138
N+ A+++W +++ ++ N K +++ ++A KQ +V+ YY +W ++
Sbjct: 102 NISHRDVARDLWEQIRKRFSVSNGPKNQKMKADLATCKQEGMTVEGYYGKLNKIWDNINS 161
Query: 139 -----IIHADVPNIYLAAVQEVYNTSKQ-DQFLMIL-RPEFEVVRGSLLNRNVVPSLDTC 191
I L QE Y Q+L L +F +R SL +R +P L+
Sbjct: 162 YRPLRICKCGRCICNLGTDQEKYREDDMVHQYLYGLNETKFHTIRSSLTSRVPLPGLEE- 220
Query: 192 VSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSC 251
V ++R+E+ ++ + + + + A+A Q R + + + +F + +
Sbjct: 221 VYNIVRQEEDMVNNRSSNEE----RTDVTAFAVQMRPRSEVISE-------KFANSEKLQ 269
Query: 252 SKKFCNYCKQRGHIITECYV---RPPPSTQSPM*ALHAYSTTN--------ATNGGVSQS 300
+KK C +C + GH C+V P P ++ +T+ NGG +
Sbjct: 270 NKKLCTHCNRGGHSPENCFVLIGYPEWWGDRPRGKSNSNGSTSRGRGRFGPGFNGGQPRP 329
Query: 301 EMIQQMVISALPSI------------------------GI-----QGKSSNASH------ 325
+ ++ PS G+ G+S N S+
Sbjct: 330 TYVNVVMTGPFPSSEHVNRVITDSDRDAVSGLTDEQWRGVVKLLNAGRSDNKSNAHETQS 389
Query: 326 -------PWFLDSGASYHMTGSLEYLHNLHSY--------DGNKKIQIADGNTLSITDVG 370
W LD+GAS+HMTG+LE L ++ S DGNK++ +++G T+ +
Sbjct: 390 GTCSLFTSWILDTGASHHMTGNLELLSDMRSMSPVLIILADGNKRVAVSEG-TVRLGS-- 446
Query: 371 DINSDFRDVLVSPGLASNLLSVGQLV 396
+ + V L S+L+SVGQ++
Sbjct: 447 --HLILKSVFYVKELESDLISVGQMM 470
>UniRef100_Q7X6S0 OSJNBb0011N17.2 protein [Oryza sativa]
Length = 1262
Score = 105 bits (262), Expect = 9e-21
Identities = 92/369 (24%), Positives = 171/369 (45%), Gaps = 33/369 (8%)
Query: 14 VRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALE--EWETKDAQIITWILST 71
++ KNY +W + + +K + L ++ G K P +++E W T ++ ++ W+L++
Sbjct: 49 IKLGVKNYLSWSRRALLILKTKGLEGYVTGEVKEPENTSSVEWKTWSTTNSLVVAWLLTS 108
Query: 72 INPQMINNLRSFSFAQEMWNYLKRIYN-QDNPAKRFQLELEIANYKQGNSSVQEYYSGFL 130
+ P + + + S A EMW L ++Y+ + N + + +I+ +QG SV EY +
Sbjct: 109 LIPAIATTVETISSASEMWKTLTKLYSGEGNVMLMVEAQEKISALRQGERSVAEYVAELK 168
Query: 131 NLWTEHS-----AIIHADVPNIYLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVV 185
+LW++ + H+D +A +++ + +FL L PEFE R ++ ++ +
Sbjct: 169 SLWSDLDHYDPLGLEHSDC----IAKMKKWVERRRVIEFLKGLNPEFEGRRDAMFHQTTL 224
Query: 186 PSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFG 245
P+LD ++ + +EE L + + A S T A +GK + +CF+C + G
Sbjct: 225 PTLDEAIAAMAQEE---LKKKVLPSAAPCSPSPTYAIV---QGK----ETRECFNCGEMG 274
Query: 246 HIARSC-SKKFCNYCKQRGHIITECYVRPPPSTQSPM*ALHAY-------STTNATNGGV 297
H+ R C + + Y + RG + +S + Y + T
Sbjct: 275 HLMRDCHAPRKPTYGRGRGVDRGGTRGGRGYAGRSNRGRGYGYRGDYKANAVTLEEGSSG 334
Query: 298 SQSEMIQQMVISALPSIGIQGKSSNASH-PWFLDSGASYHMTG-SLEYL-HNLHSYDGNK 354
+ + + S S S N SH W LDSGAS H+TG S E+ + +S+ +
Sbjct: 335 TTPDNVANFAHSTSGSFNQAFMSMNTSHSSWILDSGASRHVTGMSGEFTSYKPYSFAHKE 394
Query: 355 KIQIADGNT 363
IQ ADG +
Sbjct: 395 TIQTADGTS 403
>UniRef100_Q7Y165 Putative retrotransposon protein [Oryza sativa]
Length = 1176
Score = 104 bits (260), Expect = 2e-20
Identities = 92/364 (25%), Positives = 169/364 (46%), Gaps = 33/364 (9%)
Query: 19 KNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALE--EWETKDAQIITWILSTINPQM 76
KNY +W + + +K + L ++ G K P +++E W T ++ ++ W+L+++ P +
Sbjct: 55 KNYLSWSRRALLILKTKGLEGYVTGEVKEPENTSSVEWKTWSTTNSLVVAWLLTSLIPAI 114
Query: 77 INNLRSFSFAQEMWNYLKRIYN-QDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTE 135
+ + S A EMW L ++Y+ + N + + +I+ +QG SV EY + +LW++
Sbjct: 115 ATTVETISSASEMWKTLTKLYSGEGNVMLMVEAQEKISALRQGERSVAEYVAELKSLWSD 174
