

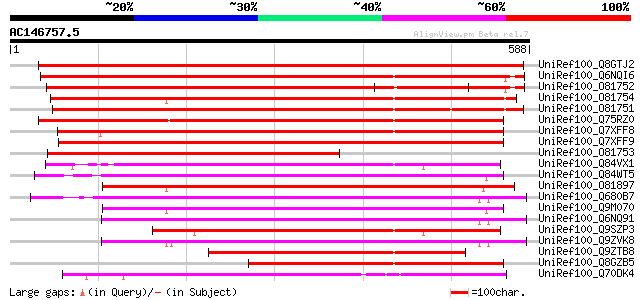
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.5 - phase: 0 /pseudo
(588 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8GTJ2 Endoxylanase precursor [Carica papaya] 761 0.0
UniRef100_Q6NQI6 At4g33840/F17I5_30 [Arabidopsis thaliana] 675 0.0
UniRef100_O81752 Hypothetical protein F17I5.30 [Arabidopsis thal... 664 0.0
UniRef100_O81754 Hypothetical protein F17I5.50 [Arabidopsis thal... 649 0.0
UniRef100_O81751 Hypothetical protein F17I5.20 [Arabidopsis thal... 644 0.0
UniRef100_Q75RZ0 Putative 1,4-beta-xylanase [Triticum aestivum] 522 e-146
UniRef100_Q7XFF8 Putative 1,4-beta-xylanase [Oryza sativa] 521 e-146
UniRef100_Q7XFF9 Putative 1,4-beta-xylanase [Oryza sativa] 509 e-142
UniRef100_O81753 Hypothetical protein F17I5.40 [Arabidopsis thal... 426 e-118
UniRef100_Q84VX1 At4g38650 [Arabidopsis thaliana] 401 e-110
UniRef100_Q84WT5 Putative glycosyl hydrolase family 10 protein [... 395 e-108
UniRef100_O81897 Beta-xylan endohydrolase-like protein [Arabidop... 393 e-108
UniRef100_Q680B7 1,4-beta-xylan endohydrolase [Arabidopsis thali... 375 e-102
UniRef100_Q9M070 Hypothetical protein AT4g33820 [Arabidopsis tha... 373 e-102
UniRef100_Q6NQ91 At2g14690 [Arabidopsis thaliana] 371 e-101
UniRef100_Q9SZP3 Hypothetical protein F20M13.210 [Arabidopsis th... 361 4e-98
UniRef100_Q9ZVK8 1,4-beta-xylan endohydrolase [Arabidopsis thali... 360 5e-98
UniRef100_Q9ZTB8 Tapetum specific protein [Zea mays] 327 6e-88
UniRef100_Q8GZB5 Anther endoxylanase [Hordeum vulgare var. disti... 306 1e-81
UniRef100_Q70DK4 Beta-1,4-xylanase [Clostridium thermocellum] 263 8e-69
>UniRef100_Q8GTJ2 Endoxylanase precursor [Carica papaya]
Length = 584
Score = 761 bits (1965), Expect = 0.0
Identities = 351/555 (63%), Positives = 446/555 (80%), Gaps = 5/555 (0%)
Query: 33 GFEAEALSYDYSANVECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKSLGNK 92
G E ALSYDY+A+++CL +PQK QY GGII NPELN GL+GW+TFGDA I+HR + N
Sbjct: 24 GLETNALSYDYTASIQCLENPQKAQYGGGIITNPELNQGLKGWSTFGDAKIQHRVAGSNS 83
Query: 93 FVVTHSRNQPHDSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKFGGAIFAE 152
F+V H+R+QPHDSVSQ +YL+ Y+ SAWI+VSE PV A+ KT G+K+ GA+ AE
Sbjct: 84 FIVAHTRSQPHDSVSQTLYLQSNKLYTFSAWIRVSEGKTPVKAIFKTKSGYKYAGAVVAE 143
Query: 153 PNCWSMLKGGLIADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRK 212
NCWSMLKGGL D +G AELYFE++NTSVEIW+D++SLQPFT+++W+SHQ+ SI+K RK
Sbjct: 144 SNCWSMLKGGLTVDASGPAELYFETDNTSVEIWIDSISLQPFTQQEWKSHQDQSIKKIRK 203
Query: 213 RKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFEN 272
+ V ++AV++ G+PLPN ++S++ K+ GFPFG AIN+NI+NNNAYQ WF+SRFTVTTFEN
Sbjct: 204 KNVRIQAVDKLGNPLPNTTVSISPKKIGFPFGCAINRNIVNNNAYQSWFSSRFTVTTFEN 263
Query: 273 EMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLND 332
EMKW + E +QG ++Y ADAM+ FA+K GIA+RGHN+FWDDP+YQ WVSSLSP+ LN
Sbjct: 264 EMKWASTEPSQGHEDYSTADAMVQFAKKNGIAIRGHNVFWDDPKYQSGWVSSLSPNDLNA 323
Query: 333 AVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMN 392
A KR+NS+++RYKGQ+IGWDVVNENLHFSFFESKLG N SA + E H D TTLFMN
Sbjct: 324 AATKRINSVMNRYKGQVIGWDVVNENLHFSFFESKLGANASAVFYGEAHKTDPSTTLFMN 383
Query: 393 EYNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLR 452
EYNT+EDSRDG +TP Y+EK++ IQS+ +GIGLESHF +SPPN+PYMR+++DTL
Sbjct: 384 EYNTVEDSRDGQATPAKYLEKLRSIQSLPGNGNMGIGLESHFSSSPPNIPYMRSAIDTLA 443
Query: 453 ATGLPIWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNF 512
ATGLP+W+TE+DV S NQA EQ+LREAHSHP ++GIV+W+AWSP GCYR+CLTDNNF
Sbjct: 444 ATGLPVWLTEVDVQSGGNQAQSLEQILREAHSHPKVRGIVIWSAWSPNGCYRMCLTDNNF 503
Query: 513 KNLPAGDVVDQLINEW-GRAEKSGTTDQNGYFEASLFHGDYEI--EINHPIKKKSNFTHH 569
NLP GDVVD+L+ EW G A G TDQNG+F++SLFHGDYEI ++NHP K S+ +HH
Sbjct: 504 HNLPTGDVVDKLLREWGGGATVKGKTDQNGFFQSSLFHGDYEIKVQVNHPSKLPSSSSHH 563
Query: 570 IKVLSK--DEFKKTK 582
L+ DE K+T+
Sbjct: 564 TFKLNSTDDESKQTR 578
>UniRef100_Q6NQI6 At4g33840/F17I5_30 [Arabidopsis thaliana]
Length = 576
Score = 675 bits (1742), Expect = 0.0
Identities = 323/557 (57%), Positives = 412/557 (72%), Gaps = 15/557 (2%)
Query: 35 EAEALSYDYSANVECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKSLGNKFV 94
E +++ YDYSA +ECL +P KPQYNGGII NP+L +G QGW+ FG+A ++ R+ GNKFV
Sbjct: 19 EEQSVPYDYSATIECLENPYKPQYNGGIIVNPDLQNGSQGWSQFGNAKVDFREFGGNKFV 78
Query: 95 VTHSRNQPHDSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKFGGAIFAEPN 154
V RNQ DS+SQK+YL KG+ Y+ SAW+QVS PV+AV K +K G++ AE
Sbjct: 79 VATQRNQSSDSISQKVYLEKGILYTFSAWLQVSIGKSPVSAVFKKNGEYKHAGSVVAESK 138
Query: 155 CWSMLKGGLIADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRK 214
CWSMLKGGL D +G AEL+FES NT VEIWVD+VSLQPFT+++W SH E SI K RK
Sbjct: 139 CWSMLKGGLTVDESGPAELFFESENTMVEIWVDSVSLQPFTQEEWNSHHEQSIGKVRKGT 198
Query: 215 VVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEM 274
V +R +N +G +PNA+IS+ K+ G+PFG A+ NIL N AYQ+WF RFTVTTF NEM
Sbjct: 199 VRIRVMNNKGETIPNATISIEQKKLGYPFGCAVENNILGNQAYQNWFTQRFTVTTFGNEM 258
Query: 275 KWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAV 334
KWY+ E +G+++Y ADAML F + GIAVRGHN+ WDDP+YQP WV+SLS + L +AV
Sbjct: 259 KWYSTERIRGQEDYSTADAMLSFFKSHGIAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAV 318
Query: 335 EKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEY 394
