

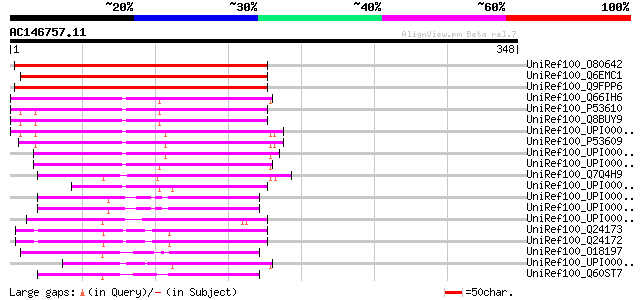
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.11 - phase: 0 /pseudo
(348 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O80642 Putative geranylgeranyl transferase type I beta... 206 6e-52
UniRef100_Q6EMC1 Geranylgeranyltransferase type I beta subunit [... 198 2e-49
UniRef100_Q9FPP6 Geranylgeranyltransferase beta subunit [Arabido... 173 6e-42
UniRef100_Q66IH6 MGC89595 protein [Xenopus tropicalis] 124 5e-27
UniRef100_P53610 Geranylgeranyl transferase type I beta subunit ... 116 8e-25
UniRef100_Q8BUY9 Mus musculus 16 days embryo head cDNA, RIKEN fu... 115 2e-24
UniRef100_UPI00001AEC1C UPI00001AEC1C UniRef100 entry 114 5e-24
UniRef100_P53609 Geranylgeranyl transferase type I beta subunit ... 113 7e-24
UniRef100_UPI00002DA721 UPI00002DA721 UniRef100 entry 108 2e-22
UniRef100_UPI00003646CA UPI00003646CA UniRef100 entry 103 7e-21
UniRef100_Q7Q4H9 ENSANGP00000019628 [Anopheles gambiae str. PEST] 96 2e-18
UniRef100_UPI00003AF00C UPI00003AF00C UniRef100 entry 94 6e-18
UniRef100_UPI0000433673 UPI0000433673 UniRef100 entry 88 4e-16
UniRef100_UPI0000433672 UPI0000433672 UniRef100 entry 88 4e-16
UniRef100_UPI00003C20A7 UPI00003C20A7 UniRef100 entry 86 2e-15
UniRef100_Q24173 CG3469-PA [Drosophila melanogaster] 85 3e-15
UniRef100_Q24172 Geranylgeranyl transferase beta-subunit type I ... 85 3e-15
UniRef100_O18197 Hypothetical protein Y48E1B.3 [Caenorhabditis e... 84 7e-15
UniRef100_UPI00003646CB UPI00003646CB UniRef100 entry 81 5e-14
UniRef100_Q60ST7 Hypothetical protein CBG20759 [Caenorhabditis b... 78 4e-13
>UniRef100_O80642 Putative geranylgeranyl transferase type I beta subunit
[Arabidopsis thaliana]
Length = 375
Score = 206 bits (525), Expect = 6e-52
Identities = 103/174 (59%), Positives = 125/174 (71%)
Query: 4 SEEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVE 63
S S + +KD H+ + +MMY LLP Y+SQEIN LTLA+F+IS L L + V+
Sbjct: 24 SPPVQSSPSANFEKDRHLMYLEMMYELLPYHYQSQEINRLTLAHFIISGLHFLGARDRVD 83
Query: 64 KEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALA 123
K+ VA WVLSFQ +G+FYGF GS++SQFP DENG HN SHLASTYCALA
Sbjct: 84 KDVVAKWVLSFQAFPTNRVSLKDGEFYGFFGSRSSQFPIDENGDLKHNGSHLASTYCALA 143
Query: 124 ILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
ILK++G+DLS++DS+S+ SM NLQQ DGSFMPIHIGGETDLRFVYCA I M
Sbjct: 144 ILKVIGHDLSTIDSKSLLISMINLQQDDGSFMPIHIGGETDLRFVYCAAAICYM 197
Score = 34.3 bits (77), Expect = 5.2
Identities = 29/100 (29%), Positives = 45/100 (45%), Gaps = 27/100 (27%)
Query: 43 LTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPP 102
L Y + +L+S ++KE+ N++L+ Q G GF
Sbjct: 185 LRFVYCAAAICYMLDSWSGMDKESAKNYILNCQSYDG-----------GF---------- 223
Query: 103 DENGVFHHNNSHLASTYCALAILKIVGY---DLSSLDSES 139
G+ + SH +TYCA+A L+++GY DL S DS S