Query: 136 HS-----AIIHADVPNIYLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDT 190
+ H+D +A +++ + +FL L PEFE R ++ ++ +P+LD
Sbjct: 175 LDHYDPLGLEHSDC----IAKMKKWVERRRVIEFLKGLNPEFEGRRDAMFHQTTLPTLDE 230
Query: 191 CVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARS 250
++ + +EE L + + A S T A +GK + +CF+C + GH+ R
Sbjct: 231 AIAAMAQEE---LKKKVLPSAAPCSPSPTYAIV---QGK----ETRECFNCGEMGHLMRD 280
Query: 251 C-SKKFCNYCKQRGHIITECYVRPPPSTQSPM*ALHAY-------STTNATNGGVSQSEM 302
C + + Y + RG + +S + Y + T + +
Sbjct: 281 CHAPRKPTYGRGRGVDRGGTRGGRGYAGRSNRGRGYGYRGDYKANAVTLEEGSSGTTPDN 340
Query: 303 IQQMVISALPSIGIQGKSSNASH-PWFLDSGASYHMTG-SLEYL-HNLHSYDGNKKIQIA 359
+ S S S N SH W LDSGAS H+TG S E+ + +S+ + IQ A
Sbjct: 341 VANFAHSTSGSFNQAFMSMNTSHSSWILDSGASRHVTGMSGEFTSYKPYSFAHKETIQTA 400
Query: 360 DGNT 363
DG +
Sbjct: 401 DGTS 404
Score = 48.5 bits (114), Expect = 0.001
Identities = 23/40 (57%), Positives = 30/40 (74%), Gaps = 1/40 (2%)
Query: 570 KQVLKFF-TDSGGEYMSHEFQEYLQHKGILSQ*SCPNTPP 608
K+V +F TD+GGEYM+ EF +L +GIL Q SCP+TPP
Sbjct: 444 KEVTEFIRTDNGGEYMNSEFGHFLSLEGILHQTSCPDTPP 483
>UniRef100_O23741 SLG-Sc and SLA-Sc genes and Melmoth retrotransposon sequence
[Brassica oleracea]
Length = 1131
Score = 101 bits (252), Expect = 1e-19
Identities = 97/438 (22%), Positives = 177/438 (40%), Gaps = 61/438 (13%)
Query: 14 VRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAP-TEKAALEEWETKDAQIITWILSTI 72
++ G NY W ++ + + ++G P T W ++ + +W+L+++
Sbjct: 30 LKLDGSNYDDWNAAMKIALDAKNKIGFVDGTLTRPDTSDPTFRLWSRCNSMVKSWLLNSV 89
Query: 73 NPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNL 132
+PQ+ ++ + A ++W L ++ N + F L EI + KQG+ S+ +YY+ L
Sbjct: 90 SPQIYRSILRLNDAADIWRDLHGRFHMTNLPRTFNLTQEIQDLKQGSMSLSDYYTTLKTL 149
Query: 133 WTEHSAIIHADVPNIYLAA--VQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDT 190
W ++ D P + A +Q+ + +K +FL L + ++R ++ + V+PSL
Sbjct: 150 WDNLESVDEPDTPCVCGNAEKLQKKVDRAKIVKFLAGLNDSYAIIRRQIIMKKVLPSLVE 209
Query: 191 CVSELLREEQR-----LLTQGAMSRDALI---FESTTVAYAAQRRGKGHDMQQVQCFSCK 242
+ L +++ + +T A + + + Y KG + C C
Sbjct: 210 VYNILDQDDSQKGFSTAITPAAFNVSENVPPPMAEAGICYVQTGPNKGRPI----CSFCN 265
Query: 243 QFGHIARSCSKK------FCNYCKQRG---------HIITECYVRPPPSTQSPM------ 281
+ GHIA C KK F + K + + + PP S QSPM
Sbjct: 266 RVGHIAERCYKKHGFPPGFVSKYKSQSSGDRLQKPKQVAAQVSFSPPNSGQSPMTMDHLV 325
Query: 282 -----------*ALHAYSTTNATNGGVSQSEMIQQMVIS-------ALPSIGIQGKSSN- 322
AL + N T G S Q M S L IG+ S +
Sbjct: 326 GNHSKEQLQQFIALFSSQLPNVTMGSNEASSSKQPMDNSGISFNPTTLVFIGLLTVSRHT 385
Query: 323 -ASHPWFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDINSD----FR 377
A+ W +DSGA++H+ ++ + + +G + I+ VG + +
Sbjct: 386 LANETWIIDSGATHHVCHD-RSMYTSIDITTTSNVNLPNGMIVKISGVGIVQLNEHITLH 444
Query: 378 DVLVSPGLASNLLSVGQL 395
+VL P NLLS+ L
Sbjct: 445 NVLYIPEFRLNLLSISSL 462
>UniRef100_Q9FWZ5 Putative retroelement polyprotein [Arabidopsis thaliana]
Length = 1404
Score = 100 bits (248), Expect = 4e-19
Identities = 101/409 (24%), Positives = 174/409 (41%), Gaps = 39/409 (9%)
Query: 5 SQKQIDNMCVRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALEEWET----- 59
SQK I + ++ G NY W + + G LWSH+ S+AP E EE ET
Sbjct: 4 SQKVITTVILQ--GGNYLTWSRTTKTVLCGRGLWSHVIS-SQAPKEDKEEEETETISPEE 60
Query: 60 -----KDAQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIY-NQDNPAKRFQLELEIA 113
