

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.7 - phase: 0
(367 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
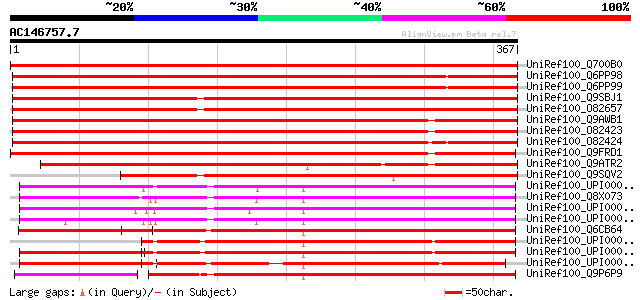
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q700B0 Pyruvate dehydrogenase kinase [Cicer arietinum] 690 0.0
UniRef100_Q6PP98 Mitochondrial pyruvate dehydrogenase kinase iso... 659 0.0
UniRef100_Q6PP99 Mitochondrial pyruvate dehydrogenase kinase iso... 653 0.0
UniRef100_Q9SBJ1 Pyruvate dehydrogenase kinase [Arabidopsis thal... 617 e-175
UniRef100_O82657 Pyruvate dehydrogenase kinase [Arabidopsis thal... 611 e-173
UniRef100_Q9AWB1 Pyruvate dehydrogenase kinase 1 [Oryza sativa] 558 e-157
UniRef100_O82423 Pyruvate dehydrogenase kinase isoform 1 [Zea mays] 543 e-153
UniRef100_O82424 Pyruvate dehydrogenase kinase isoform 2 [Zea mays] 540 e-152
UniRef100_Q9FRD1 Putative pyruvate dehydrogenase kinase [Oryza s... 536 e-151
UniRef100_Q9ATR2 Pyruvate dehydrogenase kinase [Oryza sativa] 494 e-138
UniRef100_Q9SQV2 Putative pyruvate dehydrogenase kinase, 5' part... 481 e-134
UniRef100_UPI000023574E UPI000023574E UniRef100 entry 305 1e-81
UniRef100_Q8X073 Related to pyruvate dehydrogenase kinase isofor... 305 1e-81
UniRef100_UPI000023D197 UPI000023D197 UniRef100 entry 302 1e-80
UniRef100_UPI000021955D UPI000021955D UniRef100 entry 296 6e-79
UniRef100_Q6CB64 Similar to tr|Q8X073 Neurospora crassa Related ... 257 3e-67
UniRef100_UPI000042E26C UPI000042E26C UniRef100 entry 254 2e-66
UniRef100_UPI000042CAFB UPI000042CAFB UniRef100 entry 254 2e-66
UniRef100_UPI000042F396 UPI000042F396 UniRef100 entry 246 9e-64
UniRef100_Q9P6P9 SPAC644.11c protein [Schizosaccharomyces pombe] 243 6e-63
>UniRef100_Q700B0 Pyruvate dehydrogenase kinase [Cicer arietinum]
Length = 367
Score = 690 bits (1780), Expect = 0.0
Identities = 339/367 (92%), Positives = 357/367 (96%)
Query: 1 MAAIEAFSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIAR 60
MAAIEAFSKSL++EVHKWGCLKQTGVSLRYMMEFGS+PTDKNLLISAQFLQKELAIRIAR
Sbjct: 1 MAAIEAFSKSLIEEVHKWGCLKQTGVSLRYMMEFGSKPTDKNLLISAQFLQKELAIRIAR 60
Query: 61 RAIELESLPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNN 120
RAIELE+LPYGLS+KPA+LKVRDWYVDSFRDIRS PE+K++ DE+EFT+VIKAIKVRHNN
Sbjct: 61 RAIELETLPYGLSQKPAVLKVRDWYVDSFRDIRSFPEIKNINDEKEFTEVIKAIKVRHNN 120
Query: 121 VVPTMALGVQQLKKELKTKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPH 180
VVPTMALGVQQLKK LK + +ED VEIH+FLDRFYLSRIG+RMLIGQHVELHNPNPPP+
Sbjct: 121 VVPTMALGVQQLKKGLKPNMVNEDFVEIHQFLDRFYLSRIGIRMLIGQHVELHNPNPPPY 180
Query: 181 VVGYIHTKMSPVSVARNASEDARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFEL 240
VVGYIHTKMSPV VARNASEDARSIC+REYGSAP+INIYGDPDFTFPYVPAHLHLMVFEL
Sbjct: 181 VVGYIHTKMSPVEVARNASEDARSICLREYGSAPDINIYGDPDFTFPYVPAHLHLMVFEL 240
Query: 241 VKNSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTA 300
VKNSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGI RSGLPKIFTYLYSTA
Sbjct: 241 VKNSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIARSGLPKIFTYLYSTA 300
Query: 301 RNPLDEHADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLG 360
RNPLDEH DLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLG
Sbjct: 301 RNPLDEHEDLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLG 360
Query: 361 DSQEPLP 367
DSQEPLP
Sbjct: 361 DSQEPLP 367
>UniRef100_Q6PP98 Mitochondrial pyruvate dehydrogenase kinase isoform 2 [Glycine max]
Length = 369
Score = 659 bits (1699), Expect = 0.0
Identities = 318/365 (87%), Positives = 349/365 (95%), Gaps = 1/365 (0%)
Query: 3 AIEAFSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRA 62
A E FSKSL++EVH+WGCLKQTGVSLRYMMEFGS+PTDKNLLISAQFL KELAIRIARRA
Sbjct: 6 ACETFSKSLIEEVHRWGCLKQTGVSLRYMMEFGSKPTDKNLLISAQFLHKELAIRIARRA 65
Query: 63 IELESLPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNNVV 122
IELE+LPYGLS+KPA+LKVRDWYVDSFRD+R+ P +K++ DE++FT++IKAIKVRHNNVV
Sbjct: 66 IELENLPYGLSQKPAVLKVRDWYVDSFRDLRAFPNIKNVNDEQDFTEMIKAIKVRHNNVV 125
Query: 123 PTMALGVQQLKKELKTKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHVV 182
PTMALGVQQLKK + KI EDLVEIH+FLDRFY+SRIG+RMLIGQHVELHNPNPPPHVV
Sbjct: 126 PTMALGVQQLKKRMDPKIVYEDLVEIHQFLDRFYMSRIGIRMLIGQHVELHNPNPPPHVV 185
Query: 183 GYIHTKMSPVSVARNASEDARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFELVK 242