Sbjct: 224 ---GLIPGSESHGGATYCAIASLRLMGYIGVDLLSNDSSS 260
>UniRef100_Q6EMC1 Geranylgeranyltransferase type I beta subunit [Catharanthus roseus]
Length = 359
Score = 198 bits (503), Expect = 2e-49
Identities = 99/170 (58%), Positives = 121/170 (70%)
Query: 8 SSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAV 67
S E D+D HV F +MMY LLP+ Y+SQEINH+TLA+F I LDIL +L ++K+ V
Sbjct: 10 SDSELQFFDRDRHVQFLEMMYDLLPSRYQSQEINHITLAHFAIVGLDILGALDRIDKQEV 69
Query: 68 ANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKI 127
NWVLS Q ++ +NGQFYGFHGS++SQF ++ G N SHLAS+YCAL IL+
Sbjct: 70 INWVLSLQAHPKNADELDNGQFYGFHGSRSSQFQSNDEGDTVPNVSHLASSYCALTILRT 129
Query: 128 VGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
VGYD S L+S+ + SMKNLQQ DGSFMPIH G ETDLRFVYCA I M
Sbjct: 130 VGYDFSLLNSKLILESMKNLQQQDGSFMPIHSGAETDLRFVYCAAAICFM 179
>UniRef100_Q9FPP6 Geranylgeranyltransferase beta subunit [Arabidopsis thaliana]
Length = 376
Score = 173 bits (439), Expect = 6e-42
Identities = 94/175 (53%), Positives = 113/175 (63%), Gaps = 1/175 (0%)
Query: 4 SEEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVE 63
S S + +KD H+ + +MMY LLP Y+SQEIN LTLA+F+IS L L + V+
Sbjct: 24 SPPVQSSPSANFEKDRHLMYLEMMYELLPYHYQSQEINRLTLAHFIISGLHFLGARDRVD 83
Query: 64 KEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSK-TSQFPPDENGVFHHNNSHLASTYCAL 122
K VA WVLSFQ + F K FP +NG HN SHLASTYCAL
Sbjct: 84 KYVVAKWVLSFQAFPTNRVSLKRWRILWFLWFKGVLSFPLMKNGDLKHNGSHLASTYCAL 143
Query: 123 AILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
AILK++G+DLS++DS+S+ SM NLQQ DGSFMPIHIGGETDLRFVYCA I M
Sbjct: 144 AILKVIGHDLSTIDSKSLLISMINLQQDDGSFMPIHIGGETDLRFVYCAAAICYM 198
Score = 34.3 bits (77), Expect = 5.2
Identities = 29/100 (29%), Positives = 45/100 (45%), Gaps = 27/100 (27%)
Query: 43 LTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPP 102
L Y + +L+S ++KE+ N++L+ Q G GF
Sbjct: 186 LRFVYCAAAICYMLDSWSGMDKESAKNYILNCQSYDG-----------GF---------- 224
Query: 103 DENGVFHHNNSHLASTYCALAILKIVGY---DLSSLDSES 139
G+ + SH +TYCA+A L+++GY DL S DS S
Sbjct: 225 ---GLIPGSESHGGATYCAIASLRLMGYIGVDLLSNDSSS 261
>UniRef100_Q66IH6 MGC89595 protein [Xenopus tropicalis]
Length = 372
Score = 124 bits (310), Expect = 5e-27
Identities = 73/186 (39%), Positives = 105/186 (56%), Gaps = 8/186 (4%)
Query: 1 MSSSEEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLH 60
M+ E S E D HV + Q LLP S E N LT+A+F +S LD+L+SL+
Sbjct: 1 MAEDENGSPEERLLFLPDRHVRYFQRSLQLLPEQCASLETNRLTIAFFALSGLDMLDSLN 60
Query: 61 LVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP--PDE-NGVFH-HNNSHLA 116
++ K + W+ S QV T D +N GF GS P P + +G++H H++ H+A
Sbjct: 61 VINKSEIIEWIYSLQVL--PTEDKSNLDRCGFRGSSCLGLPFNPSKGHGLYHPHDSGHIA 118
Query: 117 STYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITG 176
TY A+A L I+G DLS ++ E+ + ++ LQ PDGSF + G E D+RFVYCA I