+D ++ + +++ ++ A+E+W+ LK +Y N+ N + F+++ I
Sbjct: 61 EKWFQEDQAVLALLQNSLETSILEGYSYCETAKELWDTLKNVYGNESNLTRVFEVKKAIN 120
Query: 114 NYKQGNSSVQEYYSGFLNLWTEHSAIIHADV-PNIYLAAVQEVYNTSKQDQFLMILRPEF 172
Q + +++ F +LW+E ++ + P I + E K L+ L P +
Sbjct: 121 ELSQEDLEFTKHFGKFRSLWSELKSLRPGTLDPKI----LHERREQDKVFGLLLTLNPGY 176
Query: 173 EVVRGSLLNRNVVPSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHD 232
+ LL +PSLD S++ +E+ G S LI + A + K D
Sbjct: 177 NDLIKHLLRSEKLPSLDEVCSKIQKEQGSTGLFGGKSE--LITANKGEVVANKGVYKNED 234
Query: 233 MQQVQCFSCKQFGHIARSC-----SKKFCNYCKQRGHIITECYVRPPPSTQSPM*ALHAY 287
+ + C CK+ GH C K + R H E + + S
Sbjct: 235 RKLLTCDHCKKKGHTKDKCWLLHPHLKPAKFKDSRAHFSQETHEEQSQAGSSK------- 287
Query: 288 STTNATNGGVSQSEMIQQMV--ISALPSIGIQGKSSNASHPWFLDSGASYHMTGSLEYLH 345
T+ + G + ++ ++ I +L GI S +S +DSGAS+HM + L
Sbjct: 288 GETSTSFGDYVRKSDLEALIKSIVSLKESGITFSSQTSSGSIVIDSGASHHMISNSNLLD 347
Query: 346 NLHSYDGNKKIQIADGNTLSITDVGDINSDFRD--VLVSPGLASNLLSV 392
N+ G+ + IA+G+ + I +G++ +D P SNLLSV
Sbjct: 348 NIEPALGH--VIIANGDKVPIEGIGNLKLFNKDSKAFFMPKFTSNLLSV 394
Score = 44.3 bits (103), Expect = 0.025
Identities = 23/56 (41%), Positives = 33/56 (58%), Gaps = 1/56 (1%)
Query: 553 KCFLYLRNF*HMLKLNFKQVLKFF-TDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
+ F NF + F +K F TD+GGEY S +F+++L +GI+ Q SCP TP
Sbjct: 552 RVFEAFTNFETYVTNQFNAKIKVFRTDNGGEYTSQKFRDHLAKRGIIHQTSCPYTP 607
>UniRef100_Q9FIC5 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1462
Score = 99.0 bits (245), Expect = 9e-19
Identities = 94/406 (23%), Positives = 171/406 (41%), Gaps = 32/406 (7%)
Query: 18 GKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTE-KAALEEWETKDAQIITWILSTINPQM 76
G NY W R+ +K + + +G P E ++W +A +++W+ TI+ +
Sbjct: 43 GPNYDEWATNLRLALKARKKFGFADGTIPQPDETNPDFDDWIANNALVVSWMKLTIHESL 102
Query: 77 INNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEH 136
++ + +MW ++++ + N + +L+ E+A +Q + ++ YY LW
Sbjct: 103 ATSMSHLDDSHDMWTHIQKRFGVKNGQRIQRLKTELATCRQKGTPIETYYGKLSQLWRSL 162
Query: 137 SAIIHADVPNIYLAAVQEVYNTSKQDQFLMIL-RPEFEVVRGSLLNRNVVPSLDTCVSEL 195
+ A + V++ K QFLM L + V+ +LL+R +PSL+ + L
Sbjct: 163 ADYQQAKT----MEEVRKEREEDKLHQFLMGLDESMYGAVKSALLSRVPLPSLEEAYNTL 218
Query: 196 LR-EEQRLLTQGAMSR-DALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSCSK 253
+ EE + L++ R D + + T + Q G V S K R S
Sbjct: 219 TQDEESKSLSRLHDERNDEKMRRNATSSSRNQSASMGRGSSVVPATSFKGKQSFGRGAS- 277
Query: 254 KFCNYCKQRGHIITECYVRPPPSTQSPM*ALHAYSTTNATNGGVS-----QSEMIQQMVI 308
N+ G ST + ++ T A G+S Q + ++QM+
Sbjct: 278 --ANHVANIGE-----------STTAATSSMSGSQLTEADRVGISGLNDEQWKQLRQMLK 324
Query: 309 SALPSIGIQGKSSNASHPWFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITD 368
+ S W +DSGA+ HMTG+LE+L ++ I++ DG + T
Sbjct: 325 ERNFNSTNTKSSKFFLESWIIDSGATNHMTGTLEFLRDVCDMP-PIMIKLPDGRLTTSTK 383
Query: 369 VGDI----NSDFRDVLVSPGLASNLLSVGQLVVILMLIFHVLVVLC 410
G + + D ++V GL +L+SV QL +F + +C
Sbjct: 384 HGRVYLGSSLDLQEVFFVDGLHCHLISVSQLTRAKSCVFQITDKVC 429
>UniRef100_Q5XWR5 Putative retroelement pol polyprotein-like [Solanum tuberosum]
Length = 1476
Score = 96.3 bits (238), Expect = 6e-18
Identities = 68/272 (25%), Positives = 123/272 (45%), Gaps = 20/272 (7%)
Query: 19 KNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALE--EWETKDAQIITWILSTINPQM 76