GYIHTKMSPV VARNASEDAR+IC REYGSAP+++IYGDPDFTFPYVPAHLHLMVFELVK
Sbjct: 186 GYIHTKMSPVEVARNASEDARAICCREYGSAPDVHIYGDPDFTFPYVPAHLHLMVFELVK 245
Query: 243 NSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARN 302
NSLRAVQER+MDSDKV+PPIRIIVADG+EDVTIK+SDEGGGI RSGLPKIFTYLYSTARN
Sbjct: 246 NSLRAVQERFMDSDKVAPPIRIIVADGIEDVTIKVSDEGGGIARSGLPKIFTYLYSTARN 305
Query: 303 PLDEHADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS 362
PLDEH+DLG+ D+V TMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS
Sbjct: 306 PLDEHSDLGIGDNV-TMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS 364
Query: 363 QEPLP 367
QEPLP
Sbjct: 365 QEPLP 369
>UniRef100_Q6PP99 Mitochondrial pyruvate dehydrogenase kinase isoform 1 [Glycine max]
Length = 369
Score = 653 bits (1685), Expect = 0.0
Identities = 315/365 (86%), Positives = 350/365 (95%), Gaps = 1/365 (0%)
Query: 3 AIEAFSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRA 62
A E FSKSL++EV++WGCLKQTGVSLRYMMEFGS+PT+KNLLISAQFL KELAIRIARRA
Sbjct: 6 ACETFSKSLIEEVNRWGCLKQTGVSLRYMMEFGSKPTNKNLLISAQFLHKELAIRIARRA 65
Query: 63 IELESLPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNNVV 122
+ELE+LPYGLS+KPA+LKVRDWYVDSFRD+R+ P++K++ DEREFT++IKAIKVRHNNVV
Sbjct: 66 VELENLPYGLSQKPAVLKVRDWYVDSFRDVRAFPDIKNVNDEREFTEMIKAIKVRHNNVV 125
Query: 123 PTMALGVQQLKKELKTKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHVV 182
PTMA+GVQQLKK + KI EDLVEIH+FLDRFY+SRIG+RMLIGQHVELHNPNPPPHVV
Sbjct: 126 PTMAMGVQQLKKGMDPKIVYEDLVEIHQFLDRFYMSRIGIRMLIGQHVELHNPNPPPHVV 185
Query: 183 GYIHTKMSPVSVARNASEDARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFELVK 242
GYIHTKMSPV VARNASEDARSIC REYGSAP+++IYGDP+FTFPYVPAHLHLMVFELVK
Sbjct: 186 GYIHTKMSPVEVARNASEDARSICCREYGSAPDVHIYGDPNFTFPYVPAHLHLMVFELVK 245
Query: 243 NSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARN 302
NSLRAVQER+M+SDKV+PPIRIIVADG+EDVTIK+SDEGGGI RSGLPKIFTYLYSTARN
Sbjct: 246 NSLRAVQERFMNSDKVAPPIRIIVADGIEDVTIKVSDEGGGIARSGLPKIFTYLYSTARN 305
Query: 303 PLDEHADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS 362
PLDEH+DLG+ D+V TMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS
Sbjct: 306 PLDEHSDLGIGDNV-TMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS 364
Query: 363 QEPLP 367
QEPLP
Sbjct: 365 QEPLP 369
>UniRef100_Q9SBJ1 Pyruvate dehydrogenase kinase [Arabidopsis thaliana]
Length = 366
Score = 617 bits (1590), Expect = e-175
Identities = 297/365 (81%), Positives = 334/365 (91%), Gaps = 4/365 (1%)
Query: 3 AIEAFSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRA 62
A E F KSL+++VHKWGC+KQTGVSLRYMMEFGS+PT++NLLISAQFL KEL IR+ARRA
Sbjct: 6 ACEMFPKSLIEDVHKWGCMKQTGVSLRYMMEFGSKPTERNLLISAQFLHKELPIRVARRA 65
Query: 63 IELESLPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNNVV 122
IEL++LPYGLS KPA+LKVRDWY++SFRD+R+ PE+KD DE++FT +IKA+KVRHNNVV
Sbjct: 66 IELQTLPYGLSDKPAVLKVRDWYLESFRDMRAFPEIKDSGDEKDFTQMIKAVKVRHNNVV 125
Query: 123 PTMALGVQQLKKELKTKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHVV 182
P MALGV QLKK + +S +L EIH+FLDRFYLSRIG+RMLIGQHVELHNPNPP H V
Sbjct: 126 PMMALGVNQLKKGM----NSGNLDEIHQFLDRFYLSRIGIRMLIGQHVELHNPNPPLHTV 181
Query: 183 GYIHTKMSPVSVARNASEDARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFELVK 242
GYIHTKMSP+ VARNASEDARSIC REYGSAPEINIYGDP FTFPYVP HLHLM++ELVK
Sbjct: 182 GYIHTKMSPMEVARNASEDARSICFREYGSAPEINIYGDPSFTFPYVPTHLHLMMYELVK 241
Query: 243 NSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARN 302
NSLRAVQER++DSD+V+PPIRIIVADG+EDVTIK+SDEGGGI RSGLP+IFTYLYSTARN
Sbjct: 242 NSLRAVQERFVDSDRVAPPIRIIVADGIEDVTIKVSDEGGGIARSGLPRIFTYLYSTARN 301
Query: 303 PLDEHADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS 362
PL+E DLG+AD TMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS
Sbjct: 302 PLEEDVDLGIADVPVTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS 361
Query: 363 QEPLP 367
QEPLP
Sbjct: 362 QEPLP 366
>UniRef100_O82657 Pyruvate dehydrogenase kinase [Arabidopsis thaliana]
Length = 366
Score = 611 bits (1575), Expect = e-173
Identities = 295/365 (80%), Positives = 332/365 (90%), Gaps = 4/365 (1%)
Query: 3 AIEAFSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRA 62
A E F KSL+++VHKWGC+KQTGVSLRYMMEFGS+PT++NLLISAQFL KEL IR+ARRA