Sbjct: 119 MTYTAIASLLILGDDLSRVNKEACLAGLRALQLPDGSFCAVPEGSENDMRFVYCAACICY 178
Query: 177 M--EWT 180
M +W+
Sbjct: 179 MLNDWS 184
>UniRef100_P53610 Geranylgeranyl transferase type I beta subunit [Rattus norvegicus]
Length = 377
Score = 116 bits (291), Expect = 8e-25
Identities = 74/186 (39%), Positives = 106/186 (56%), Gaps = 11/186 (5%)
Query: 1 MSSSEE---ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDI 55
M+++E+ A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+
Sbjct: 1 MAATEDDRLAGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDM 60
Query: 56 LNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP--PDEN-GVFH-HN 111
L+SL +V K+ + W+ S QV T D +N GF GS P P +N G H ++
Sbjct: 61 LDSLDVVNKDDIIEWIYSLQVL--PTEDRSNLDRCGFRGSSYLGIPFNPSKNPGTAHPYD 118
Query: 112 NSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
+ H+A TY L+ L I+G DLS +D E+ + ++ LQ DGSF + G E D+RFVYCA
Sbjct: 119 SGHIAMTYTGLSCLIILGDDLSRVDKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCA 178
Query: 172 GWITGM 177
I M
Sbjct: 179 SCICYM 184
>UniRef100_Q8BUY9 Mus musculus 16 days embryo head cDNA, RIKEN full-length enriched
library, clone:C130072G06 product:GERANYLGERANYL
TRANSFERASE TYPE I BETA SUBUNIT (EC 2.5.1.-) (TYPE I
PROTEIN GERANYL-GERANYLTRANSFERASE BETA SUBUNIT)
(GGTASE-I-BETA) homolog [Mus musculus]
Length = 377
Score = 115 bits (287), Expect = 2e-24
Identities = 73/186 (39%), Positives = 105/186 (56%), Gaps = 11/186 (5%)
Query: 1 MSSSEE---ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDI 55
M+++E+ A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+
Sbjct: 1 MATTEDDRLAGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDM 60
Query: 56 LNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP--PDEN-GVFH-HN 111
L+SL +V K+ + W+ S QV T D +N GF GS P P +N G H ++
Sbjct: 61 LDSLDVVNKDDIIEWIYSLQVL--PTEDRSNLSRCGFRGSSYLGIPFNPSKNPGAAHPYD 118
Query: 112 NSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
+ H+A TY L+ L I+G DL +D E+ + ++ LQ DGSF + G E D+RFVYCA
Sbjct: 119 SGHIAMTYTGLSCLIILGDDLGRVDKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCA 178
Query: 172 GWITGM 177
I M
Sbjct: 179 SCICYM 184
>UniRef100_UPI00001AEC1C UPI00001AEC1C UniRef100 entry
Length = 377
Score = 114 bits (284), Expect = 5e-24
Identities = 76/201 (37%), Positives = 111/201 (54%), Gaps = 15/201 (7%)
Query: 1 MSSSEE---ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDI 55
M+++E+ A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+
Sbjct: 1 MAATEDERLAGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDM 60
Query: 56 LNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDEN---GVFH-HN 111
L+SL +V K+ + W+ S QV T D +N GF GS P + + G H ++
Sbjct: 61 LDSLDVVNKDDIIEWIYSLQVL--PTEDRSNLNRCGFRGSSYLGIPFNPSKAPGTAHPYD 118