++RV S+VSRYKGQL+GWDVVNENLHFSFFESK G S + H +D +T +FMNEY
Sbjct: 319 KRRVYSVVSRYKGQLLGWDVVNENLHFSFFESKFGPKASYNTYTMAHAVDPRTPMFMNEY 378
Query: 395 NTIEDSRDGLSTPPTYIEKIKEIQSVN--SQLPLGIGLESHFPNSPPNLPYMRASLDTLR 452
NT+E +D S+P Y+ K++E+QS+ ++PL IGLESHF S PN+PYMR++LDT
Sbjct: 379 NTLEQPKDLTSSPARYLGKLRELQSIRVAGKIPLAIGLESHF--STPNIPYMRSALDTFG 436
Query: 453 ATGLPIWITELDVASQPN-QALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNN 511
ATGLPIW+TE+DV + PN +A YFEQVLRE H+HP + G+VMWT +SP GCYR+CLTD N
Sbjct: 437 ATGLPIWLTEIDVDAPPNVRANYFEQVLREGHAHPKVNGMVMWTGYSPSGCYRMCLTDGN 496
Query: 512 FKNLPAGDVVDQLINEWG--RAEKSGTTDQNGYFEASLFHGDYEIEINHPI---KKKSNF 566
FKNLP GDVVD+L+ EWG R++ +G TD NG FEA LFHGDY++ I+HP+ K NF
Sbjct: 497 FKNLPTGDVVDKLLREWGGLRSQTTGVTDANGLFEAPLFHGDYDLRISHPLTNSKASYNF 556
Query: 567 THHIKVLSKDEFKKTKQ 583
T L+ D+ KQ
Sbjct: 557 T-----LTSDDDSSQKQ 568
>UniRef100_O81752 Hypothetical protein F17I5.30 [Arabidopsis thaliana]
Length = 669
Score = 664 bits (1713), Expect = 0.0
Identities = 318/550 (57%), Positives = 406/550 (73%), Gaps = 15/550 (2%)
Query: 42 DYSANVECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKSLGNKFVVTHSRNQ 101
D++A +CL +P KPQYNGGII NP+L +G QGW+ FG+A ++ R+ GNKFVV RNQ
Sbjct: 119 DFAAKQQCLENPYKPQYNGGIIVNPDLQNGSQGWSQFGNAKVDFREFGGNKFVVATQRNQ 178
Query: 102 PHDSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKFGGAIFAEPNCWSMLKG 161
DS+SQK+YL KG+ Y+ SAW+QVS PV+AV K +K G++ AE CWSMLKG
Sbjct: 179 SSDSISQKVYLEKGILYTFSAWLQVSIGKSPVSAVFKKNGEYKHAGSVVAESKCWSMLKG 238
Query: 162 GLIADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVVVRAVN 221
GL D +G AEL+FES NT VEIWVD+VSLQPFT+++W SH E SI K RK V +R +N
Sbjct: 239 GLTVDESGPAELFFESENTMVEIWVDSVSLQPFTQEEWNSHHEQSIGKVRKGTVRIRVMN 298
Query: 222 EQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEY 281
+G +PNA+IS+ K+ G+PFG A+ NIL N AYQ+WF RFTVTTF NEMKWY+ E
Sbjct: 299 NKGETIPNATISIEQKKLGYPFGCAVENNILGNQAYQNWFTQRFTVTTFGNEMKWYSTER 358
Query: 282 AQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSI 341
+G+++Y ADAML F + GIAVRGHN+ WDDP+YQP WV+SLS + L +AV++RV S+
Sbjct: 359 IRGQEDYSTADAMLSFFKSHGIAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAVKRRVYSV 418
Query: 342 VSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSR 401
VSRYKGQL+GWDVVNENLHFSFFESK G S + H +D +T +FMNEYNT+E +
Sbjct: 419 VSRYKGQLLGWDVVNENLHFSFFESKFGPKASYNTYTMAHAVDPRTPMFMNEYNTLEQPK 478
Query: 402 DGLSTPPTYIEKIKEIQSVN--SQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIW 459
D S+P Y+ K++E+QS+ ++PL IGLESHF S PN+PYMR++LDT ATGLPIW
Sbjct: 479 DLTSSPARYLGKLRELQSIRVAGKIPLAIGLESHF--STPNIPYMRSALDTFGATGLPIW 536
Query: 460 ITELDVASQPN-QALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLPAG 518
+TE+DV + PN +A YFEQVLRE H+HP + G+VMWT +SP GCYR+CLTD NFKNLP G
Sbjct: 537 LTEIDVDAPPNVRANYFEQVLREGHAHPKVNGMVMWTGYSPSGCYRMCLTDGNFKNLPTG 596
Query: 519 DVVDQLINEWG--RAEKSGTTDQNGYFEASLFHGDYEIEINHPI---KKKSNFTHHIKVL 573
DVVD+L+ EWG R++ +G TD NG FEA LFHGDY++ I+HP+ K NFT L
Sbjct: 597 DVVDKLLREWGGLRSQTTGVTDANGLFEAPLFHGDYDLRISHPLTNSKASYNFT-----L 651
Query: 574 SKDEFKKTKQ 583
+ D+ KQ
Sbjct: 652 TSDDDSSQKQ 661
Score = 138 bits (347), Expect = 5e-31
Identities = 65/110 (59%), Positives = 86/110 (78%), Gaps = 5/110 (4%)
Query: 414 IKEIQSV--NSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITELDVASQPN- 470
+KE+QS+ + + L IGLESHF PN+PYMR++LD L ATGL IW+TE+DV + P+
Sbjct: 2 LKELQSIRISGYIRLAIGLESHFKT--PNIPYMRSALDILAATGLLIWLTEIDVEAPPSV 59
Query: 471 QALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLPAGDV 520
QA YFEQVLR+ H+HP ++G+V+W +SP GCYR+CLTD NF+NLP GDV
Sbjct: 60 QAKYFEQVLRDGHAHPQVKGMVVWGGYSPSGCYRMCLTDGNFRNLPTGDV 109
>UniRef100_O81754 Hypothetical protein F17I5.50 [Arabidopsis thaliana]
Length = 574
Score = 649 bits (1673), Expect = 0.0
Identities = 312/546 (57%), Positives = 400/546 (73%), Gaps = 21/546 (3%)
Query: 47 VECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKSLGNKFVVTHSRNQPHDSV 106
V+CL P KPQYNGGII +P++ DG GWT FG+A ++ RK + F V R QP DSV
Sbjct: 16 VQCLEIPLKPQYNGGIIVSPDVRDGTLGWTPFGNAKVDFRKIGNHNFFVARDRKQPFDSV 75
Query: 107 SQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKFGGAIFAEPNCWSMLKGGLIAD 166
SQK+YL KGL Y+ SAW+QVS+ PV AV K +K G++ AE CWSMLKGGL D
Sbjct: 76 SQKVYLEKGLLYTFSAWLQVSKGKAPVKAVFKKNGEYKLAGSVVAESKCWSMLKGGLTVD 135
Query: 167 TTGVAELYFE-------------SNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKR 213
+G AELYFE S +T+VEIWVD+VSLQPFT+++W SH E SI+K+RKR
Sbjct: 136 ESGPAELYFEVYFSVNCCQFLRSSEDTTVEIWVDSVSLQPFTQEEWNSHHEQSIQKERKR 195
Query: 214 KVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENE 273
V +RAVN +G P+P A+IS+ ++ GFPFG + KNIL N AYQ+WF RFTVTTF NE
Sbjct: 196 TVRIRAVNSKGEPIPKATISIEQRKLGFPFGCEVEKNILGNKAYQNWFTQRFTVTTFANE 255
Query: 274 MKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDA 333
MKWY+ E +GK++Y ADAML F ++ G+AVRGHNI W+DP+YQP WV++LS + L +A
Sbjct: 256 MKWYSTEVVRGKEDYSTADAMLRFFKQHGVAVRGHNILWNDPKYQPKWVNALSGNDLYNA 315
Query: 334 VEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNE 393
V++RV S+VSRYKGQL GWDVVNENLHFS+FE K+G S +F D TT+FMNE
Sbjct: 316 VKRRVFSVVSRYKGQLAGWDVVNENLHFSYFEDKMGPKASYNIFKMAQAFDPTTTMFMNE 375
Query: 394 YNTIEDSRDGLSTPPTYIEKIKEIQSVN--SQLPLGIGLESHFPNSPPNLPYMRASLDTL 451