Sbjct: 6 ACEMFPKSLIEDVHKWGCMKQTGVSLRYMMEFGSKPTERNLLISAQFLHKELPIRVARRA 65
Query: 63 IELESLPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNNVV 122
IEL++LPYGLS KPA+LKVRDWY++SFRD+R+ PE+KD DE++FT +IKA+KVRHNNVV
Sbjct: 66 IELQTLPYGLSDKPAVLKVRDWYLESFRDMRAFPEIKDSGDEKDFTQMIKAVKVRHNNVV 125
Query: 123 PTMALGVQQLKKELKTKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHVV 182
P MALGV QLKK + +S +L EIH+FLDRFYLSRIG+RMLIGQHVELHNPNPP H V
Sbjct: 126 PMMALGVNQLKKGM----NSGNLDEIHQFLDRFYLSRIGIRMLIGQHVELHNPNPPLHTV 181
Query: 183 GYIHTKMSPVSVARNASEDARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFELVK 242
GYIHTKMSP+ VARNASEDARSIC REYGSAPEINIYGDP FTFPYVP HL LM++ELVK
Sbjct: 182 GYIHTKMSPMEVARNASEDARSICFREYGSAPEINIYGDPSFTFPYVPTHLDLMMYELVK 241
Query: 243 NSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARN 302
NSLRAVQER++DSD+V+PPIRIIVADG+EDVTIK+SDEGGGI RSGLP+IFTYLYSTARN
Sbjct: 242 NSLRAVQERFVDSDRVAPPIRIIVADGIEDVTIKVSDEGGGIARSGLPRIFTYLYSTARN 301
Query: 303 PLDEHADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS 362
PL+E DLG+AD TM GYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS
Sbjct: 302 PLEEDVDLGIADVPGTMGGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS 361
Query: 363 QEPLP 367
QEPLP
Sbjct: 362 QEPLP 366
>UniRef100_Q9AWB1 Pyruvate dehydrogenase kinase 1 [Oryza sativa]
Length = 363
Score = 558 bits (1437), Expect = e-157
Identities = 277/366 (75%), Positives = 318/366 (86%), Gaps = 5/366 (1%)
Query: 3 AIEAFSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRA 62
A E ++++ +EV +WG +KQTGVSLRYMMEFGS+PT++NLL+SAQFLQKEL IRIARRA
Sbjct: 2 ASEPVARAVAEEVGRWGSMKQTGVSLRYMMEFGSRPTERNLLLSAQFLQKELPIRIARRA 61
Query: 63 IELESLPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNNVV 122
+ELESLP+GLS+KPAILKVRDWY+DSFRDIR PEV++ DE FT +IK IKVRHNNVV
Sbjct: 62 LELESLPFGLSRKPAILKVRDWYLDSFRDIRYFPEVRNRNDELAFTQMIKMIKVRHNNVV 121
Query: 123 PTMALGVQQLKKE-LKTKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHV 181
PTMALGVQQLK E +T+ EIHEFLDRFY+SRIG+RMLIGQHV LH+P+P P V
Sbjct: 122 PTMALGVQQLKNEQYRTRKIPTAFDEIHEFLDRFYMSRIGIRMLIGQHVALHDPDPEPGV 181
Query: 182 VGYIHTKMSPVSVARNASEDARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFELV 241
+G I+T++SP+ VA+ ASEDARSIC+REYGSAPEI+IYGDP FTFPYV +HLHLM+FELV
Sbjct: 182 IGLINTELSPIQVAQAASEDARSICLREYGSAPEIDIYGDPTFTFPYVSSHLHLMLFELV 241
Query: 242 KNSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTAR 301
KNSLRAVQERYM+SDK PP+RIIVADG EDVTIK+SDEGGGIPRSGLP+IFTYLYSTA+
Sbjct: 242 KNSLRAVQERYMNSDKDVPPVRIIVADGAEDVTIKVSDEGGGIPRSGLPRIFTYLYSTAK 301
Query: 302 NPLDEHADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGD 361
NP D+ TMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGD
Sbjct: 302 NP----PDMDCPSEGVTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGD 357
Query: 362 SQEPLP 367
S+EPLP
Sbjct: 358 SEEPLP 363
>UniRef100_O82423 Pyruvate dehydrogenase kinase isoform 1 [Zea mays]
Length = 363
Score = 543 bits (1400), Expect = e-153
Identities = 267/366 (72%), Positives = 317/366 (85%), Gaps = 5/366 (1%)
Query: 3 AIEAFSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRA 62
A E ++++ +EV +WG ++QTGVSLRYMMEFG++PT++ LL++AQFL KEL IRIARRA
Sbjct: 2 ASEPVARAVAEEVARWGAMRQTGVSLRYMMEFGARPTERTLLLAAQFLHKELPIRIARRA 61
Query: 63 IELESLPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNNVV 122
++L+SLP+GLS KPAILKV+DWYV+SFR+IRS PEV++ KDE FT +IK I+VRH NVV
Sbjct: 62 LDLDSLPFGLSTKPAILKVKDWYVESFREIRSFPEVRNQKDELAFTQMIKMIRVRHTNVV 121
Query: 123 PTMALGVQQLKKELK-TKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHV 181
P +ALGVQQLKK+L K + EIH+FLDRFY+SRIG+RMLIGQHV LH+P+P P V
Sbjct: 122 PAIALGVQQLKKDLGGPKAFPPGIHEIHQFLDRFYMSRIGIRMLIGQHVALHDPDPEPGV 181
Query: 182 VGYIHTKMSPVSVARNASEDARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFELV 241
+G I+TKMSP++VAR ASEDAR+ICMREYGS+P+++IYGDP FTFPYV HLHLM+FELV
Sbjct: 182 IGLINTKMSPMTVARIASEDARAICMREYGSSPDVDIYGDPGFTFPYVTPHLHLMIFELV 241
Query: 242 KNSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTAR 301
KNSLRAVQERYMDSDK++PP+RIIVADG EDVTIKISDEGGGIPRSGL +IFTYLYSTA
Sbjct: 242 KNSLRAVQERYMDSDKLAPPVRIIVADGAEDVTIKISDEGGGIPRSGLSRIFTYLYSTAE 301
Query: 302 NPLDEHADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGD 361