Query: 112 NSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
+ H+A TY L+ L I+G DLS ++ E+ + ++ LQ DGSF + G E D+RFVYCA
Sbjct: 119 SGHIAMTYTGLSCLVILGDDLSRVNKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCA 178
Query: 172 GWITGM--EWT--RKKSRITY 188
I M W+ K ITY
Sbjct: 179 SCICYMLNNWSGMDMKKAITY 199
>UniRef100_P53609 Geranylgeranyl transferase type I beta subunit [Homo sapiens]
Length = 377
Score = 113 bits (283), Expect = 7e-24
Identities = 74/192 (38%), Positives = 105/192 (54%), Gaps = 12/192 (6%)
Query: 7 ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEK 64
A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+L+SL +V K
Sbjct: 10 AGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDMLDSLDVVNK 69
Query: 65 EAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDEN---GVFH-HNNSHLASTYC 120
+ + W+ S QV T D +N GF GS P + + G H +++ H+A TY
Sbjct: 70 DDIIEWIYSLQVL--PTEDRSNLNRCGFRGSSYLGIPFNPSKAPGTAHPYDSGHIAMTYT 127
Query: 121 ALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM--E 178
L+ L I+G DLS ++ E+ + ++ LQ DGSF + G E D+RFVYCA I M
Sbjct: 128 GLSCLVILGDDLSRVNKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCASCICYMLNN 187
Query: 179 WT--RKKSRITY 188
W+ K ITY
Sbjct: 188 WSGMDMKKAITY 199
>UniRef100_UPI00002DA721 UPI00002DA721 UniRef100 entry
Length = 611
Score = 108 bits (270), Expect = 2e-22
Identities = 64/174 (36%), Positives = 94/174 (53%), Gaps = 7/174 (4%)
Query: 17 KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQV 76
KD HV F Q +LP Y S E LT+ +F +S LD+L++L V+K + W+ S QV
Sbjct: 18 KDRHVRFFQRTLQVLPERYASLETTRLTIVFFALSGLDVLDALDAVDKNVMIEWIYSLQV 77
Query: 77 QRGTTNDPNNGQFYGFHGSKTSQFPPDEN--GVFH-HNNSHLASTYCALAILKIVGYDLS 133
T D +N GF GS P GV H +++ H+A TY L L I+G +LS
Sbjct: 78 L--PTEDQSNLSRCGFRGSSYMGIPYSTKGPGVSHPYDSGHVAMTYTGLCSLLILGDNLS 135
Query: 134 SLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM--EWTRKKSR 185
++ ++ + ++ LQ DGSF + G E D+RF+YCA I M +W+ +R
Sbjct: 136 RVNKQACLAGLRALQMEDGSFYALPEGSENDIRFIYCAASICYMLDDWSGMDTR 189
>UniRef100_UPI00003646CA UPI00003646CA UniRef100 entry
Length = 348
Score = 103 bits (257), Expect = 7e-21
Identities = 62/169 (36%), Positives = 92/169 (53%), Gaps = 7/169 (4%)
Query: 17 KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQV 76
KD HV F Q +LP Y S E LT+ +F +S LD+L++L +V+K + W+ S QV
Sbjct: 9 KDRHVRFFQRTLQVLPERYASLETTRLTIVFFALSGLDVLDALDVVDKNVMIEWIYSLQV 68
Query: 77 QRGTTNDPNNGQFYGFHGSKTSQFP--PDENGVFH-HNNSHLASTYCALAILKIVGYDLS 133
T +N GF GS P GV H +++ H+A TY L L I+G +LS
Sbjct: 69 L--PTEAQSNLSRCGFRGSSHIGIPYRTKGPGVSHPYDSGHVAMTYTGLCSLLILGDNLS 126
Query: 134 SLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM--EWT 180
++ ++ + ++ LQ DGSF + G E D+RF+YCA I M +W+
Sbjct: 127 RVNKQACLAGLRALQLEDGSFYALPEGSENDIRFIYCAASICYMLDDWS 175