YNT+E+S D S+ Y++K++EI+S+ + LGIGLESHF PN+PYMR++LDTL
Sbjct: 376 YNTLEESSDSDSSLARYLQKLREIRSIRVCGNISLGIGLESHF--KTPNIPYMRSALDTL 433
Query: 452 RATGLPIWITELDVASQPN-QALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDN 510
ATGLPIW+TE+DV + PN QA YFEQVLRE H+HP ++GIV W+ +SP GCYR+CLTD
Sbjct: 434 AATGLPIWLTEVDVEAPPNVQAKYFEQVLREGHAHPQVKGIVTWSGYSPSGCYRMCLTDG 493
Query: 511 NFKNLPAGDVVDQLINEWG--RAEKSGTTDQNGYFEASLFHGDYEIEINHPIKKKSNFTH 568
NFKN+P GDVVD+L++EWG R + +G TD +GYFEASLFHGDY+++I HP+ S +H
Sbjct: 494 NFKNVPTGDVVDKLLHEWGGFRRQTTGVTDADGYFEASLFHGDYDLKIAHPL-TNSKASH 552
Query: 569 HIKVLS 574
K+ S
Sbjct: 553 SFKLTS 558
>UniRef100_O81751 Hypothetical protein F17I5.20 [Arabidopsis thaliana]
Length = 544
Score = 644 bits (1660), Expect = 0.0
Identities = 315/539 (58%), Positives = 398/539 (73%), Gaps = 9/539 (1%)
Query: 49 CLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKSLGNKFVVTHSRNQPHDSVSQ 108
CL P KPQYNGGII NP++ +G QGW+ F +A + R+ GNKFVV RNQ DSVSQ
Sbjct: 2 CLEIPYKPQYNGGIIVNPDMQNGSQGWSQFENAKVNFREFGGNKFVVATQRNQSSDSVSQ 61
Query: 109 KIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKFGGAIFAEPNCWSMLKGGLIADTT 168
K+YL KG+ Y+ SAW+QVS PV+AV K +K G++ AE CWSMLKGGL D +
Sbjct: 62 KVYLEKGILYTFSAWLQVSTGKAPVSAVFKKNGEYKHAGSVVAESKCWSMLKGGLTVDES 121
Query: 169 GVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVVVRAVNEQGHPLP 228
G AEL+ ES +T+VEIWVD+VSLQPFT+ +W +HQE SI+ RK V +R VN +G +P
Sbjct: 122 GPAELFVESEDTTVEIWVDSVSLQPFTQDEWNAHQEQSIDNSRKGPVRIRVVNNKGEKIP 181
Query: 229 NASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNY 288
NASI++ KR GFPFGSA+ +NIL N AYQ+WF RFTVTTFENEMKWY+ E +G +NY
Sbjct: 182 NASITIEQKRLGFPFGSAVAQNILGNQAYQNWFTQRFTVTTFENEMKWYSTESVRGIENY 241
Query: 289 FDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVSRYKGQ 348
ADAML F + GIAVRGHN+ WD P+YQ WV+SLS + L +AV++RV S+VSRYKGQ
Sbjct: 242 TVADAMLRFFNQHGIAVRGHNVVWDHPKYQSKWVTSLSRNDLYNAVKRRVFSVVSRYKGQ 301
Query: 349 LIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPP 408
L GWDVVNENLH SFFESK G N S +F H ID TT+FMNE+ T+ED D ++P
Sbjct: 302 LAGWDVVNENLHHSFFESKFGPNASNNIFAMAHAIDPSTTMFMNEFYTLEDPTDLKASPA 361
Query: 409 TYIEKIKEIQS--VNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITELDV- 465
Y+EK++E+QS V +PLGIGLESHF S PN+PYMR++LDTL ATGLPIW+TE+DV
Sbjct: 362 KYLEKLRELQSIRVRGNIPLGIGLESHF--STPNIPYMRSALDTLGATGLPIWLTEIDVK 419
Query: 466 ASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLPAGDVVDQLI 525
A +QA YFEQVLRE H+HP ++G+V WTA++P CY +CLTD NFKNLP GDVVD+LI
Sbjct: 420 APSSDQAKYFEQVLREGHAHPHVKGMVTWTAYAP-NCYHMCLTDGNFKNLPTGDVVDKLI 478
Query: 526 NEWG--RAEKSGTTDQNGYFEASLFHGDYEIEINHPIKKKSNFTHHIKVLSKDEFKKTK 582
EWG R++ + TD +G+FEASLFHGDY++ I+HP+ S+ +H+ + S D T+
Sbjct: 479 REWGGLRSQTTEVTDADGFFEASLFHGDYDLNISHPL-TNSSVSHNFTLTSDDSSLHTQ 536
>UniRef100_Q75RZ0 Putative 1,4-beta-xylanase [Triticum aestivum]
Length = 574
Score = 522 bits (1344), Expect = e-146
Identities = 263/535 (49%), Positives = 349/535 (65%), Gaps = 11/535 (2%)
Query: 33 GFEAEALSYDYSANVECLAHPQKPQYNGGIIKNPELNDGLQGWTT-FGDAIIEHRKSLGN 91
G+ +++ YDY+A+++CL+ P + Y GGII+N E N GL GW+ +G GN
Sbjct: 16 GWMVQSVPYDYTASIKCLSSPIRALYKGGIIENSEFNSGLTGWSVPWGVTANVSSSPSGN 75
Query: 92 KFVVTHSRN--QPHDSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKG-FKFGGA 148
F + + N QP SV QKI + HYSLSAW+QVS T V AV K G F GGA
Sbjct: 76 NFALASASNNGQPSRSVYQKIQMETAHHYSLSAWLQVSSGTAVVRAVFKDPNGAFIAGGA 135
Query: 149 IFAEPNCWSMLKGGLIADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIE 208
A CWSMLKGG+ A +G EL+FE++ V+IWVD+VSLQPF+ +W H+ LS
Sbjct: 136 TVARSGCWSMLKGGMTAFASGPGELFFEADGR-VDIWVDSVSLQPFSFPEWEEHRRLSAG 194
Query: 209 KDRKRKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVT 268
K R+ V V A G PLPNA++S+ + RPGFPFG+A+ K IL+ AY+ WFASRFTV
Sbjct: 195 KARRSVVKVVARAADGVPLPNANVSVKLLRPGFPFGNAMTKEILDIPAYEQWFASRFTVA 254
Query: 269 TFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPD 328
+FENEMKWY+ E+ + ++Y ADAML A+K GIAVRGHN+ WD Q +WV L
Sbjct: 255 SFENEMKWYSTEWMENHEDYTVADAMLRLAQKHGIAVRGHNVLWDTNDTQVSWVKPLDAQ 314
Query: 329 QLNDAVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTT 388
+L A++KR++S+VSRY G++I WDVVNENLH FFES+LG+N S+ ++ V ID
Sbjct: 315 RLKAALQKRISSVVSRYAGKVIAWDVVNENLHGQFFESRLGRNASSEVYQRVARIDRTAR 374
Query: 389 LFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSV--NSQLPLGIGLESHFPNSPPNLPYMRA 446
LFMNE+ T+E+ D + Y+ K+K+I+S N + L +GLESHF PN+PYMRA
Sbjct: 375 LFMNEFGTLEEPLDAAAISSKYVAKLKQIRSYPGNRGIKLAVGLESHF--GTPNIPYMRA 432
Query: 447 SLDTLRATGLPIWITELDVASQ--PNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYR 504
+LD L +PIW+TE+DV + P +Y E+VLRE + HP ++G+VMW AW QGC+
Sbjct: 433 TLDMLAQLRVPIWLTEVDVGPKGAPYVPVYLEEVLREGYGHPNVEGMVMWAAWHAQGCWV 492
Query: 505 ICLTDNNFKNLPAGDVVDQLINEWGRAEKSGTTDQNGYFEASLFHGDYEIEINHP 559
+CLTDNNF NLPAGD VD+LI EW + T D NG E L HG+Y + HP
Sbjct: 493 MCLTDNNFNNLPAGDRVDKLIAEWRAHPEGATMDANGVTELDLVHGEYNFTVTHP 547
>UniRef100_Q7XFF8 Putative 1,4-beta-xylanase [Oryza sativa]
Length = 541
Score = 521 bits (1342), Expect = e-146
Identities = 261/515 (50%), Positives = 345/515 (66%), Gaps = 13/515 (2%)
Query: 55 KPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKS-LGNKFVVTHSRNQ------PHDSVS 107
KP YNGGII+N E N GL GW+T D S GNKF V + P SV