NP DL + TMAGYGYG+PISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGD
Sbjct: 302 NP----PDLDGHNEGVTMAGYGYGIPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGD 357
Query: 362 SQEPLP 367
S+EPLP
Sbjct: 358 SEEPLP 363
>UniRef100_O82424 Pyruvate dehydrogenase kinase isoform 2 [Zea mays]
Length = 364
Score = 540 bits (1391), Expect = e-152
Identities = 269/366 (73%), Positives = 313/366 (85%), Gaps = 4/366 (1%)
Query: 3 AIEAFSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRA 62
A E ++++ +EV +WG +KQTGV+LRYMMEFGS+PT +NLL+SAQFL KEL IR ARRA
Sbjct: 2 ASEPVARAVAEEVGRWGSMKQTGVTLRYMMEFGSRPTQRNLLLSAQFLHKELPIRFARRA 61
Query: 63 IELESLPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNNVV 122
+EL+SLP+GLS KPAILKVRDWY+DSFRDIR PEV+ DE FT +I +KVRHNNVV
Sbjct: 62 LELDSLPFGLSNKPAILKVRDWYLDSFRDIRYFPEVRSRNDELAFTQMINMVKVRHNNVV 121
Query: 123 PTMALGVQQLKKEL-KTKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHV 181
PTMALGVQQLKKEL +++ + EI EFLDRFY+SRIG+RMLIGQHV LH+P P P V
Sbjct: 122 PTMALGVQQLKKELGRSRKVPFEFDEIDEFLDRFYMSRIGIRMLIGQHVALHDPKPEPGV 181
Query: 182 VGYIHTKMSPVSVARNASEDARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFELV 241
+G I+T++SP+ VA+ A EDARS+C+REYGSAP+INIYGDP+FTFPYV HLHLM+FELV
Sbjct: 182 IGLINTRLSPIQVAQAACEDARSVCLREYGSAPDINIYGDPNFTFPYVTLHLHLMLFELV 241
Query: 242 KNSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTAR 301
KNSLRAVQERYM+SDK PP+RIIVADG EDVTIK+SDEGGGIPRSGLP+IFTYLYSTA+
Sbjct: 242 KNSLRAVQERYMNSDKDVPPVRIIVADGEEDVTIKVSDEGGGIPRSGLPRIFTYLYSTAK 301
Query: 302 NPLDEHADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGD 361
NP D + V TMAGYG+GLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGD
Sbjct: 302 NP--PELDRPNTERV-TMAGYGFGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGD 358
Query: 362 SQEPLP 367
S+EPLP
Sbjct: 359 SEEPLP 364
>UniRef100_Q9FRD1 Putative pyruvate dehydrogenase kinase [Oryza sativa]
Length = 365
Score = 536 bits (1380), Expect = e-151
Identities = 265/367 (72%), Positives = 316/367 (85%), Gaps = 5/367 (1%)
Query: 1 MAAIEAFSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIAR 60
MA E ++++ +EV +WG ++QTGV+LRYMMEFG++PT++NLL SAQFL++EL IRIAR
Sbjct: 2 MAYSEPAARAVAEEVARWGGMRQTGVTLRYMMEFGARPTERNLLRSAQFLRRELPIRIAR 61
Query: 61 RAIELESLPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNN 120
RA++L+SLP+GLS KPAILKVRDWY+DSFRD+R PEV++ DE FT++IK I+VRHNN
Sbjct: 62 RALDLDSLPFGLSTKPAILKVRDWYLDSFRDLRCFPEVRNRDDELAFTEMIKMIRVRHNN 121
Query: 121 VVPTMALGVQQLKKELK-TKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPP 179
VVPTMALGV+QLKK+L TK + EIH+FLDRFY+SRIG+RMLIGQHV LH P+P P
Sbjct: 122 VVPTMALGVRQLKKDLGGTKAFPPGIDEIHQFLDRFYMSRIGIRMLIGQHVALHEPDPEP 181
Query: 180 HVVGYIHTKMSPVSVARNASEDARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFE 239
V+G I ++SP+ VA++A+EDAR+ICMREYGSAP++NIYGDPDFTFPYV HL LM+FE
Sbjct: 182 GVIGLISKRLSPMLVAQHATEDARAICMREYGSAPDVNIYGDPDFTFPYVKLHLQLMMFE 241
Query: 240 LVKNSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYST 299
LVKNSLRAVQERYM+SDK +PP+RIIVADG EDVTIKISDEGGGIPRSGL +IFTYLYST
Sbjct: 242 LVKNSLRAVQERYMNSDKHAPPVRIIVADGAEDVTIKISDEGGGIPRSGLSRIFTYLYST 301
Query: 300 ARNPLDEHADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRL 359
A NP DL + TMAGYGYG+PISRLYARYFGGDLQIISMEGYGTDAYLHLSRL
Sbjct: 302 AENP----PDLDGRNEGVTMAGYGYGIPISRLYARYFGGDLQIISMEGYGTDAYLHLSRL 357
Query: 360 GDSQEPL 366
GDS+EPL
Sbjct: 358 GDSEEPL 364
>UniRef100_Q9ATR2 Pyruvate dehydrogenase kinase [Oryza sativa]
Length = 343
Score = 494 bits (1272), Expect = e-138
Identities = 255/349 (73%), Positives = 290/349 (83%), Gaps = 10/349 (2%)
Query: 23 QTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRAIELESLPYGLSKKPAILKVR 82
QTGVSLRYMMEFGS+PT++NLL+SAQFLQKEL IRIARRA+ELESLP+GLS+KPAILKVR
Sbjct: 1 QTGVSLRYMMEFGSRPTERNLLLSAQFLQKELPIRIARRALELESLPFGLSRKPAILKVR 60
Query: 83 DWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNNVVPTMALGVQQLKKE-LKTKID 141
DWY+DSFRDIR PEV++ DE FT +IK IKVRHNNVVPTMALGVQQLK E +T+
Sbjct: 61 DWYLDSFRDIRYFPEVRNRNDELAFTQMIKMIKVRHNNVVPTMALGVQQLKNEQYRTRKI 120
Query: 142 SEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHVVGYIHTKMSPVSVARNASED 201
EIHEFLDRFY+SRIG+RMLIGQHV LH+P+P P V+G I+T++SP+ V + ASED
Sbjct: 121 PTAFDEIHEFLDRFYMSRIGIRMLIGQHVALHDPDPEPGVIGLINTELSPIQVGQAASED 180