+NYS W ++ + + ++G + K LE +W+ +A +++W+++ ++ +
Sbjct: 25 ENYSLWSRAMQLTLLTKNKMGFIDGSLRRDDFKEELEKKQWDRCNAMVLSWLMNNVSTDL 84
Query: 77 INNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEH 136
++ + S A +WN LK +++ N ++ F L I + QG S V YYS +LW E+
Sbjct: 85 VSGILFRSNATLVWNDLKERFDKVNMSRIFHLHKAIVTHVQGVSPVSVYYSKLKDLWDEY 144
Query: 137 SAIIHADVPNIYLAA-VQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTCVSEL 195
+I+ + + + K QFLM L + R +L N PS++ C + +
Sbjct: 145 DSILPPPSCDCEKSVDYTDSMLRQKLLQFLMGLNDNYGQARSQILMMNPSPSVNQCYAMI 204
Query: 196 LREE-QRLLTQGAMSRDALIF------ESTTVAYAAQRRGKG---------HDMQQVQCF 239
+++E QR L+ + D S + +Q G G H + C
Sbjct: 205 VQDESQRSLSGSGQTIDPTALFTHRPGGSGFGSQGSQGSGNGSSNGNSHRFHKGGNIYCD 264
Query: 240 SCKQFGHIARSCSK-KFCNYCKQRGHIITECY 270
C GHI +C+K K C +C +GH CY
Sbjct: 265 FCNMKGHIRANCNKLKHCTHCNMQGHTKDTCY 296
Score = 38.1 bits (87), Expect = 1.8
Identities = 25/80 (31%), Positives = 39/80 (48%), Gaps = 4/80 (5%)
Query: 320 SSNASHPWFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDI----NSD 375
S+ S W +DSGA+ HM + L++ S K+Q+ G++ +T G
Sbjct: 411 SNTHSSAWIVDSGATDHMVSNTTLLNHGLSVSHPGKVQLPTGDSAVVTHSGSSQLTGGDV 470
Query: 376 FRDVLVSPGLASNLLSVGQL 395
++VL P NLLSV +L
Sbjct: 471 VKNVLCVPTFQFNLLSVSKL 490
Score = 36.2 bits (82), Expect = 6.8
Identities = 19/51 (37%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Query: 558 LRNF*HMLKLNFKQVLKFF-TDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
L+NF M+ F Q +K F +D+G E+ + + + GI+ Q SCP+TP
Sbjct: 654 LQNFILMIDTQFGQKIKIFRSDNGTEFFNAQCDGLFKSHGIVHQSSCPHTP 704
>UniRef100_Q9LGZ8 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1098
Score = 96.3 bits (238), Expect = 6e-18
Identities = 103/424 (24%), Positives = 168/424 (39%), Gaps = 58/424 (13%)
Query: 20 NYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALEEWETKDAQIITWILSTINPQMINN 79
NYS W + ++ ++ L+G PT + AL W+ ++ II WI ++I+P + +
Sbjct: 35 NYSEWAEELMNSLQAKQKLGFLDGTIPKPTTEPALSSWKAANSMIIGWIRTSIDPTIRST 94
Query: 80 LRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEHSAI 139
+ S A+++W+ LK+ ++ N ++ L+ EI KQ SV YY LW E
Sbjct: 95 VAFVSDAKDLWDSLKQRFSNGNGVRKQLLKDEILACKQDGQSVLVYYGRLTKLWEELQNY 154
Query: 140 IHADVPNIYLAA-VQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTCVSELLRE 198
+ A + + K QFL+ L F +R ++ ++ +P+L+ S ++ E
Sbjct: 155 KTSRTCTCEAAPDIAKEREDDKVHQFLLNLDERFRPIRSTITVQDPLPALNQVYSRVIHE 214
Query: 199 EQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSCSKKFCNY 258
EQ L SR ++ V + Q QV S +F R S C +
Sbjct: 215 EQNL----NASRIKDDIKTEAVGFTVQATPL-PPTPQVAAVSAPRF----RDRSSLTCTH 265
Query: 259 CKQRGHIITECYV-------------------------------------RPPPSTQSPM 281
++GH ITEC++ R S
Sbjct: 266 YHRQGHDITECFLVHGYPDWWLEQNGSNGSAGRGTSGRGNNGRGNNNRGGRSSSSGSRGK 325
Query: 282 *ALHAYSTTNATNGGVSQSEMIQQMVISALPSIGIQGKSSNASHPWF-----LDSGASYH 336
+A ST S ++ I Q+ IS L + S S F +D+GAS+H
Sbjct: 326 GRANAASTHPPPTSTPSNADQINQL-ISLLQAQNPATSSQKLSGKTFTTYVIIDTGASHH 384
Query: 337 MTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDINSD----FRDVLVSPGLASNLLSV 392
MTG + L N+ + + DG T G + DVL P L+SV
Sbjct: 385 MTGDITLLTNVEDIIPS-PVTKPDGTASRATKRGTLALHNAYVLPDVLFVPDFNCTLISV 443
Query: 393 GQLV 396
+L+
Sbjct: 444 AKLL 447
>UniRef100_Q6ZI27 Hypothetical protein OJ1004_H01.32 [Oryza sativa]