Sbjct: 2 KPLYNGGIIQNGEFNSGLMGWSTHRDIKAGLSSSPSGNKFAVVQRADSLSGAAVPSRSVY 61
Query: 108 QKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKF-GGAIFAEPNCWSMLKGGLIAD 166
QKI L+ HYSLSAW+QVS V A VKT G + G++ A+ CWSMLKGG+ A
Sbjct: 62 QKIQLQGDTHYSLSAWLQVSAGAAHVKAFVKTPNGERVVAGSVSAQSGCWSMLKGGMTAY 121
Query: 167 TTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVVVRAVNEQGHP 226
++G +++FES+ V+IW+D+VSLQPFT +W +H++ S K R+ V V G P
Sbjct: 122 SSGPGQIFFESD-APVDIWMDSVSLQPFTFDEWDAHRQQSAAKVRRSTVRVVVRGADGAP 180
Query: 227 LPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKD 286
+ NA++ + + R GFPFG+A+ K IL+ AY+ WF SRFTV TFENEMKWY+NE+AQ +
Sbjct: 181 MANATVIVELLRAGFPFGNALTKEILDLPAYEKWFTSRFTVATFENEMKWYSNEWAQNNE 240
Query: 287 NYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVSRYK 346
+Y ADAML A+K I +RGHN+FWDD Q WV+ L+ DQL A++KR+ S+V+RY
Sbjct: 241 DYRVADAMLKLAQKYNIKIRGHNVFWDDQNSQMKWVTPLNLDQLKAAMQKRLKSVVTRYA 300
Query: 347 GQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLST 406
G++I WDVVNENLHF+FFE+KLG N S ++N+V +D LFMNE+NT+E D
Sbjct: 301 GKVIHWDVVNENLHFNFFETKLGPNASPMIYNQVGALDKNAILFMNEFNTLEQPGDPNPV 360
Query: 407 PPTYIEKIKEIQSV--NSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITELD 464
P Y+ K+K+IQS NS L LG+GLESHF S PN+PYMR++LDTL LP+W+TE+D
Sbjct: 361 PSKYVAKMKQIQSYPGNSALKLGVGLESHF--STPNIPYMRSALDTLAQLKLPMWLTEVD 418
Query: 465 VASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLPAGDVVDQL 524
V PNQ + EQVLRE ++HP + G++MW AW +GCY +CLTDN+FKNLP G +VD+L
Sbjct: 419 VVKGPNQVKFLEQVLREGYAHPSVNGMIMWAAWHAKGCYVMCLTDNSFKNLPVGTLVDKL 478
Query: 525 INEWGRAEKSGTTDQNGYFEASLFHGDYEIEINHP 559
I EW + + TT +G E L HGDY + ++HP
Sbjct: 479 IAEWKTHKTAATTGADGAVELDLPHGDYNLTVSHP 513
>UniRef100_Q7XFF9 Putative 1,4-beta-xylanase [Oryza sativa]
Length = 539
Score = 509 bits (1310), Expect = e-142
Identities = 258/512 (50%), Positives = 343/512 (66%), Gaps = 10/512 (1%)
Query: 56 PQYNGGIIKNPELNDGLQGWTT-FGDAIIEHRKSLGNKFVVTHSRNQPHDSVSQKIYLRK 114
P Y+GG+IKN E N GL WT G + S GNKF + QP +V Q + ++
Sbjct: 3 PLYSGGVIKNSEFNVGLTDWTVPLGVQATVNSSSSGNKFAEARTDGQPSRTVYQTVQIQP 62
Query: 115 GLHYSLSAWIQVSEETVPVTAVVKTTKG-FKFGGAIFAEPNCWSMLKGGLIADTTGVAEL 173
HYSLSAW+QVS T V AVV+T G F GA A+ CWSM+KGG+ + ++G +L
Sbjct: 63 NTHYSLSAWLQVSAGTANVMAVVRTPDGQFVAAGATVAKSGCWSMIKGGMTSYSSGQGQL 122
Query: 174 YFESNNTSVEIWVDNVSLQPFTEKQWRSH-QELSIEKDRKRKV-VVRAVNEQGHPLPNAS 231
YFE++ +V IWVD+VSLQPFT +W +H Q+ S + R+ + VV A G P+PNA+
Sbjct: 123 YFEAD-AAVAIWVDSVSLQPFTFDEWDAHRQQQSAGRARRSTLGVVVARGTDGAPVPNAT 181
Query: 232 ISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNYFDA 291
++ + RPGFPFG+A+ + IL+N AY+ WFASRFTV TFENEMKWY E QG ++Y
Sbjct: 182 VTAELLRPGFPFGNAMTREILDNPAYEQWFASRFTVATFENEMKWYATEGRQGHEDYRVP 241
Query: 292 DAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVSRY-KGQLI 350
DAML AE+ G+ VRGHN+FWDD Q WV SL PD+L A++KR+ S+VSRY G++I
Sbjct: 242 DAMLALAERHGVRVRGHNVFWDDQSTQMAWVRSLGPDELRAAMDKRLRSVVSRYGGGRVI 301
Query: 351 GWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTY 410
GWDVVNENLH+SF++ KLG + S ++++V IDG+T LFMNE+NT+E D + Y
Sbjct: 302 GWDVVNENLHWSFYDGKLGPDASPAIYHQVGKIDGETPLFMNEFNTVEQPVDMAAMASKY 361
Query: 411 IEKIKEIQSV--NSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITELDVASQ 468
+ K+ +I+S N L L +GLESHF + PN+P+MRA+LDTL LPIW+TE+DVA+
Sbjct: 362 VAKMNQIRSFPGNGGLKLAVGLESHF-GATPNIPFMRATLDTLAQLKLPIWLTEIDVANG 420
Query: 469 PNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLPAGDVVDQLINEW 528
NQA + E+VLRE H HP + G+VMW AW CY +CLTD+ FKNL GDVVD+LI EW
Sbjct: 421 TNQAQHLEEVLREGHGHPNVDGMVMWAAWHATACYVMCLTDDEFKNLAVGDVVDKLIAEW 480
Query: 529 GRAEKS-GTTDQNGYFEASLFHGDYEIEINHP 559
+ TTD +G E L HG+Y + + HP
Sbjct: 481 RTHPVAVATTDADGVVELDLAHGEYNVTVTHP 512
>UniRef100_O81753 Hypothetical protein F17I5.40 [Arabidopsis thaliana]
Length = 371
Score = 426 bits (1095), Expect = e-118
Identities = 201/330 (60%), Positives = 249/330 (74%)
Query: 44 SANVECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKSLGNKFVVTHSRNQPH 103
S ++ECL +P KPQYNGGII NPEL +G QGW+ FG+A +E R+ N +VV RNQ
Sbjct: 39 STSLECLENPYKPQYNGGIIVNPELQNGSQGWSKFGNAKVEFREFGDNHYVVARQRNQSF 98
Query: 104 DSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKFGGAIFAEPNCWSMLKGGL 163
DSVSQ +YL K L Y+ SAW+QVSE PV A+ K +K G++ AE CWSMLKGGL
Sbjct: 99 DSVSQTVYLEKELLYTFSAWLQVSEGKAPVRAIFKKNGEYKNAGSVVAESKCWSMLKGGL 158
Query: 164 IADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVVVRAVNEQ 223
D +G AELYFES++T VEIWVD+VSLQPFT+K+W HQE SI K RK V +RAV+ +
Sbjct: 159 TVDESGPAELYFESDDTMVEIWVDSVSLQPFTQKEWNFHQEQSIYKARKGAVRIRAVDSE 218
Query: 224 GHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQ 283
G P+PNA+IS+ KR GFPFG + KNIL N AY++WF RFTVTTF NEMKWY+ E +
Sbjct: 219 GQPIPNATISIQQKRLGFPFGCEVEKNILGNQAYENWFTQRFTVTTFANEMKWYSTEVVR 278
Query: 284 GKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVS 343
GK++Y ADAML F ++ G+AVRGHN+ WDDP+YQP WV+SLS + L +AV++RV S+VS
Sbjct: 279 GKEDYSTADAMLRFFKQHGVAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAVKRRVFSVVS 338
Query: 344 RYKGQLIGWDVVNENLHFSFFESKLGQNFS 373
RYKGQL GWDVVNENLHFSFFE+K+G S
Sbjct: 339 RYKGQLAGWDVVNENLHFSFFENKMGPKAS 368
>UniRef100_Q84VX1 At4g38650 [Arabidopsis thaliana]
Length = 562
Score = 401 bits (1031), Expect = e-110