Query: 202 ARSICMREYGSAP--EINIYGDPDFTFPYVPAHLHLMVFELVKNSLR-AVQERYMDSDKV 258
ARSIC+REYGS ++IY DP FTFPYV +HLHLM ELVKNSL +ERYM SD+
Sbjct: 181 ARSICLREYGSTSSWRLDIYEDPTFTFPYVSSHLHLMNLELVKNSLAWQYRERYMSSDED 240
Query: 259 SPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDEHADLGVADSVTT 318
PP+RIIVADG TIK+SDEGGGIPRSGLP+IFTYLYSTA+NP D+ T
Sbjct: 241 VPPVRIIVADG--GRTIKVSDEGGGIPRSGLPRIFTYLYSTAKNP----PDMDCPSEGVT 294
Query: 319 MAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDSQEPLP 367
MAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS+EPLP
Sbjct: 295 MAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDSEEPLP 343
>UniRef100_Q9SQV2 Putative pyruvate dehydrogenase kinase, 5' partial [Arabidopsis
thaliana]
Length = 297
Score = 481 bits (1237), Expect = e-134
Identities = 236/301 (78%), Positives = 262/301 (86%), Gaps = 18/301 (5%)
Query: 81 VRDWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNNVVPTMALGVQQLKKELKTKI 140
VRDWY++SFRD+R+ PE+KD DE++FT +IKA+KVRHNNVVP MALGV QLKK +
Sbjct: 1 VRDWYLESFRDMRAFPEIKDSGDEKDFTQMIKAVKVRHNNVVPMMALGVNQLKKGM---- 56
Query: 141 DSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHVVGYIHTKMSPVSVARNASE 200
+S +L EIH+FLDRFYLSRIG+RMLIGQHVELHNPNPP H VGYIHTKMSP+ VARNASE
Sbjct: 57 NSGNLDEIHQFLDRFYLSRIGIRMLIGQHVELHNPNPPLHTVGYIHTKMSPMEVARNASE 116
Query: 201 DARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFELVKNSLRAVQERYMDSDKVSP 260
DARSIC REYGSAPEINIYGDP FTFPYVP HLHLM++ELVKNSLRAVQER++DSD+V+P
Sbjct: 117 DARSICFREYGSAPEINIYGDPSFTFPYVPTHLHLMMYELVKNSLRAVQERFVDSDRVAP 176
Query: 261 PIRIIVADGLEDVTIK--------------ISDEGGGIPRSGLPKIFTYLYSTARNPLDE 306
PIRIIVADG+EDVTIK +SDEGGGI RSGLP+IFTYLYSTARNPL+E
Sbjct: 177 PIRIIVADGIEDVTIKPFRSLLHRFDPIIVVSDEGGGIARSGLPRIFTYLYSTARNPLEE 236
Query: 307 HADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDSQEPL 366
DLG+AD TMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDSQEPL
Sbjct: 237 DVDLGIADVPVTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDSQEPL 296
Query: 367 P 367
P
Sbjct: 297 P 297
>UniRef100_UPI000023574E UPI000023574E UniRef100 entry
Length = 405
Score = 305 bits (782), Expect = 1e-81
Identities = 176/406 (43%), Positives = 235/406 (57%), Gaps = 53/406 (13%)
Query: 8 SKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRAIELES 67
S+ L+ + + TGVSLR M++FG +P+ L ++QFL +EL IR+A R +L
Sbjct: 5 SERLMDTIRHYASFPATGVSLRQMVQFGDRPSTGTLFRASQFLSEELPIRLAHRVQDLGE 64
Query: 68 LPYGLSKKPAILKVRDWYVDSFRDIRSC-------------------------------- 95
LP GLS+ P+I KV+DWY SF +
Sbjct: 65 LPDGLSEMPSIKKVQDWYAQSFEILAETTQNPSVREGQYRSAMTNGNGNGKAAAAARRYF 124
Query: 96 ----------PEVKDMKDEREFTDVIKAIKVRHNNVVPTMALGVQQLK-KELKTKIDSED 144
PE+ D + F ++ IK RH++VV T+A G+ + K K + +IDS
Sbjct: 125 VPSDDQGNWPPELNDYNER--FAKTLQQIKRRHDSVVTTVAQGILEWKRKRQRLQIDST- 181
Query: 145 LVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPP--PHVVGYIHTKMSPVSVARNASEDA 202
I FLDRFY+SRIG+RMLIGQH+ L P+ VG I TK + VA A E+A
Sbjct: 182 ---IQSFLDRFYMSRIGIRMLIGQHIALTEQTHVRHPNYVGIICTKTNVREVALEAIENA 238
Query: 203 RSICMREYG--SAPEINIYGDPDFTFPYVPAHLHLMVFELVKNSLRAVQERYMDSDKVSP 260
R +C YG AP++ + D F YVP HL M+FE +KNSLRAV ER+ + P
Sbjct: 239 RFVCEDYYGLFEAPKVQLVCKEDLNFMYVPGHLSHMLFETLKNSLRAVVERHGADKEAFP 298
Query: 261 PIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDEHADLGVADSVTTMA 320
++I+A+G ED+TIK+SDEGGGIPRS +P ++TY+Y+T + D +D MA
Sbjct: 299 VTKVIIAEGKEDITIKVSDEGGGIPRSAIPLVWTYMYTTVEQTPNLDPDFDKSDFKAPMA 358
Query: 321 GYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDSQEPL 366
G+GYGLPISRLYARYFGGDL++ISMEGYGTD YLHL+RL S EPL
Sbjct: 359 GFGYGLPISRLYARYFGGDLKLISMEGYGTDVYLHLNRLSSSSEPL 404
>UniRef100_Q8X073 Related to pyruvate dehydrogenase kinase isoform 2, mitochondrial
[Neurospora crassa]
Length = 405
Score = 305 bits (781), Expect = 1e-81
Identities = 176/404 (43%), Positives = 240/404 (58%), Gaps = 50/404 (12%)
Query: 8 SKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRAIELES 67
S+ L+ + + TGVSLR M++FG +P+ L ++QFL +EL IR+A R EL++
Sbjct: 6 SEKLMDTIRHYASFPATGVSLRQMVQFGEKPSTGTLFRASQFLAEELPIRLAHRVQELDN 65
Query: 68 LPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKD--------------------------- 100
LP GL++ P++ KV+ WY SF + P +++