Length = 304
Score = 91.7 bits (226), Expect = 1e-16
Identities = 61/227 (26%), Positives = 110/227 (47%), Gaps = 13/227 (5%)
Query: 49 TEKAALEEWETKDAQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQL 108
T+ ++ W+T D +++ +I ++ + + R + A+EMW+YLK+ Y Q++ A RF L
Sbjct: 21 TDATKIKAWKTADDRVMGFIFMSVEVPIRMSFRDHTTAKEMWDYLKQRYTQESGALRFSL 80
Query: 109 ELEIANYKQGNSSVQEYYSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSK-QDQFLMI 167
+ N +Q + S++E+Y+ F L + A +E ++ QF+M
Sbjct: 81 LQNLHNLQQQDQSIEEFYNAFTRLSGQLEAQTPKGASGCAQCKAREKHDQENLVYQFVMR 140
Query: 168 LRPEFEVVRGSLLNRNVVPSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRR 227
L +FE +R LL R P++ +++L+ EE RL + + + V A QR
Sbjct: 141 LSSQFESIRVQLLGRPTRPTMAEALADLIAEETRLCSLDTTPTPVV---THNVMAAPQRV 197
Query: 228 GKGHDMQQVQCFSCKQFGHIARSCSKKFCNYCKQRGHIITECYVRPP 274
G +F IA+ S + C++CK+ GHI +C+ P
Sbjct: 198 G-------APMGGIPEFSSIAK--SDRICSHCKKVGHISDDCFYLHP 235
>UniRef100_O82331 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1149
Score = 91.3 bits (225), Expect = 2e-16
Identities = 105/427 (24%), Positives = 174/427 (40%), Gaps = 77/427 (18%)
Query: 1 YTMDSQKQIDNMCVRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPT-------EKAA 53
YT+ S + + V+ + +NY W+ QF ++ G+ L +NG APT +
Sbjct: 8 YTLPSLNISNCVTVKLTDRNYILWKSQFESFLSGQGLLGFVNGAYAAPTGTVSGPQDAGV 67
Query: 54 LEEWETKDAQIITWILS---TINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLEL 110
E D Q W S ++ +++ + + E+W L + +N+ + ++ F+L+
Sbjct: 68 TEAIPNPDYQ--AWFRSDQVVMSEDILSVVVGSKTSHEVWMNLAKHFNRISSSRIFELQR 125
Query: 111 EIANYKQGNSSVQEYYSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQDQFLMI--L 168
+ + + +++EY +L + A + + V K F M+ L
Sbjct: 126 RLHSLSKEGKTMEEYLR-YLKTICDQLASVGSPV-------------AEKMKIFAMVHGL 171
Query: 169 RPEFEVVRGSL---LNRNVVPSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQ 225
E+E + SL L+ PS + V L + RL QG D + ++
Sbjct: 172 TREYEPLITSLEGTLDAFPGPSYEDVVYRLKNFDDRL--QGYTVTDVSPHLAFNTFRSSN 229
Query: 226 R-------RGKGHDMQQVQCFSCKQFGHIARSCS---KKFCNYCKQRGHIITECYVRPPP 275
R RGKG+ + + F +QF + S S K C C +RGH +C+ R
Sbjct: 230 RGRGGRNNRGKGNFSTRGRGFQ-QQFSSSSSSVSASEKPMCQICGKRGHYALQCWHRFDD 288
Query: 276 STQSPM*ALHAYSTTNATNGGVSQSEMIQQMVISALPSIGIQGKSSNASHPWFLDSGASY 335
S Q A A+S + T+ VS W DS A+
Sbjct: 289 SYQHSEAAAAAFSALHITD--VSDDS------------------------GWVPDSAATA 322
Query: 336 HMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDI-------NSDFRDVLVSPGLASN 388
H+T + L + Y GN + +DGN L IT +G N +DVLV P +A +
Sbjct: 323 HITNNSSRLQQMQPYLGNDTVMASDGNFLPITHIGSANLPSTSGNLPLKDVLVCPNIAKS 382
Query: 389 LLSVGQL 395
LLSV +L
Sbjct: 383 LLSVSKL 389
Score = 38.1 bits (87), Expect = 1.8
Identities = 19/54 (35%), Positives = 31/54 (57%)
Query: 554 CFLYLRNF*HMLKLNFKQVLKFFTDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
C L+++ + L ++ F +D GGE+ S+ F ++LQ GI SCP+TP
Sbjct: 549 CSLFMKFQSFVENLLQTKIGTFQSDGGGEFTSNRFLQHLQESGIQHYISCPHTP 602
>UniRef100_Q851Y3 Putative polyprotein [Oryza sativa]
Length = 1299
Score = 89.4 bits (220), Expect = 7e-16
Identities = 83/347 (23%), Positives = 159/347 (44%), Gaps = 33/347 (9%)
Query: 19 KNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALE--EWETKDAQIITWILSTINPQM 76
KNY +W + + +K + L ++ G K P +++E W T ++ ++ W+L+++ P +