Identities = 214/527 (40%), Positives = 302/527 (56%), Gaps = 35/527 (6%)
Query: 41 YDYSANVECLAHPQKPQYNGGIIKNPELN----DGLQGWTTFGDAIIEHRKSLGNKFVVT 96
YD +A EC A +KP YNGG++K+ + + D L G +G + T
Sbjct: 34 YDSTAYTECRAEAEKPLYNGGMLKDQKPSVPGKDSLTG--------------IGAHYTPT 79
Query: 97 HSRNQPHDSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKG-FKFGGAIFAEPNC 155
+ H+ IY +S+ I+ + V A ++ G++ A+ C
Sbjct: 80 YIL---HNLTQNTIYC-----FSIWVKIEAGAASAHVRARLRADNATLNCVGSVTAKHGC 131
Query: 156 WSMLKGGLIADTTGVAE-LYFESNNTS--VEIWVDNVSLQPFTEKQWRSHQELSIEKDRK 212
WS LKGG + D+ L+FE++ +++ V + SLQPFT++QWR++Q+ I RK
Sbjct: 132 WSFLKGGFLLDSPCKQSILFFETSEDDGKIQLQVTSASLQPFTQEQWRNNQDYFINTARK 191
Query: 213 RKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFEN 272
R V + E G + A +++ F GSAI+K IL N YQ+WF RF T FEN
Sbjct: 192 RAVTIHVSKENGESVEGAEVTVEQISKDFSIGSAISKTILGNIPYQEWFVKRFDATVFEN 251
Query: 273 EMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLND 332
E+KWY E QGK NY AD M+ F I RGHNIFW+DP+Y P+WV +L+ + L
Sbjct: 252 ELKWYATEPDQGKLNYTLADKMMNFVRANRIIARGHNIFWEDPKYNPDWVRNLTGEDLRS 311
Query: 333 AVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMN 392
AV +R+ S+++RY+G+ + WDV NE LHF F+E++LG+N S F ID TLF N
Sbjct: 312 AVNRRIKSLMTRYRGEFVHWDVSNEMLHFDFYETRLGKNASYGFFAAAREIDSLATLFFN 371
Query: 393 EYNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLR 452
++N +E D ST YI +++E+Q + GIGLE HF + PN+ MRA LD L
Sbjct: 372 DFNVVETCSDEKSTVDEYIARVRELQRYDGVRMDGIGLEGHF--TTPNVALMRAILDKLA 429
Query: 453 ATGLPIWITELDVAS---QPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTD 509
LPIW+TE+D++S +QA+Y EQVLRE SHP + GI++WTA P GCY++CLTD
Sbjct: 430 TLQLPIWLTEIDISSSLDHRSQAIYLEQVLREGFSHPSVNGIMLWTALHPNGCYQMCLTD 489
Query: 510 NNFKNLPAGDVVDQLINEWGRAEKSGTTDQNGYFEASLFHGDYEIEI 556
+ F+NLPAGDVVDQ + EW E TTD +G F F G+Y + I
Sbjct: 490 DKFRNLPAGDVVDQKLLEWKTGEVKATTDDHGSFSFFGFLGEYRVGI 536
>UniRef100_Q84WT5 Putative glycosyl hydrolase family 10 protein [Arabidopsis
thaliana]
Length = 570
Score = 395 bits (1014), Expect = e-108
Identities = 207/541 (38%), Positives = 309/541 (56%), Gaps = 28/541 (5%)
Query: 29 YITTGFEAEALSYDYSANVECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKS 88
++ +G + S+ +S N EC+ P + G+ LQ + D E K
Sbjct: 19 HVDSGVSIDPFSHSHSLNTECVMKPPRSSETKGL---------LQFSRSLEDDSDEEWKI 69
Query: 89 LGNKFVVTHSRNQPHDSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKF--G 146
GN F+ ++Q+I L +G YS SAW+++ E VV T+ + G
Sbjct: 70 DGNGFI---------REMAQRIQLHQGNIYSFSAWVKLREGNDKKVGVVFRTENGRLVHG 120
Query: 147 GAIFAEPNCWSMLKGGLIADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELS 206
G + A CW++LKGG++ D +G +++FES N +I NV L+ F++++W+ Q+
Sbjct: 121 GEVRANQECWTLLKGGIVPDFSGPVDIFFESENRGAKISAHNVLLKQFSKEEWKLKQDQL 180
Query: 207 IEKDRKRKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFT 266
IEK RK KV E + ISL + F G +N IL + Y+ WFASRF
Sbjct: 181 IEKIRKSKVRFEVTYENKTAVKGVVISLKQTKSSFLLGCGMNFRILQSQGYRKWFASRFK 240
Query: 267 VTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSL- 325
+T+F NEMKWY E A+G++NY AD+ML FAE GI VRGH + WD+P+ QP+WV ++
Sbjct: 241 ITSFTNEMKWYATEKARGQENYTVADSMLKFAEDNGILVRGHTVLWDNPKMQPSWVKNIK 300
Query: 326 SPDQLNDAVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDG 385
P+ + + R+NS++ RYKG+L GWDVVNENLH+ +FE LG N S +N ID
Sbjct: 301 DPNDVMNVTLNRINSVMKRYKGKLTGWDVVNENLHWDYFEKMLGANASTSFYNLAFKIDP 360
Query: 386 QTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSV--NSQLPLGIGLESHFPNSPPNLPY 443
LF+NEYNTIE++++ +TP + ++EI + N + IG + HF + PNL Y
Sbjct: 361 DVRLFVNEYNTIENTKEFTATPIKVKKMMEEILAYPGNKNMKGAIGAQGHFGPTQPNLAY 420
Query: 444 MRASLDTLRATGLPIWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCY 503
+R++LDTL + GLPIW+TE+D+ PNQA Y E +LREA+SHP ++GI+++ G
Sbjct: 421 IRSALDTLGSLGLPIWLTEVDMPKCPNQAQYVEDILREAYSHPAVKGIIIFGGPEVSGFD 480
Query: 504 RICLTDNNFKNLPAGDVVDQLINEWGRAEKSGTTD-----QNGYFEASLFHGDYEIEINH 558
++ L D +F N GDV+D+L+ EW + T+ N E SL HG Y + ++H
Sbjct: 481 KLTLADKDFNNTQTGDVIDKLLKEWQQKSSEIQTNFTADSDNEEEEVSLLHGHYNVNVSH 540
Query: 559 P 559
P
Sbjct: 541 P 541
>UniRef100_O81897 Beta-xylan endohydrolase-like protein [Arabidopsis thaliana]
Length = 536
Score = 393 bits (1009), Expect = e-108
Identities = 196/483 (40%), Positives = 297/483 (60%), Gaps = 16/483 (3%)
Query: 106 VSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKF--GGAIFAEPNCWSMLKGGL 163
++Q+I L +G YS SAW+++ E VV T+ +F GG + A+ CW++LKGG+
Sbjct: 38 MTQRIQLHEGNIYSFSAWVKLREGNNKKVGVVFRTENGRFVHGGEVRAKKRCWTLLKGGI 97
Query: 164 IADTTGVAELYFE-------SNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVV 216
+ D +G +++FE S++ +I +VSL+ F++++W+ Q+ IEK RK KV
Sbjct: 98 VPDVSGSVDIFFEVQQLAIYSDDKEAKISASDVSLKQFSKQEWKLKQDQLIEKIRKSKVR 157
Query: 217 VRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKW 276
+ + A IS+ +P F G A+N IL + Y++WFASRF +T+F NEMKW
Sbjct: 158 FEVTYQNKTAVKGAVISIEQTKPSFLLGCAMNFRILQSEGYRNWFASRFKITSFTNEMKW 217
Query: 277 YTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSL-SPDQLNDAVE 335
YT E +G +NY AD+ML FAE+ GI VRGH + WDDP QP WV + P+ L +
Sbjct: 218 YTTEKERGHENYTAADSMLKFAEENGILVRGHTVLWDDPLMQPTWVPKIEDPNDLMNVTL 277
Query: 336 KRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYN 395
R+NS+++RYKG+L GWDVVNEN+H+ +FE LG N S+ +N +D T+F+NEYN