>UniRef100_Q7Q4H9 ENSANGP00000019628 [Anopheles gambiae str. PEST]
Length = 360
Score = 95.5 bits (236), Expect = 2e-18
Identities = 60/183 (32%), Positives = 91/183 (48%), Gaps = 13/183 (7%)
Query: 20 HVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVE---KEAVANWVLSFQV 76
H + H LP S + +T+A+F +S LD+L+SLH++ ++ + NW+ QV
Sbjct: 10 HAKYFIRFLHTLPGRLASHDSTRVTIAFFAVSGLDVLDSLHMLTDTFQQDICNWIYKLQV 69
Query: 77 QRGTTNDPNNGQFYGFHGSKTSQF--PPDENGVFHHNNSHLASTYCALAILKIVGYDLSS 134
P + G GS T PD G+ + HLA TY +A+L +G DLS
Sbjct: 70 ----VPKPGERGYGGIQGSSTFDVIGTPDSCGLQLYRWGHLAITYTGIAVLVALGDDLSR 125
Query: 135 LDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM--EWTR--KKSRITY*I 190
L+ ++ + +Q+ DGSF G E D+RFVYCA I M +W R +K Y +
Sbjct: 126 LNRRAIIEGVAAVQREDGSFSATIEGSEQDMRFVYCAAAICAMLNDWGRVDRKKMADYIL 185
Query: 191 ASL 193
S+
Sbjct: 186 KSI 188
>UniRef100_UPI00003AF00C UPI00003AF00C UniRef100 entry
Length = 305
Score = 94.0 bits (232), Expect = 6e-18
Identities = 54/139 (38%), Positives = 80/139 (56%), Gaps = 6/139 (4%)
Query: 43 LTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP- 101
LT+A+F +S LD+L+SL +V K+ + W+ S QV T D +N GF GS P
Sbjct: 2 LTIAFFALSGLDMLDSLDVVNKDDIIEWIYSLQVL--PTEDKSNLNRCGFRGSSYLGMPF 59
Query: 102 -PDENGVFHH--NNSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIH 158
P + H ++ H+A TY L+ L I+G DLS ++ +++ + ++ LQ DGSF +
Sbjct: 60 NPSKGSDVSHPYDSGHIAMTYTGLSCLVILGDDLSRVNKDALLAGLRALQLEDGSFCAVL 119
Query: 159 IGGETDLRFVYCAGWITGM 177
G E D+RFVYCA I M
Sbjct: 120 EGSENDMRFVYCASCICYM 138
>UniRef100_UPI0000433673 UPI0000433673 UniRef100 entry
Length = 300
Score = 87.8 bits (216), Expect = 4e-16
Identities = 56/155 (36%), Positives = 78/155 (50%), Gaps = 15/155 (9%)
Query: 20 HVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEA---VANWVLSFQV 76
H + Q + ++PN + + L +A+F IS LDILN L+ + +E NW+ Q+
Sbjct: 11 HAKYFQRLLRIMPNCSAEFDCSRLAVAFFAISGLDILNCLNDLSEETKLEAINWIYRLQI 70
Query: 77 QRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYDLSSLD 136
G GF S T P D + HLA TY L L I+G DLS +D
Sbjct: 71 TGA-------GPRSGFQPSTT--IPKDAP---KYQCGHLAMTYIGLVTLLILGDDLSRVD 118
Query: 137 SESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
+S+ M+ Q PDGSF I G E+D+RF+YCA
Sbjct: 119 KKSIIEGMRACQNPDGSFTAIITGCESDMRFLYCA 153
>UniRef100_UPI0000433672 UPI0000433672 UniRef100 entry
Length = 293
Score = 87.8 bits (216), Expect = 4e-16
Identities = 56/155 (36%), Positives = 78/155 (50%), Gaps = 15/155 (9%)
Query: 20 HVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEA---VANWVLSFQV 76
H + Q + ++PN + + L +A+F IS LDILN L+ + +E NW+ Q+
Sbjct: 3 HAKYFQRLLRIMPNCSAEFDCSRLAVAFFAISGLDILNCLNDLSEETKLEAINWIYRLQI 62
Query: 77 QRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYDLSSLD 136
G GF S T P D + HLA TY L L I+G DLS +D