Sbjct: 55 KNYLSWSRRALLILKTKGLEGYVTGEIKEPENISSVEWKTWSTTNSLVVAWLLTSLIPAI 114
Query: 77 INNLRSFSFAQEMWNYLKRIYN-QDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWT- 134
+ + S A EMW L +Y+ + N + + +I+ +QG SV EY + +LW+
Sbjct: 115 ATTVETISSASEMWKTLTNLYSGEGNVMLMVEAQEKISVLRQGERSVAEYVAELKHLWSD 174
Query: 135 -EHSAIIHADVPNIYLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTCVS 193
+H + + P+ +A +++ + +FL L EFE R ++ ++ +PSLD ++
Sbjct: 175 LDHYDPLGLEHPDC-IAKMRKWIERRRVIEFLKGLNSEFEGRRDAMFHQTTLPSLDEAIA 233
Query: 194 ELLREE--QRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSC 251
+ +EE +++L S + + VA + + R +CF+C + GH+ R
Sbjct: 234 AMAQEELKKKVLPSATPSSPSPTY---VVAQSKETR---------ECFNCGEMGHLIRDY 281
Query: 252 -SKKFCNYCKQRGHIITECYVRPPPSTQSPM*ALHAYSTTNATNGGV---------SQSE 301
+ + +Y RG R Y + V S +
Sbjct: 282 RAPRKPSY--GRGRFGDRGGARGGRGYAGRGNRGRGYEYRSDHRANVVTLEESCSGSTNV 339
Query: 302 MIQQMVISALPSIGIQGKSSNASHP-WFLDSGASYHMTGSLEYLHNL 347
+ +V S+ + S N+SH W LDSGAS H+T +L + +L
Sbjct: 340 DVANLVHSSSGNSNQAFMSINSSHSNWILDSGASRHVTVNLVSISSL 386
Score = 47.4 bits (111), Expect = 0.003
Identities = 19/32 (59%), Positives = 26/32 (80%)
Query: 577 TDSGGEYMSHEFQEYLQHKGILSQ*SCPNTPP 608
TD+GGEY+++EF +L +GIL Q SCP+TPP
Sbjct: 451 TDNGGEYINNEFSSFLSSEGILHQTSCPDTPP 482
>UniRef100_Q9SLL4 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1402
Score = 88.6 bits (218), Expect = 1e-15
Identities = 98/445 (22%), Positives = 173/445 (38%), Gaps = 91/445 (20%)
Query: 1 YTMDSQKQIDNMCVRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALEE---- 56
Y++ S + + V + KNY W+ QF ++ G+ L + G AP++ + + +
Sbjct: 4 YSVPSLNISNCVTVTLTAKNYILWKSQFESFLDGQGLLGFVTGSIPAPSQTSVVSDIDGS 63
Query: 57 -----------WETKDAQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKR 105
W D + +W+L + +++ + + + + E+W + +N+ + ++
Sbjct: 64 TSASPNPEYYTWFKTDRVVKSWLLGSFLEDILSVVVNCNTSHEVWISVANHFNRVSSSRL 123
Query: 106 FQLELEIANYKQGNSSVQEYYSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQDQFL 165
F+L+ + N + + S+ EY + + LA+V T K F
Sbjct: 124 FELQRRLQNVSKRDKSMDEYLKDLKTICDQ-------------LASVGSPV-TEKMKIFA 169
Query: 166 MI--LRPEFEVVRGSLLNRNVV---PSLDTCVSELLREEQRLLTQGAMSRDA----LIFE 216
+ L E+E ++ ++ N PSL+ + +L + RL QG + A + F
Sbjct: 170 ALNGLGREYEPIKTTIENSMDALPGPSLEDVIPKLTGYDDRL--QGYLEETAVSPHVAFN 227
Query: 217 STTV-------AYAAQRRGKGHDMQQVQCFSCKQFGHIARSCSKK------------FCN 257
TT + A RGKG + FS + G + S C
Sbjct: 228 ITTSDDSNASGYFNAYNRGKGKSNRGRNSFSTRGRGFHQQISSTNSSSGSQSGGTSVVCQ 287
Query: 258 YCKQRGHIITECYVRPPPSTQSPM*ALHAYSTTNATNGGVSQSEMIQQMVISALPSIGIQ 317
C + GH +C+ R N E+ + AL ++ I
Sbjct: 288 ICGKMGHPALKCWHR--------------------FNNSYQYEELPR-----ALAAMRIT 322
Query: 318 GKSSNASHPWFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDI----- 372
+ + W DS A+ H+T S L Y G+ + +ADGN L IT G
Sbjct: 323 DITDQHGNEWLPDSAATAHVTNSPRSLQQSQPYHGSDAVMVADGNFLPITHTGSTNLASS 382
Query: 373 --NSDFRDVLVSPGLASNLLSVGQL 395
N DVLV P + +LLSV +L
Sbjct: 383 SGNVPLTDVLVCPSITKSLLSVSKL 407
Score = 37.0 bits (84), Expect = 4.0
Identities = 18/42 (42%), Positives = 28/42 (65%), Gaps = 1/42 (2%)
Query: 566 KLNFKQVLKFFTDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
+LN K + F D GGE+++H+F ++LQ+ GI S P+TP
Sbjct: 580 QLNHK-ISVFQCDGGGEFVNHKFLQHLQNHGIQQHISYPHTP 620
>UniRef100_Q94IU9 Copia-like polyprotein [Arabidopsis thaliana]