Sbjct: 278 NRINSVMTRYKGKLTGWDVVNENVHWDYFEKMLGANASSSFYNLAFKLDPDVTMFVNEYN 337
Query: 396 TIEDSRDGLSTPPTYIEKIKEIQSV--NSQLPLGIGLESHFPNSPPNLPYMRASLDTLRA 453
TIE+ + +TP EK++EI + N + IG + HF + PNL YMR++LDTL +
Sbjct: 338 TIENRVEVTATPVKVKEKMEEILAYPGNMNIKGAIGAQGHFRPTQPNLAYMRSALDTLGS 397
Query: 454 TGLPIWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFK 513
GLPIW+TE+D+ PNQ +Y E++LREA+SHP ++GI+++ G ++ L D F
Sbjct: 398 LGLPIWLTEVDMPKCPNQEVYIEEILREAYSHPAVKGIIIFAGPEVSGFDKLTLADKYFN 457
Query: 514 NLPAGDVVDQLINEWGRAEKSG----TTDQNGYFEASLFHGDYEIEINHPIKKKSNFTHH 569
N GDV+D+L+ EW ++ + T +N E SL HG Y + ++HP K + +
Sbjct: 458 NTATGDVIDKLLKEWQQSSEIPKIFMTDSENDEEEVSLLHGHYNVNVSHPWMKNMSTSFS 517
Query: 570 IKV 572
++V
Sbjct: 518 LEV 520
>UniRef100_Q680B7 1,4-beta-xylan endohydrolase [Arabidopsis thaliana]
Length = 570
Score = 375 bits (963), Expect = e-102
Identities = 205/580 (35%), Positives = 325/580 (55%), Gaps = 45/580 (7%)
Query: 24 LIT*SYITTGFEAEALSYDYSANVECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAII 83
LI+ + +G + SYD S ECL P + N G G+ +
Sbjct: 15 LISLLLLGSGICMDPFSYDQSLKSECLMEPPQTTANTG-----------------GEGVK 57
Query: 84 EHRKSLGNKFVVTHSRNQPHDSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGF 143
E + + N +V + + LR+G Y SAW+++ E+ + + K
Sbjct: 58 E----------LKINENGGIRNVVEGVDLREGNIYITSAWVKLRNESQRKVGMTFSEKNG 107
Query: 144 K--FGGAIFAEPNCWSMLKGGLIADTTGVAELYFESNNTS-VEIWVDNVSLQPFTEKQWR 200
+ FGG + A+ CWS+LKGG+ AD +G +++FES+ + +EI V NV +Q F + QWR
Sbjct: 108 RNVFGGEVMAKRGCWSLLKGGITADFSGPIDIFFESDGLAGLEISVQNVRMQRFHKTQWR 167
Query: 201 SHQELSIEKDRKRKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDW 260
Q+ IEK RK KV + + L + IS+ +P F G A+N IL +++Y++W
Sbjct: 168 LQQDQVIEKIRKNKVRFQMSFKNKSALEGSVISIEQIKPSFLLGCAMNYRILESDSYREW 227
Query: 261 FASRFTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPN 320
F SRF +T+F NEMKWY E +G++NY AD+M+ AE+ I V+GH + WDD +QPN
Sbjct: 228 FVSRFRLTSFTNEMKWYATEAVRGQENYKIADSMMQLAEENAILVKGHTVLWDDKYWQPN 287
Query: 321 WVSSLS-PDQLNDAVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNE 379
WV +++ P+ L + R+NS++ RYKG+LIGWDV+NEN+HF++FE+ LG N SA +++
Sbjct: 288 WVKTITDPEDLKNVTLNRMNSVMKRYKGRLIGWDVMNENVHFNYFENMLGGNASAIVYSL 347
Query: 380 VHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSV--NSQLPLGIGLESHFPNS 437
+D LF+NE+NT+E +D + +P ++K++EI S N+ + GIG + HF
Sbjct: 348 ASKLDPDIPLFLNEFNTVEYDKDRVVSPVNVVKKMQEIVSFPGNNNIKGGIGAQGHFAPV 407
Query: 438 PPNLPYMRASLDTLRATGLPIWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWTAW 497
PNL YMR +LDTL + P+W+TE+D+ P+Q Y E +LREA+SHP ++ I+++
Sbjct: 408 QPNLAYMRYALDTLGSLSFPVWLTEVDMFKCPDQVKYMEDILREAYSHPAVKAIILYGGP 467
Query: 498 SPQGCYRICLTDNNFKNLPAGDVVDQLINEWGR------AEKSGTTDQNG------YFEA 545
G ++ L D +FKN AGD++D+L+ EW + + D+ G E
Sbjct: 468 EVSGFDKLTLADKDFKNTQAGDLIDKLLQEWKQEPVEIPIQHHEHNDEEGGRIIGFSPEI 527
Query: 546 SLFHGDYEIEINHPIKKKSNFTHHIKVLSKDEFKKTKQFI 585
SL HG Y + + +P K + ++V + + Q +
Sbjct: 528 SLLHGHYRVTVTNPSMKNLSTRFSVEVTKESGHLQEVQLV 567
>UniRef100_Q9M070 Hypothetical protein AT4g33820 [Arabidopsis thaliana]
Length = 546
Score = 373 bits (958), Expect = e-102
Identities = 191/480 (39%), Positives = 282/480 (57%), Gaps = 26/480 (5%)
Query: 106 VSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKF--GGAIFAEPNCWSMLKGGL 163
++Q+I L +G YS SAW+++ E VV T+ + GG + A CW++LKGG+
Sbjct: 38 MAQRIQLHQGNIYSFSAWVKLREGNDKKVGVVFRTENGRLVHGGEVRANQECWTLLKGGI 97
Query: 164 IADTTGVAELYFE----------------SNNTSVEIWVDNVSLQPFTEKQWRSHQELSI 207
+ D +G +++FE S N +I NV L+ F++++W+ Q+ I
Sbjct: 98 VPDFSGPVDIFFEIHTYILCVNVVLMRKQSENRGAKISAHNVLLKQFSKEEWKLKQDQLI 157
Query: 208 EKDRKRKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTV 267
EK RK KV E + ISL + F G +N IL + Y+ WFASRF +
Sbjct: 158 EKIRKSKVRFEVTYENKTAVKGVVISLKQTKSSFLLGCGMNFRILQSQGYRKWFASRFKI 217
Query: 268 TTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSL-S 326
T+F NEMKWY E A+G++NY AD+ML FAE GI VRGH + WD+P+ QP+WV ++
Sbjct: 218 TSFTNEMKWYATEKARGQENYTVADSMLKFAEDNGILVRGHTVLWDNPKMQPSWVKNIKD 277
Query: 327 PDQLNDAVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQ 386
P+ + + R+NS++ RYKG+L GWDVVNENLH+ +FE LG N S +N ID
Sbjct: 278 PNDVMNVTLNRINSVMKRYKGKLTGWDVVNENLHWDYFEKMLGANASTSFYNLAFKIDPD 337
Query: 387 TTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSV--NSQLPLGIGLESHFPNSPPNLPYM 444
LF+NEYNTIE++++ +TP + ++EI + N + IG + HF + PNL Y+
Sbjct: 338 VRLFVNEYNTIENTKEFTATPIKVKKMMEEILAYPGNKNMKGAIGAQGHFGPTQPNLAYI 397
Query: 445 RASLDTLRATGLPIWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYR 504
R++LDTL + GLPIW+TE+D+ PNQA Y E +LREA+SHP ++GI+++ G +
Sbjct: 398 RSALDTLGSLGLPIWLTEVDMPKCPNQAQYVEDILREAYSHPAVKGIIIFGGPEVSGFDK 457
Query: 505 ICLTDNNFKNLPAGDVVDQLINEWGRAEKSGTTD-----QNGYFEASLFHGDYEIEINHP 559
+ L D +F N GDV+D+L+ EW + T+ N E SL HG Y + ++HP
Sbjct: 458 LTLADKDFNNTQTGDVIDKLLKEWQQKSSEIQTNFTADSDNEEEEVSLLHGHYNVNVSHP 517
>UniRef100_Q6NQ91 At2g14690 [Arabidopsis thaliana]
Length = 529
Score = 371 bits (952), Expect = e-101
Identities = 189/499 (37%), Positives = 300/499 (59%), Gaps = 18/499 (3%)
Query: 105 SVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFK--FGGAIFAEPNCWSMLKGG 162
+V + + LR+G Y SAW+++ E+ + + K + FGG + A+ CWS+LKGG