Sbjct: 66 LPDGLNEMPSVKKVQAWYAQSFEATPN-PSIEEGQYASHNGASYASGLNHKKFSASRRYF 124
Query: 101 -MKDE------------REFTDVIKAIKVRHNNVVPTMALGVQQLK-KELKTKIDSEDLV 146
M D+ ++F + IK RH++VV TMA G+ + K K + +ID
Sbjct: 125 AMVDDTGDWPPDLHLYNQKFAQTLHKIKRRHDSVVTTMAQGILEYKRKRQRMQIDHN--- 181
Query: 147 EIHEFLDRFYLSRIGVRMLIGQHVELHNPNP--PPHVVGYIHTKMSPVSVARNASEDARS 204
I FLDRFY+SRIG+RMLIGQH+ L + N P VG I TK +A+ A E+AR
Sbjct: 182 -IQSFLDRFYMSRIGIRMLIGQHIALTDQNHYRDPSYVGIICTKTYVKDLAQEAIENARF 240
Query: 205 ICMREYG--SAPEINIYGDPDFTFPYVPAHLHLMVFELVKNSLRAVQERYMDSDKVSPPI 262
+C YG AP+I + +P+ F YVP HL M+FE +KNSLRAV E + + P
Sbjct: 241 VCEDHYGLFEAPKIQLVCNPNLNFMYVPGHLSHMLFETLKNSLRAVVETHGQDKQEFPVT 300
Query: 263 RIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDEHADLGVADSVTTMAGY 322
++IVA+G ED+TIKISDEGGGIPRS +P ++TY+Y+T + D +D MAG+
Sbjct: 301 KVIVAEGKEDITIKISDEGGGIPRSAIPLVWTYMYTTVDRTPNLDPDFDKSDFKAPMAGF 360
Query: 323 GYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDSQEPL 366
GYGLPISRLYARYFGGDL++ISMEGYGTD YLHL+RL S EPL
Sbjct: 361 GYGLPISRLYARYFGGDLKLISMEGYGTDVYLHLNRLSSSSEPL 404
>UniRef100_UPI000023D197 UPI000023D197 UniRef100 entry
Length = 414
Score = 302 bits (773), Expect = 1e-80
Identities = 176/412 (42%), Positives = 239/412 (57%), Gaps = 57/412 (13%)
Query: 8 SKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRAIELES 67
S+ L+ + + TGVSLR M++FG +P+ L ++QFL +EL IR+A R EL+
Sbjct: 6 SERLMDTIRHYARFPATGVSLRQMVQFGEKPSVGTLFRASQFLAEELPIRLAHRVQELDE 65
Query: 68 LPYGLSKKPAILKVRDWYVDSF-----RDIRSCPE------------------------- 97
LP GL++ P+++KV+DWY SF R+ P
Sbjct: 66 LPDGLNEMPSVIKVKDWYAQSFEVSIGRNAFRLPAATPNPSIDEGESDGWGGLQNNNSKN 125
Query: 98 ----------VKDMKD--------EREFTDVIKAIKVRHNNVVPTMALGVQQLKKEL-KT 138
V D D + F + IK RH++VV TMA G+ + K+ +
Sbjct: 126 KGLTRRYFAVVDDSSDWPADLHLYNQRFAQTLHQIKRRHDSVVTTMAQGILEYKRRRQRM 185
Query: 139 KIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVEL--HNPNPPPHVVGYIHTKMSPVSVAR 196
+IDS I FLDRFY+SRIG+RMLIGQH+ L + + P VG I T+ + +A+
Sbjct: 186 QIDST----IQSFLDRFYMSRIGIRMLIGQHIALTDQSHHRDPTYVGIICTRTNVQDLAQ 241
Query: 197 NASEDARSICMREYG--SAPEINIYGDPDFTFPYVPAHLHLMVFELVKNSLRAVQERYMD 254
A E+AR +C YG AP++ + +P+ F YVP HL M+FE +KNSLRAV E +
Sbjct: 242 EAIENARFVCEDHYGLFEAPKVQLVCNPNLNFMYVPGHLSHMLFETLKNSLRAVVETHGM 301
Query: 255 SDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDEHADLGVAD 314
+ P ++IVA+G ED+TIKISDEGGGIPRS +P ++TY+Y+T D +D
Sbjct: 302 EKQAFPVTKVIVAEGKEDITIKISDEGGGIPRSAIPLVWTYMYTTVDRTPSLDPDFDKSD 361
Query: 315 SVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDSQEPL 366
MAG+GYGLPISRLYARYFGGDL++ISMEGYGTD YLHL+RL S EPL
Sbjct: 362 FKAPMAGFGYGLPISRLYARYFGGDLKLISMEGYGTDVYLHLNRLSSSSEPL 413
>UniRef100_UPI000021955D UPI000021955D UniRef100 entry
Length = 416
Score = 296 bits (758), Expect = 6e-79
Identities = 173/414 (41%), Positives = 243/414 (57%), Gaps = 59/414 (14%)
Query: 8 SKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPT-------DKNLLISAQFLQKELAIRIAR 60
S+ L+ + + TGVSLR M++FG +P+ + L ++QFL +EL IR+A
Sbjct: 6 SERLMDTIRHYAKFPATGVSLRQMVQFGEKPSVGKYQSPGRTLFRASQFLAEELPIRLAH 65
Query: 61 RAIELESLPYGLSKKPAILKVRDWYVDSFRDIRSC---PEVKD----------------- 100
R EL++LP GL++ P++ KV DWY SF + P + +
Sbjct: 66 RVHELDTLPDGLNEMPSVKKVLDWYAQSFELLSEATPNPSIAEGQYASQAVTSQNGNGWT 125
Query: 101 -----------MKDE------------REFTDVIKAIKVRHNNVVPTMALGVQQLKKEL- 136
M D+ + F+ + IK RH+ VV TMA G+ + K++
Sbjct: 126 KKQHAARRYFAMVDDYGDWPPELQLYNQRFSQTLNKIKRRHDGVVTTMAQGILEYKRQRQ 185
Query: 137 KTKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNP--PPHVVGYIHTKMSPVSV 194
+ +ID+ + FLDRFY+SRIG+RMLIGQH+ L + + P VG I TK + +
Sbjct: 186 RMQIDNN----MQSFLDRFYMSRIGIRMLIGQHIALTDQSHYRDPTYVGIICTKTNVRDL 241
Query: 195 ARNASEDARSICMREYG--SAPEINIYGDPDFTFPYVPAHLHLMVFELVKNSLRAVQERY 252
A+ A E+AR +C YG AP+I + +P+ F YVP HL M+FE +KNSLRAV E +
Sbjct: 242 AQEAIENARFVCEDHYGLFEAPKIQLVCNPNINFMYVPGHLSHMLFETLKNSLRAVVETH 301
Query: 253 MDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDEHADLGV 312
+ P ++IVA+G ED+TIKI+DEGGGIPRS +P ++TY+Y+T + + D
Sbjct: 302 GQDKQEFPVTKVIVAEGKEDITIKITDEGGGIPRSAIPLVWTYMYTTVDSTPNLDPDFDK 361
Query: 313 ADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDSQEPL 366
+D MAG+GYGLPISRLYARYFGGDL++ISMEGYGTD YLHL+RL S EPL