Length = 1466
Score = 87.4 bits (215), Expect = 3e-15
Identities = 94/430 (21%), Positives = 170/430 (38%), Gaps = 91/430 (21%)
Query: 11 NMCVRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAAL--------------EE 56
++ ++ + NY W+ QF + ++L +NGV P + + E+
Sbjct: 16 SVTLKLNDSNYLLWKTQFESLLSSQKLIGFVNGVVTPPAQTRLVVNDDVTSEVPNPQYED 75
Query: 57 WETKDAQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYK 116
W D + +W+ T++ +++ ++ + + ++++W L +N+ + A+ F L +
Sbjct: 76 WFCTDQLVRSWLFGTLSEEVLGHVHNLTTSRQIWISLAENFNKSSIAREFSLRRNLQLLT 135
Query: 117 QGNSSVQEYYSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQDQFLMILRPEFE--- 173
+ + S+ Y F + S+I + V + K FL L E++
Sbjct: 136 KKDKSLSVYCRDFKIICDSLSSI------------GKPVEESMKIFGFLNGLGREYDPIT 183
Query: 174 VVRGSLLNRNVVPSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAY----------- 222
V S L++ P+ + +SE+ + +L S D + + +A+
Sbjct: 184 TVIQSSLSKLPAPTFNDVISEVQGFDSKL-----QSYDDTVSVNPHLAFNTERSNSGAPQ 238
Query: 223 ----------AAQRRGKGHDMQQVQCFSCKQFGHIARSCSKKFCNYCKQRGHIITECYVR 272
+ Q RG+G + + FS Q S + C C + GH +CY R
Sbjct: 239 YNSNSRGRGRSGQNRGRGGYSTRGRGFSQHQSAS-PSSGQRPVCQICGRIGHTAIKCYNR 297
Query: 273 PPPSTQSPM*ALHAYSTTNATNGGVSQSEMIQQMVISALPSIGIQGKSSNASHPWFLDSG 332
+ QS + SAL GK W+ DS
Sbjct: 298 FDNNYQSE----------------------VPTQAFSALRVSDETGKE------WYPDSA 329
Query: 333 ASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGD--INSD-----FRDVLVSPGL 385
A+ H+T S L N +Y+GN + + DG L IT VG I+S +VLV P +
Sbjct: 330 ATAHITASTSGLQNATTYEGNDAVLVGDGTYLPITHVGSTTISSSKGTIPLNEVLVCPAI 389
Query: 386 ASNLLSVGQL 395
+LLSV +L
Sbjct: 390 QKSLLSVSKL 399
>UniRef100_Q6I5B6 Putative polyprotein [Oryza sativa]
Length = 1204
Score = 86.3 bits (212), Expect = 6e-15
Identities = 97/439 (22%), Positives = 168/439 (38%), Gaps = 80/439 (18%)
Query: 14 VRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAP------------------------- 48
V F G NYS W R++++G+RLW L+ P
Sbjct: 15 VLFDGCNYSHWAQHMRLHMRGQRLWDVLSSELPCPPCPIAPTMPSLASQATDDDREKAKE 74
Query: 49 ----------TEKAALEEWETKDAQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIYN 98
++ A + W +DA+ +++++ + + + + A MW +L Y
Sbjct: 75 QFDDAMENYQSQFALYKAWLDEDARASAILVASMEIHLTGEVVTLTSAHLMWTHLHDRYA 134
Query: 99 QDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEHSAI---------------IHAD 143
+ A + + + +QG+S+V E+Y+ ++W + ++ H D
Sbjct: 135 PTSDALYLAMVRQEQSLQQGDSTVDEFYTQLSSIWRQLDSLGPTICHTYPCCQRQRSHMD 194
Query: 144 VPNIYLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTCVSELLREEQRLL 203
+ IY FL LR E+E R LL+R+ ++ ++E+ EE RL
Sbjct: 195 LRRIY--------------DFLTRLRSEYESTRAQLLSRHPRVTIMEALTEIRSEEIRLR 240
Query: 204 TQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSC-SKKFCNYCKQR 262
G + + + TVA +A H S + + S C YC +
Sbjct: 241 EAGILPLPSSVLAVRTVASSASSTPAVHSTVSSSSSSARPPTTVVPSTRGHLHCTYCDKD 300
Query: 263 GHIITECYVRPPPSTQSPM*ALHAYSTTNATNGGVSQSEMIQQM----VISALPSIGIQG 318
GH+ + C+ R + + ++ ++GG E++ + +A S+G
Sbjct: 301 GHVESFCF-RKKKDLRRGNSSKGTSGSSQKSSGGSDSQEILMLLRRLTASAATGSVGSVA 359
Query: 319 KSSNASHPWFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDINSDFRD 378
S S L S +S + S +H+ DG + I TLSI S F
Sbjct: 360 LPSAQSGSAVLGSSSSTEGSSSASVPTTVHTADGT-PLAIVGRGTLSI-------SSFSV 411
Query: 379 VLVS--PGLASNLLSVGQL 395
VS P LA L+S GQL