Sbjct: 28 NVVEGVDLREGNIYITSAWVKLRNESQRKVGMTFSEKNGRNVFGGEVMAKRGCWSLLKGG 87
Query: 163 LIADTTGVAELYFESNNTS-VEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVVVRAVN 221
+ AD +G +++FES+ + +EI V NV +Q F + QWR Q+ IEK RK KV +
Sbjct: 88 ITADFSGPIDIFFESDGLAGLEISVQNVRMQRFHKTQWRLQQDQVIEKIRKNKVRFQMSF 147
Query: 222 EQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEY 281
+ L + IS+ +P F G A+N IL +++Y++WF SRF +T+F NEMKWY E
Sbjct: 148 KNKSALEGSVISIEQIKPSFLLGCAMNYRILESDSYREWFVSRFRLTSFTNEMKWYATEA 207
Query: 282 AQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLS-PDQLNDAVEKRVNS 340
+G++NY AD+M+ AE+ I V+GH + WDD +QPNWV +++ P+ L + R+NS
Sbjct: 208 VRGQENYKIADSMMQLAEENAILVKGHTVLWDDKYWQPNWVKTITDPEDLKNVTLNRMNS 267
Query: 341 IVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDS 400
++ RYKG+LIGWDV+NEN+HF++FE+ LG N SA +++ +D LF+NE+NT+E
Sbjct: 268 VMKRYKGRLIGWDVMNENVHFNYFENMLGGNASAIVYSLASKLDPDIPLFLNEFNTVEYD 327
Query: 401 RDGLSTPPTYIEKIKEIQSV--NSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPI 458
+D + +P ++K++EI S N+ + GIG + HF PNL YMR +LDTL + P+
Sbjct: 328 KDRVVSPVNVVKKMQEIVSFPGNNNIKGGIGAQGHFAPVQPNLAYMRYALDTLGSLSFPV 387
Query: 459 WITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLPAG 518
W+TE+D+ P+Q Y E +LREA+SHP ++ I+++ G ++ L D +FKN AG
Sbjct: 388 WLTEVDMFKCPDQVKYMEDILREAYSHPAVKAIILYGGPEVSGFDKLTLADKDFKNTQAG 447
Query: 519 DVVDQLINEWGR------AEKSGTTDQNG------YFEASLFHGDYEIEINHPIKKKSNF 566
D++D+L+ EW + + D+ G E SL HG Y + + +P K +
Sbjct: 448 DLIDKLLQEWKQEPVEIPIQHHEHNDEEGGRIIGFSPEISLLHGHYRVTVTNPSMKNLST 507
Query: 567 THHIKVLSKDEFKKTKQFI 585
++V + + Q +
Sbjct: 508 RFSVEVTKESGHLQEVQLV 526
>UniRef100_Q9SZP3 Hypothetical protein F20M13.210 [Arabidopsis thaliana]
Length = 433
Score = 361 bits (926), Expect = 4e-98
Identities = 179/406 (44%), Positives = 245/406 (60%), Gaps = 13/406 (3%)
Query: 162 GLIADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIE--------KDRKR 213
GL + L ++ +++ V + SLQPFT++QWR++Q+ I RKR
Sbjct: 4 GLSSKEDSFLILLTSEDDGKIQLQVTSASLQPFTQEQWRNNQDYFINTVIHTLISNARKR 63
Query: 214 KVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENE 273
V + E G + A +++ F GSAI+K IL N YQ+WF RF T FENE
Sbjct: 64 AVTIHVSKENGESVEGAEVTVEQISKDFSIGSAISKTILGNIPYQEWFVKRFDATVFENE 123
Query: 274 MKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDA 333
+KWY E QGK NY AD M+ F I RGHNIFW+DP+Y P+WV +L+ + L A
Sbjct: 124 LKWYATEPDQGKLNYTLADKMMNFVRANRIIARGHNIFWEDPKYNPDWVRNLTGEDLRSA 183
Query: 334 VEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNE 393
V +R+ S+++RY+G+ + WDV NE LHF F+E++LG+N S F ID TLF N+
Sbjct: 184 VNRRIKSLMTRYRGEFVHWDVSNEMLHFDFYETRLGKNASYGFFAAAREIDSLATLFFND 243
Query: 394 YNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRA 453
+N +E D ST YI +++E+Q + GIGLE HF + PN+ MRA LD L
Sbjct: 244 FNVVETCSDEKSTVDEYIARVRELQRYDGVRMDGIGLEGHF--TTPNVALMRAILDKLAT 301
Query: 454 TGLPIWITELDVAS---QPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDN 510
LPIW+TE+D++S +QA+Y EQVLRE SHP + GI++WTA P GCY++CLTD+
Sbjct: 302 LQLPIWLTEIDISSSLDHRSQAIYLEQVLREGFSHPSVNGIMLWTALHPNGCYQMCLTDD 361
Query: 511 NFKNLPAGDVVDQLINEWGRAEKSGTTDQNGYFEASLFHGDYEIEI 556
F+NLPAGDVVDQ + EW E TTD +G F F G+Y + I
Sbjct: 362 KFRNLPAGDVVDQKLLEWKTGEVKATTDDHGSFSFFGFLGEYRVGI 407
>UniRef100_Q9ZVK8 1,4-beta-xylan endohydrolase [Arabidopsis thaliana]
Length = 552
Score = 360 bits (925), Expect = 5e-98
Identities = 190/522 (36%), Positives = 299/522 (56%), Gaps = 41/522 (7%)
Query: 105 SVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFK--FGGAIFAEPNCWSMLKGG 162
+V + + LR+G Y SAW+++ E+ + + K + FGG + A+ CWS+LKGG
Sbjct: 28 NVVEGVDLREGNIYITSAWVKLRNESQRKVGMTFSEKNGRNVFGGEVMAKRGCWSLLKGG 87
Query: 163 LIADTTGVAELYFE-------------SNNTSV-----------EIWVDNVSLQPFTEKQ 198
+ AD +G +++FE S N + EI V NV +Q F + Q
Sbjct: 88 ITADFSGPIDIFFEVKELIVCSFVEISSTNVGIYTKQSDGLAGLEISVQNVRMQRFHKTQ 147
Query: 199 WRSHQELSIEKDRKRKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQ 258
WR Q+ IEK RK KV + + L + IS+ +P F G A+N IL +++Y+
Sbjct: 148 WRLQQDQVIEKIRKNKVRFQMSFKNKSALEGSVISIEQIKPSFLLGCAMNYRILESDSYR 207
Query: 259 DWFASRFTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQ 318
+WF SRF +T+F NEMKWY E +G++NY AD+M+ AE+ I V+GH + WDD +Q
Sbjct: 208 EWFVSRFRLTSFTNEMKWYATEAVRGQENYKIADSMMQLAEENAILVKGHTVLWDDKYWQ 267
Query: 319 PNWVSSLS-PDQLNDAVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMF 377
PNWV +++ P+ L + R+NS++ RYKG+LIGWDV+NEN+HF++FE+ LG N SA ++
Sbjct: 268 PNWVKTITDPEDLKNVTLNRMNSVMKRYKGRLIGWDVMNENVHFNYFENMLGGNASAIVY 327
Query: 378 NEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSV--NSQLPLGIGLESHFP 435
+ +D LF+NE+NT+E +D + +P ++K++EI S N+ + GIG + HF
Sbjct: 328 SLASKLDPDIPLFLNEFNTVEYDKDRVVSPVNVVKKMQEIVSFPGNNNIKGGIGAQGHFA 387
Query: 436 NSPPNLPYMRASLDTLRATGLPIWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWT 495
PNL YMR +LDTL + P+W+TE+D+ P+Q Y E +LREA+SHP ++ I+++
Sbjct: 388 PVQPNLAYMRYALDTLGSLSFPVWLTEVDMFKCPDQVKYMEDILREAYSHPAVKAIILYG 447
Query: 496 AWSPQGCYRICLTDNNFKNLPAGDVVDQLINEWGR------AEKSGTTDQNG------YF 543
G ++ L D +FKN AGD++D+L+ EW + + D+ G
Sbjct: 448 GPEVSGFDKLTLADKDFKNTQAGDLIDKLLQEWKQEPVEIPIQHHEHNDEEGGRIIGFSP 507
Query: 544 EASLFHGDYEIEINHPIKKKSNFTHHIKVLSKDEFKKTKQFI 585
E SL HG Y + + +P K + ++V + + Q +