Sbjct: 362 SDFKAPMAGFGYGLPISRLYARYFGGDLKLISMEGYGTDVYLHLNRLSSSSEPL 415
>UniRef100_Q6CB64 Similar to tr|Q8X073 Neurospora crassa Related to pyruvate
dehydrogenase kinase [Yarrowia lipolytica]
Length = 461
Score = 257 bits (657), Expect = 3e-67
Identities = 144/288 (50%), Positives = 184/288 (63%), Gaps = 8/288 (2%)
Query: 82 RDWYVDSFRDIRSCP-EVKDMKDEREFTDVIKAIKVRHNNVVPTMALGVQQLKKELKTKI 140
R +Y + ++ S P EV + + T ++ IK RH+ VV T+A G+ + K+ K
Sbjct: 178 RRYYANVSPEVSSWPPEVYEFN--KTITATLQKIKQRHDPVVTTVAQGITEWKQVYKKSA 235
Query: 141 DSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHVVGYIHTKMSPVSVARNASE 200
S + I FLDRFY+SRIG+RMLIGQH+ L+ VG I TK + V ++A
Sbjct: 236 AS---LSIQSFLDRFYMSRIGIRMLIGQHIALNLHAKQEDYVGIICTKTNVREVVQDAIA 292
Query: 201 DARSICMREYG--SAPEINIYGDPDFTFPYVPAHLHLMVFELVKNSLRAVQERYMDSDKV 258
+AR IC YG AP++ I PD F YVP HL M+FE +KNSLRAV E +
Sbjct: 293 NARFICEDWYGLFEAPKVEIVCQPDINFMYVPGHLSHMLFETLKNSLRAVVETHGVDADY 352
Query: 259 SPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDEHADLGVADSVTT 318
PP+++IVA+G ED+TIKISDEGGGIPRS +P I+TYLY+T D +D
Sbjct: 353 YPPVKVIVAEGHEDITIKISDEGGGIPRSAIPLIWTYLYTTVEATPSLEPDFNKSDFKAP 412
Query: 319 MAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDSQEPL 366
MAG+GYGLPISRLYARYFGGDL++ISMEGYGTD YLHL+RL S EPL
Sbjct: 413 MAGFGYGLPISRLYARYFGGDLKLISMEGYGTDVYLHLNRLSSSSEPL 460
Score = 85.5 bits (210), Expect = 2e-15
Identities = 41/97 (42%), Positives = 65/97 (66%)
Query: 7 FSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRAIELE 66
F+ L +++ + TGVSLR M++FGS+P+ L ++QF+ +EL IR+A R +LE
Sbjct: 12 FTPELAEKIKHYARFPATGVSLRQMVQFGSKPSAGTLFRASQFISEELPIRLAHRVRDLE 71
Query: 67 SLPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKD 103
LP GLS++P+IL+VR+WY SF ++ S + K K+
Sbjct: 72 DLPDGLSEQPSILRVRNWYAQSFDELTSLKQPKISKE 108
>UniRef100_UPI000042E26C UPI000042E26C UniRef100 entry
Length = 388
Score = 254 bits (650), Expect = 2e-66
Identities = 137/274 (50%), Positives = 186/274 (67%), Gaps = 9/274 (3%)
Query: 96 PEVKDMKDEREFTDVIKAIKVRHNNVVPTMALGVQQLKKELKTKIDSEDLVEIHEFLDRF 155
PEV + + FT +++ IK RH+ V T+A GV + K++ KT V I E+LDRF
Sbjct: 120 PEVHEYNER--FTHLLENIKKRHDPTVTTVAQGVLEWKRKRKT---GRIGVPIQEWLDRF 174
Query: 156 YLSRIGVRMLIGQHVELHNPNPPPHVVGYIHTKMSPVSVARNASEDARSICMREYG--SA 213
Y+SRIG+R LIGQHV L+ P P VG I T+ + + A E+AR +C YG
Sbjct: 175 YMSRIGIRFLIGQHVALNTLQPHPDYVGIICTRANVHDICHEAIENARYVCEEHYGLFKG 234
Query: 214 PEINIYGDPDFTFPYVPAHLHLMVFELVKNSLRAVQERY-MDSDKVSPPIRIIVADGLED 272
P I + D TFPYVP HL + FEL+KNSLRAV ER+ +++++ PPI+++V +G ED
Sbjct: 235 PPIQLLCPKDLTFPYVPGHLSHICFELLKNSLRAVVERFGVENEEAFPPIKVVVVEGRED 294
Query: 273 VTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDEHADLGVADSVTTMAGYGYGLPISRLY 332
+TIKISDEGGGIPRS +P I+TYLY+T + E A + +D MAG+GYGLP++RLY
Sbjct: 295 ITIKISDEGGGIPRSAIPMIWTYLYTTMSDEGLE-ATIEQSDFKAPMAGFGYGLPLARLY 353
Query: 333 ARYFGGDLQIISMEGYGTDAYLHLSRLGDSQEPL 366
AR+FGGDL++ISM+GYGTD Y+ L++L S EPL
Sbjct: 354 ARFFGGDLRLISMDGYGTDVYISLNKLSSSCEPL 387
>UniRef100_UPI000042CAFB UPI000042CAFB UniRef100 entry
Length = 462
Score = 254 bits (650), Expect = 2e-66
Identities = 137/274 (50%), Positives = 186/274 (67%), Gaps = 9/274 (3%)
Query: 96 PEVKDMKDEREFTDVIKAIKVRHNNVVPTMALGVQQLKKELKTKIDSEDLVEIHEFLDRF 155
PEV + + FT +++ IK RH+ V T+A GV + K++ KT V I E+LDRF
Sbjct: 194 PEVHEYNER--FTHLLENIKKRHDPTVTTVAQGVLEWKRKRKT---GRIGVPIQEWLDRF 248
Query: 156 YLSRIGVRMLIGQHVELHNPNPPPHVVGYIHTKMSPVSVARNASEDARSICMREYG--SA 213
Y+SRIG+R LIGQHV L+ P P VG I T+ + + A E+AR +C YG
Sbjct: 249 YMSRIGIRFLIGQHVALNTLQPHPDYVGIICTRANVHDICHEAIENARYVCEEHYGLFKG 308
Query: 214 PEINIYGDPDFTFPYVPAHLHLMVFELVKNSLRAVQERY-MDSDKVSPPIRIIVADGLED 272
P I + D TFPYVP HL + FEL+KNSLRAV ER+ +++++ PPI+++V +G ED
Sbjct: 309 PPIQLLCPKDLTFPYVPGHLSHICFELLKNSLRAVVERFGVENEEAFPPIKVVVVEGRED 368
Query: 273 VTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDEHADLGVADSVTTMAGYGYGLPISRLY 332
+TIKISDEGGGIPRS +P I+TYLY+T + E A + +D MAG+GYGLP++RLY
Sbjct: 369 ITIKISDEGGGIPRSAIPMIWTYLYTTMSDEGLE-ATIEQSDFKAPMAGFGYGLPLARLY 427
Query: 333 ARYFGGDLQIISMEGYGTDAYLHLSRLGDSQEPL 366
AR+FGGDL++ISM+GYGTD Y+ L++L S EPL
Sbjct: 428 ARFFGGDLRLISMDGYGTDVYISLNKLSSSCEPL 461
Score = 85.9 bits (211), Expect = 2e-15
Identities = 40/90 (44%), Positives = 60/90 (66%)