Sbjct: 63 TGA-------GPRSGFQPSTT--IPKDAP---KYQCGHLAMTYIGLVTLLILGDDLSRVD 110
Query: 137 SESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
+S+ M+ Q PDGSF I G E+D+RF+YCA
Sbjct: 111 KKSIIEGMRACQNPDGSFTAIITGCESDMRFLYCA 145
>UniRef100_UPI00003C20A7 UPI00003C20A7 UniRef100 entry
Length = 490
Score = 85.9 bits (211), Expect = 2e-15
Identities = 55/173 (31%), Positives = 85/173 (48%), Gaps = 18/173 (10%)
Query: 12 TTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLV---EKEAVA 68
T+ ++ H++F +LP PY S + +TL YF IS LD+LN+ + + EK +
Sbjct: 97 TSRLETKKHISFLLRCLRMLPQPYTSADDQRMTLGYFAISGLDLLNATNKIPTEEKVELI 156
Query: 69 NWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIV 128
+WV + Q+ G GF GS ++ P + +++A TY AL +L I+
Sbjct: 157 DWVYAQQLPTG-----------GFRGSPSTTSPCSSSTTSASGGANIAMTYAALLVLAIL 205
Query: 129 GYDLSSLDSESMSSSMKNLQQPDGSFMPIH--IGG--ETDLRFVYCAGWITGM 177
D + LD E + + +LQ DG F +GG + D RF YCA I M
Sbjct: 206 RDDFARLDREPLKRFISSLQHRDGGFAAEQAVVGGIVDRDPRFTYCAVAICSM 258
>UniRef100_Q24173 CG3469-PA [Drosophila melanogaster]
Length = 395
Score = 84.7 bits (208), Expect = 3e-15
Identities = 59/181 (32%), Positives = 88/181 (48%), Gaps = 15/181 (8%)
Query: 5 EEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVE- 63
E+ + E + K H +LLP S + T+ +F + LD+LNSLHLV
Sbjct: 2 EDRTDTEPVLLSK--HAKNLLRFLNLLPARMASHDNTRSTIVFFAVCGLDVLNSLHLVPP 59
Query: 64 --KEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVF-----HHNNSHLA 116
++ + +W+ V N+ N G F G + P E+ ++ HLA
Sbjct: 60 QLRQDIIDWIYGGLVVP-RDNEKNCGGFMGCR----AMVPKTEDAEILECMRNYQWGHLA 114
Query: 117 STYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITG 176
TY +LA+L +G DLS LD +S+ + +Q+P+GSF G E D+RFVYCA I
Sbjct: 115 MTYTSLAVLVTLGDDLSRLDRKSIVDGVAAVQKPEGSFSACIDGSEDDMRFVYCAATICY 174
Query: 177 M 177
M
Sbjct: 175 M 175
>UniRef100_Q24172 Geranylgeranyl transferase beta-subunit type I [Drosophila
melanogaster]
Length = 395
Score = 84.7 bits (208), Expect = 3e-15
Identities = 59/181 (32%), Positives = 88/181 (48%), Gaps = 15/181 (8%)
Query: 5 EEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVE- 63
E+ + E + K H +LLP S + T+ +F + LD+LNSLHLV
Sbjct: 2 EDRTDTEPVLLSK--HAKNLLRFLNLLPARMASHDNTRSTIVFFAVCGLDVLNSLHLVPP 59
Query: 64 --KEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVF-----HHNNSHLA 116
++ + +W+ V N+ N G F G + P E+ ++ HLA
Sbjct: 60 QLRQDIIDWIYGGLVVP-RDNEKNCGGFMGCR----AMVPKTEDAEILECMRNYQWGHLA 114
Query: 117 STYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITG 176
TY +LA+L +G DLS LD +S+ + +Q+P+GSF G E D+RFVYCA I
Sbjct: 115 MTYTSLAVLVTLGDDLSRLDRKSIVDGVAAVQKPEGSFSACIDGSEDDMRFVYCAATICY 174
Query: 177 M 177
M
Sbjct: 175 M 175
>UniRef100_O18197 Hypothetical protein Y48E1B.3 [Caenorhabditis elegans]
Length = 360
Score = 83.6 bits (205), Expect = 7e-15
Identities = 52/168 (30%), Positives = 86/168 (50%), Gaps = 19/168 (11%)