Sbjct: 508 EISLLHGHYRVTVTNPSMKNLSTRFSVEVTKESGHLQEVQLV 549
>UniRef100_Q9ZTB8 Tapetum specific protein [Zea mays]
Length = 329
Score = 327 bits (838), Expect = 6e-88
Identities = 154/293 (52%), Positives = 202/293 (68%), Gaps = 4/293 (1%)
Query: 226 PLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGK 285
P+ +A++S+ + R GFPFG+A+ K IL AY+ WF SRF+V TFENEMKWY+ E+ Q
Sbjct: 7 PMAHANVSIELLRLGFPFGNAVTKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNH 66
Query: 286 DNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVSRY 345
++Y DAM+ K I VRGHN+FWDD Q WV L+ QL A++KR+ S+VS Y
Sbjct: 67 EDYRVPDAMMSLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPY 126
Query: 346 KGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLS 405
G++I WDVVNENLHF+FFE+KLG SA+++ +V +D LFMNE+NT+E D
Sbjct: 127 AGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNP 186
Query: 406 TPPTYIEKIKEIQSV--NSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITEL 463
P Y+ K+ +I+ N L LG+GLESHF S PN+PYMR+SLDTL LP+W+TE+
Sbjct: 187 VPAKYVAKMNQIRGYAGNGGLKLGVGLESHF--STPNIPYMRSSLDTLAKLKLPMWLTEV 244
Query: 464 DVASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLP 516
DV PNQ Y EQVLRE +HP + GIVMW W +GCY +CLT+N+FKNLP
Sbjct: 245 DVVKSPNQVKYLEQVLREGFAHPNVDGIVMWAGWHAKGCYVMCLTNNSFKNLP 297
>UniRef100_Q8GZB5 Anther endoxylanase [Hordeum vulgare var. distichum]
Length = 318
Score = 306 bits (784), Expect = 1e-81
Identities = 144/293 (49%), Positives = 201/293 (68%), Gaps = 6/293 (2%)
Query: 271 ENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQL 330
ENEMKWY+ E+ + +++Y DAML A++ GI VRGHN+FWD Q WV+ LS D+L
Sbjct: 1 ENEMKWYSTEWKRNREDYSVPDAMLALAQRHGIKVRGHNVFWDTNNMQMAWVNPLSADEL 60
Query: 331 NDAVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLF 390
A++KR++S+V+RY G++I WDVVNENLH F+ES+LG N SA ++ +V ID TLF
Sbjct: 61 KAAMQKRLSSLVTRYAGKVIAWDVVNENLHGQFYESRLGPNVSAELYQQVAKIDTNATLF 120
Query: 391 MNEYNTIEDSRDGLSTPPTYIEKIKEIQSV--NSQLPLGIGLESHFPNSPPNLPYMRASL 448
MNEY+T+E + D + Y K+++I+S N + L +GLESHF PN+PYMRA+L
Sbjct: 121 MNEYDTLEWALDVTAMASKYAAKMEQIRSYPGNDGIKLAVGLESHF--ETPNIPYMRATL 178
Query: 449 DTLRATGLPIWITELDVA--SQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRIC 506
D L +PIW+TE+DV+ ++P Q Y E VLRE + HP ++G+V+W AW GC+ +C
Sbjct: 179 DMLAQLKVPIWLTEVDVSPKTRPYQVEYLEDVLREGYGHPNVEGMVLWAAWHKHGCWVMC 238
Query: 507 LTDNNFKNLPAGDVVDQLINEWGRAEKSGTTDQNGYFEASLFHGDYEIEINHP 559
LTDN+F NLP G+VVD+LI+EW + TTD +G E L HG+Y + HP
Sbjct: 239 LTDNSFTNLPTGNVVDKLIDEWKTHPVAATTDAHGVAELDLVHGEYRFTVTHP 291
>UniRef100_Q70DK4 Beta-1,4-xylanase [Clostridium thermocellum]
Length = 639
Score = 263 bits (673), Expect = 8e-69
Identities = 170/523 (32%), Positives = 265/523 (50%), Gaps = 29/523 (5%)
Query: 60 GGIIKNPELNDG-LQGWTTFGDAIIEH---RKSLGNKFVVTHSRNQPHDSVSQKIY--LR 113
G ++ NP G +GW +G+ IE GN V R Q + V+Q + L
Sbjct: 29 GNLLFNPGFELGSTEGWYPYGECTIEAVGTEAHSGNYSVFVTDRTQDWNGVAQDMLDKLT 88
Query: 114 KGLHYSLSAWIQVS-----EETVPVTAVVKTTKGFKFGG--AIFAEPNCWSMLKGGLIAD 166
G+ Y +SAW++V+ + +P+ V +T K + +I E + W L G
Sbjct: 89 VGMTYQVSAWVKVAGTGSHQVKIPMKKV-ETGKEPVYDNIPSITVEGSEWYRLSGPYSYT 147
Query: 167 TTGVA--ELYFESNNTSVEIWVDNVSLQPF-TEKQWRSHQELSIEKDRKRKVVVRAVNEQ 223
T V ELY E V +VD+V++ + W+ IE+ RKR +R V+
Sbjct: 148 GTNVTNLELYIEGPQPGVSYYVDDVTVTEVGSAATWKEEANARIEQIRKRDPKIRIVDSN 207
Query: 224 GHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQ 283
P+ SI + + F FGSAI N +++ Y ++F + + FENE KWY+NE +Q
Sbjct: 208 NKPVSGVSIDVRQVKHEFGFGSAITMNGIHDPRYTEFFKNNYEWAVFENEAKWYSNESSQ 267
Query: 284 GKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVS 343
G +Y +AD + + + GI VRGH IFW+ ++QP+W+ L+ D L A++ R+ S+V
Sbjct: 268 GNVSYANADYLYNWCAENGIKVRGHCIFWEPEEWQPSWLKGLTGDALMKAIDARLESVVP 327
Query: 344 RYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDG 403
++G+ + WDV NE LH FF+S+LG++ MF +D LF+N+YN I
Sbjct: 328 HFRGKFLHWDVNNEMLHGDFFKSRLGESIWPYMFKRARELDPDAKLFVNDYNIIT----- 382
Query: 404 LSTPPTYIEKIKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITEL 463
YI +I+ + +++ GIG++ HF L ++A LD L G+PIW+TE
Sbjct: 383 YVEGDAYIRQIEWLLQNGAEID-GIGVQGHFDEDVEPL-VVKARLDNLATLGIPIWVTEY 440
Query: 464 DVASQP--NQALYFEQVLREAHSHPGIQGIVMWTAWSPQGC--YRICLTDNNFKNLPAGD 519
D + +A E + R A SHP ++GI+MW W+ + D+++ AG
Sbjct: 441 DSKTPDVNKRAENLENLYRIAFSHPAVEGIIMWGFWAGNHWRGQDAAIVDHDWTVNEAGK 500
Query: 520 VVDQLINEWGRAEKSGTTDQNGYFEASLFHGDYEIEINHPIKK 562
L+ EW SGTTD G F+ FHG YEI ++ P K+
Sbjct: 501 RYQALLKEW-TTITSGTTDSTGAFDFRGFHGTYEITVSVPGKE 542
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,057,909,695
Number of Sequences: 2790947
Number of extensions: 47228834
Number of successful extensions: 103667
Number of sequences better than 10.0: 325
Number of HSP's better than 10.0 without gapping: 206
Number of HSP's successfully gapped in prelim test: 119
Number of HSP's that attempted gapping in prelim test: 102886
Number of HSP's gapped (non-prelim): 362
length of query: 588
length of database: 848,049,833
effective HSP length: 133
effective length of query: 455
effective length of database: 476,853,882
effective search space: 216968516310
effective search space used: 216968516310
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 78 (34.7 bits)
Medicago: description of AC146757.5