Query: 8 SKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRAIELES 67
S +L +H + QTGVSL+ M+ FG PT LL ++QFL +EL IR++ R +EL +
Sbjct: 7 SGALWDSIHHYSSFPQTGVSLQQMIHFGHNPTPGTLLKASQFLSEELPIRLSHRVVELNA 66
Query: 68 LPYGLSKKPAILKVRDWYVDSFRDIRSCPE 97
LP GL+K P+I KV++WY SF ++ + P+
Sbjct: 67 LPDGLAKMPSINKVKEWYAQSFEELVTFPK 96
>UniRef100_UPI000042F396 UPI000042F396 UniRef100 entry
Length = 432
Score = 246 bits (627), Expect = 9e-64
Identities = 134/267 (50%), Positives = 178/267 (66%), Gaps = 18/267 (6%)
Query: 96 PEVKDMKDEREFTDVIKAIKVRHNNVVPTMALGVQQLKKELKTKIDSEDLVEIHEFLDRF 155
PEV D ++ FT +++ IK RH+ V T+A GV + K+ KT + I EFLDRF
Sbjct: 178 PEVHDYNEK--FTQLLQVIKHRHDPTVTTVAQGVLEWKRMQKTSVIG---TPIQEFLDRF 232
Query: 156 YLSRIGVRMLIGQHVELHNPNPPPHVVGYIHTKMSPVSVARNASEDARSICMREYG--SA 213
Y+SRIG+R LIGQH+ L+ P P VG I T+ A +AR +C Y +
Sbjct: 233 YMSRIGIRFLIGQHIALNTLPPHPDYVGIICTR---------AVHNARYVCEEHYALFKS 283
Query: 214 PEINIYGDPDFTFPYVPAHLHLMVFELVKNSLRAVQERY-MDSDKVSPPIRIIVADGLED 272
P I + P+ TFPY+P HL + FEL+KNSLRAV ERY +D+D PPI+++V +G ED
Sbjct: 284 PNIKLVCPPNLTFPYIPGHLSHICFELLKNSLRAVVERYGVDNDDEYPPIKVVVVEGRED 343
Query: 273 VTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDEHADLGVADSVTTMAGYGYGLPISRLY 332
+TIKISDEGGGIPRS +P I+TYLY+T + E G +D MAG+GYGLP+SRLY
Sbjct: 344 ITIKISDEGGGIPRSAIPHIWTYLYTTMSDEGLEDTIQG-SDFKAPMAGFGYGLPLSRLY 402
Query: 333 ARYFGGDLQIISMEGYGTDAYLHLSRL 359
AR+FGGDL++ISM+GYGTD Y+ L++L
Sbjct: 403 ARFFGGDLRLISMDGYGTDVYISLNKL 429
Score = 78.6 bits (192), Expect = 3e-13
Identities = 41/99 (41%), Positives = 63/99 (63%), Gaps = 1/99 (1%)
Query: 8 SKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRAIELES 67
S +L +++ + QTGVSL+ M+ FG P+ LL ++QFL +EL IR++ R +ELE
Sbjct: 7 SSALWDKIYHFASFPQTGVSLQQMILFGQNPSQGTLLKASQFLSEELPIRLSHRVVELEG 66
Query: 68 LPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKDERE 106
LP GL+K +I V++WY SF ++ S P + +K E E
Sbjct: 67 LPDGLNKMTSINIVKEWYAQSFDELVSFPRPR-LKPELE 104
>UniRef100_Q9P6P9 SPAC644.11c protein [Schizosaccharomyces pombe]
Length = 425
Score = 243 bits (620), Expect = 6e-63
Identities = 128/269 (47%), Positives = 181/269 (66%), Gaps = 8/269 (2%)
Query: 101 MKDEREFTDVIKAIKVRHNNVVPTMALGVQQLKKELKTKIDSEDLVEIHEFLDRFYLSRI 160
+K F ++ I+ RH+NV +AL +Q+ +++ +ID+ I FLDRFY+SRI
Sbjct: 161 LKFNSNFAYLLNTIRTRHDNVAVEIALDIQEYRRKTN-QIDNS----IQIFLDRFYMSRI 215
Query: 161 GVRMLIGQHVELHNPNPPPHVVGYIHTKMSPVSVARNASEDARSICMREYG--SAPEINI 218
G+RML+GQ++ L + P + VG I T+ + + A+E+A+ IC YG APEI I
Sbjct: 216 GIRMLLGQYIALVSEPPRENYVGVISTRANIYQIIEGAAENAKYICRLAYGLFEAPEIQI 275
Query: 219 YGDPDFTFPYVPAHLHLMVFELVKNSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKIS 278
DP YV +HL+ VFE++KNSLRA E + PPI++IVA G ED+TIKIS
Sbjct: 276 ICDPSLEMMYVESHLNHAVFEILKNSLRATVEFHGVDSDFFPPIKVIVAKGQEDITIKIS 335
Query: 279 DEGGGIPRSGLPKIFTYLYSTARNPL-DEHADLGVADSVTTMAGYGYGLPISRLYARYFG 337
DEGGGI R +P +++Y+++TA L D+ D+ A+S T MAG+G+GLP++RLY RYFG
Sbjct: 336 DEGGGISRRNIPLVWSYMFTTASPTLTDDPHDIVSANSTTPMAGFGFGLPLARLYTRYFG 395
Query: 338 GDLQIISMEGYGTDAYLHLSRLGDSQEPL 366
GDL++ISMEGYGTD Y+HL+RL +S EPL
Sbjct: 396 GDLELISMEGYGTDVYIHLNRLCESAEPL 424
Score = 49.3 bits (116), Expect = 2e-04
Identities = 30/89 (33%), Positives = 47/89 (52%)
Query: 4 IEAFSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRAI 63
+ K+L ++V+ QTG+SL+ ++ FG PT L + FL+ EL IR+ARR
Sbjct: 1 MSVLGKTLQEKVNLLAQYPQTGLSLKQLVYFGKNPTPGTLFRAGLFLRDELPIRLARRIQ 60
Query: 64 ELESLPYGLSKKPAILKVRDWYVDSFRDI 92
+L++L L I V+ Y S +I
Sbjct: 61 DLQNLSPMLRSMKRISSVKAAYGRSMEEI 89
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 620,610,853
Number of Sequences: 2790947
Number of extensions: 26688549
Number of successful extensions: 64124
Number of sequences better than 10.0: 1024
Number of HSP's better than 10.0 without gapping: 228
Number of HSP's successfully gapped in prelim test: 796
Number of HSP's that attempted gapping in prelim test: 62917
Number of HSP's gapped (non-prelim): 1102
length of query: 367
length of database: 848,049,833
effective HSP length: 129
effective length of query: 238
effective length of database: 488,017,670
effective search space: 116148205460
effective search space used: 116148205460
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146757.7