Query: 8 SSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLV----E 63
S +T T + H+ F ++ P PY + E + T+ F ISSLD+L L +
Sbjct: 20 SGDQTQTFEFKKHIGFLIRHLNVFPQPYNTLETSRNTIFLFAISSLDLLGELDNLLTPER 79
Query: 64 KEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALA 123
++A +W+ Q NG GF GS + + +G ++ ++LA TY AL
Sbjct: 80 RQAYIDWIYGLQF--------TNGNVCGFRGSHSCE----NSG---YDEANLAQTYSALL 124
Query: 124 ILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
L I+G DL +D +++ ++K Q+ +G F +G E+D+RFV+CA
Sbjct: 125 SLAILGDDLKKVDRKAILKTVKTAQRDNGCFWSQGVGSESDMRFVFCA 172
>UniRef100_UPI00003646CB UPI00003646CB UniRef100 entry
Length = 311
Score = 80.9 bits (198), Expect = 5e-14
Identities = 46/148 (31%), Positives = 81/148 (54%), Gaps = 8/148 (5%)
Query: 37 SQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSK 96
+ ++ LT+ +F +S LD+L++L +V+K + W+ S QV + + + F S+
Sbjct: 1 AHSLSRLTIVFFALSGLDVLDALDVVDKNVMIEWIYSLQV---LPTEARKWEAFSFT-SQ 56
Query: 97 TSQFPPDENGVFHH--NNSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSF 154
S+ N H ++ H+A TY L L I+G +LS ++ ++ + ++ LQ DGSF
Sbjct: 57 DSRIKGTNNECVSHPYDSGHVAMTYTGLCSLLILGDNLSRVNKQACLAGLRALQLEDGSF 116
Query: 155 MPIHIGGETDLRFVYCAGWITGM--EWT 180
+ G E D+RF+YCA I M +W+
Sbjct: 117 YALPEGSENDIRFIYCAASICYMLDDWS 144
>UniRef100_Q60ST7 Hypothetical protein CBG20759 [Caenorhabditis briggsae]
Length = 358
Score = 77.8 bits (190), Expect = 4e-13
Identities = 46/156 (29%), Positives = 79/156 (50%), Gaps = 19/156 (12%)
Query: 20 HVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLV----EKEAVANWVLSFQ 75
H+ F ++ P PY + E + T+ F IS+LD+L L + ++ +W+ Q
Sbjct: 31 HIGFLIRHLNVFPEPYNTLETSRNTIFLFAISALDLLGELDNLLTPERRQGYIDWIYDLQ 90
Query: 76 VQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYDLSSL 135
+ NG GF GS + + ++ ++LA TY AL L I+G DL +
Sbjct: 91 L--------TNGNVCGFRGSHSCEGS-------EYDEANLAQTYSALLSLAILGDDLKRV 135
Query: 136 DSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
D +++ ++K Q+ +G F +G E+D+RFV+CA
Sbjct: 136 DRKAILKTVKESQRDNGCFWSQGVGSESDMRFVFCA 171
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.335 0.144 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 521,662,929
Number of Sequences: 2790947
Number of extensions: 19588717
Number of successful extensions: 64372
Number of sequences better than 10.0: 115
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 85
Number of HSP's that attempted gapping in prelim test: 64164
Number of HSP's gapped (non-prelim): 221
length of query: 348
length of database: 848,049,833
effective HSP length: 128
effective length of query: 220
effective length of database: 490,808,617
effective search space: 107977895740
effective search space used: 107977895740
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146757.11