Sbjct: 412 PAVSYVPKLAMQLMSAGQL 430
Score = 40.8 bits (94), Expect = 0.28
Identities = 19/47 (40%), Positives = 30/47 (63%), Gaps = 1/47 (2%)
Query: 559 RNF*HMLKLNFKQVLKFF-TDSGGEYMSHEFQEYLQHKGILSQ*SCP 604
++F M++ +F ++ F DS GEY+S E + +L +G LSQ SCP
Sbjct: 603 KSFARMIRTHFDSPIRVFRADSAGEYLSRELRVFLSEQGTLSQFSCP 649
>UniRef100_Q9MAJ8 F27F5.19 [Arabidopsis thaliana]
Length = 1309
Score = 85.9 bits (211), Expect = 8e-15
Identities = 62/244 (25%), Positives = 113/244 (45%), Gaps = 8/244 (3%)
Query: 18 GKNYSAWEFQFRMYVKGERLWSHLNGVSKAP-TEKAALEEWETKDAQIITWILSTINPQM 76
G +Y+ W RM + + S ++G P W ++ + TW+L+ ++ ++
Sbjct: 87 GTSYNNWSIAMRMSLDAKNKLSFVDGSLPRPDVSDRMFRIWSRCNSMVKTWLLNVVSKEI 146
Query: 77 INNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTE- 135
+++ + A EMWN L + N +++QLE I KQ N + YY+ LW +
Sbjct: 147 YDSILYYEDAVEMWNDLFSRFRVSNLPRKYQLEQSIHTLKQRNLDLSTYYTKKKTLWVQL 206
Query: 136 -HSAIIHADVPNI-YLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTCVS 193
++ ++ N ++ + E TS+ QFLM L F +RG +LN P L +
Sbjct: 207 ANTRVLTVRKCNCDHVKELLEEAETSRIIQFLMGLNDNFAHIRGQILNMKPRPGLTEIYN 266
Query: 194 ELLREE-QRLLTQGAMSRDALIF--ESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARS 250
L ++E QRL+ +S F +++ V + +G ++ +C C + GH+
Sbjct: 267 MLDQDESQRLVGSTPLSNLTAAFQVQASPVIDSQVNMAQG-SYKKPKCSFCNKLGHLVDK 325
Query: 251 CSKK 254
C KK
Sbjct: 326 CYKK 329
>UniRef100_Q9FKA8 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 370
Score = 85.9 bits (211), Expect = 8e-15
Identities = 65/268 (24%), Positives = 124/268 (46%), Gaps = 27/268 (10%)
Query: 14 VRFSGKNYSAWEFQFRMYVKGERLWSHLNG-VSKAPTEKAALEEWETKDAQIITWILSTI 72
V +G NY+ W + ++ +R ++G + K ++ E W+T ++ I+ WI +I
Sbjct: 45 VVLNGDNYNEWSEEMLNALQAKRKTGFIDGTIQKPASDSPDFENWKTVNSMIVGWIRVSI 104
Query: 73 NPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNL 132
P++ + + S A +W+ L++ ++ N + Q++ ++A+ +Q +V +YY NL
Sbjct: 105 EPKVKSTVTFISDAHLLWDELRQRFSVTNNVRVHQIKAQLASCRQEGQTVIDYYGRLCNL 164
Query: 133 WTE-----HSAII-HADVPNIYLAAVQEVYNTSKQDQFLMIL-RPEFEVVRGSLLNRNVV 185
W E SA+ H V L A+ + + K QF++ L F + +L+N + +
Sbjct: 165 WDELKNYQASAVCPHGSV----LTAIVKERDDEKLHQFVLGLDSARFSGLCTNLINMDPL 220
Query: 186 PSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFG 245
PSL S+++REEQR+ + V + A+ +Q S
Sbjct: 221 PSLGVAYSQVIREEQRIHASRTQEQ-----RQEVVGFVARH-------EQSSAMSSPAQS 268
Query: 246 HIARSCSKK---FCNYCKQRGHIITECY 270
I S K C++C + GH +C+
Sbjct: 269 SIESSIVKSRPVLCSHCGRTGHEKKDCW 296
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.347 0.154 0.508
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,917,992,284
Number of Sequences: 2790947
Number of extensions: 74488889
Number of successful extensions: 346807
Number of sequences better than 10.0: 2843
Number of HSP's better than 10.0 without gapping: 186
Number of HSP's successfully gapped in prelim test: 2659
Number of HSP's that attempted gapping in prelim test: 339809
Number of HSP's gapped (non-prelim): 6551
length of query: 1309
length of database: 848,049,833
effective HSP length: 139
effective length of query: 1170
effective length of database: 460,108,200
effective search space: 538326594000
effective search space used: 538326594000
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 81 (35.8 bits)
Medicago: description of AC146758.11


