

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146756.6 - phase: 0
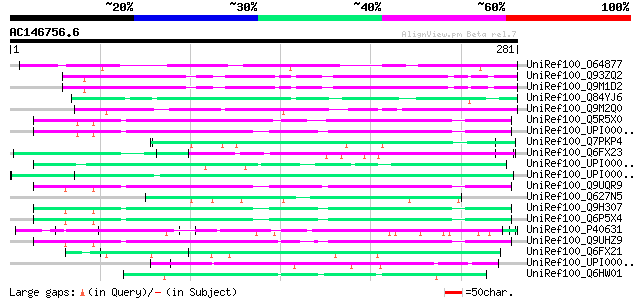
(281 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O64877 Hypothetical protein At2g44430 [Arabidopsis tha... 83 9e-15
UniRef100_Q93ZQ2 AT3g60110/T2O9_90 [Arabidopsis thaliana] 77 4e-13
UniRef100_Q9M1D2 Hypothetical protein T2O9.90 [Arabidopsis thali... 77 4e-13
UniRef100_Q84YJ6 DNA-binding protein family-like [Oryza sativa] 69 1e-10
UniRef100_Q9M2Q0 Hypothetical protein T10K17.190 [Arabidopsis th... 64 6e-09
UniRef100_Q5R5X0 Hypothetical protein DKFZp459I0427 [Pongo pygma... 63 7e-09
UniRef100_UPI0000369C76 UPI0000369C76 UniRef100 entry 63 1e-08
UniRef100_Q7PKP4 ENSANGP00000009670 [Anopheles gambiae str. PEST] 62 1e-08
UniRef100_Q6FX23 Similarities with tr|Q08294 Saccharomyces cerev... 62 2e-08
UniRef100_UPI000042E61B UPI000042E61B UniRef100 entry 61 3e-08
UniRef100_UPI000035F990 UPI000035F990 UniRef100 entry 61 4e-08
UniRef100_Q9UQR9 MEMA [Homo sapiens] 61 4e-08
UniRef100_Q627N5 Hypothetical protein CBG00596 [Caenorhabditis b... 59 1e-07
UniRef100_Q9H307 Pinin [Homo sapiens] 59 1e-07
UniRef100_Q6P5X4 Pinin, desmosome associated protein [Homo sapiens] 59 1e-07
UniRef100_P40631 Micronuclear linker histone polyprotein (MIC LH... 59 1e-07
UniRef100_Q9UHZ9 Nuclear protein SDK3 [Homo sapiens] 59 1e-07
UniRef100_Q6FX21 Similarities with tr|Q08294 Saccharomyces cerev... 59 1e-07
UniRef100_UPI000049934A UPI000049934A UniRef100 entry 58 2e-07
UniRef100_Q6HW01 Hypothetical protein [Bacillus anthracis] 58 2e-07
>UniRef100_O64877 Hypothetical protein At2g44430 [Arabidopsis thaliana]
Length = 646
Score = 82.8 bits (203), Expect = 9e-15
Identities = 86/295 (29%), Positives = 120/295 (40%), Gaps = 86/295 (29%)
Query: 6 QAQSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVK--PSTTTFSHKGDHK 63
Q S + K+D+ ++ LSR K P++VC+KR S+ K PS+++FS K D
Sbjct: 419 QEASGMRSGKADAETSDSSLSR----QKSSGPLVVCKKRRSVSAKASPSSSSFSQKDD-- 472
Query: 64 PIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGAR-SRRSNKNLSSNASNKKP 122
T EE + K+ TG R SRR+NK + A+N K
Sbjct: 473 ------------------------TKEETLSEEKDNIATGVRSSRRANKVAAVVANNTK- 507
Query: 123 PSNSTPRTGSSANKPAETPKLKNKAEGVSDKK-------------KNNGAAGFLNRIKKN 169
TG NK +T N + S K+ K A FL R+KKN
Sbjct: 508 -------TGKGRNKQKQTESKTNSSNDNSSKQDTGKTEKKTVSADKKKSVADFLKRLKKN 560
Query: 170 ESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGS 229
+ + + GG N K K K R R S S
Sbjct: 561 SPQKEAKDQNKSGG-----------------------NVKKDSKTKPRELR------SSS 591
Query: 230 GDKRNKNIESNS-KRNVGRPPKKAAETVAST--KRGRESSASGGKDKRPKKRSKK 281
K+ +E+ KR GRP KK AE AS KRGR++ ++G +K+PKKR +K
Sbjct: 592 VGKKKAEVENTPVKRAPGRPQKKTAEATASASGKRGRDTGSTGKDNKQPKKRIRK 646
>UniRef100_Q93ZQ2 AT3g60110/T2O9_90 [Arabidopsis thaliana]
Length = 641
Score = 77.4 bits (189), Expect = 4e-13
Identities = 77/255 (30%), Positives = 112/255 (43%), Gaps = 31/255 (12%)
Query: 30 SLSKHKPPILV---CRKRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYS 86
S+S+ K +L C+K+SS K S ++ S + D K +E+ + T S
Sbjct: 415 SVSRQKSSVLSLVPCKKKSSALKKTSPSSSSRQKDEKKSQEVSEEKIVTTTATTSARSSR 474
Query: 87 ETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNK 146
T +E AK+ ++ R+ N+ KK T + ++ E K + K
Sbjct: 475 RTSKEIAVVAKD-----TKTGRAKNNI------KKQTDTKTESSDDDDDEKEENSKTEKK 523
Query: 147 AEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVV 206
V+DKKK+ A FL RIKKN + G S K G QKK
Sbjct: 524 T--VADKKKS--VADFLKRIKKNSPQK------GKETTSKNQKKNDGNVKKENDHQKKSD 573
Query: 207 NNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESS 266
N K E K + G + N N +S+SKR + K+ AE VA+ KRGRES
Sbjct: 574 GNVKKENSKVKPRELRSSTGKKKVEVENNNSKSSSKR---KQTKETAE-VATGKRGRES- 628
Query: 267 ASGGKDKRPKKRSKK 281
G DK+P+KRS++
Sbjct: 629 --GKDDKQPRKRSRR 641
>UniRef100_Q9M1D2 Hypothetical protein T2O9.90 [Arabidopsis thaliana]
Length = 644
Score = 77.4 bits (189), Expect = 4e-13
Identities = 77/255 (30%), Positives = 112/255 (43%), Gaps = 31/255 (12%)
Query: 30 SLSKHKPPILV---CRKRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYS 86
S+S+ K +L C+K+SS K S ++ S + D K +E+ + T S
Sbjct: 418 SVSRQKSSVLSLVPCKKKSSALKKTSPSSSSRQKDEKKSQEVSEEKIVTTTATTSARSSR 477
Query: 87 ETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNK 146
T +E AK+ ++ R+ N+ KK T + ++ E K + K
Sbjct: 478 RTSKEIAVVAKD-----TKTGRAKNNI------KKQTDTKTESSDDDDDEKEENSKTEKK 526
Query: 147 AEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVV 206
V+DKKK+ A FL RIKKN + G S K G QKK
Sbjct: 527 T--VADKKKS--VADFLKRIKKNSPQK------GKETTSKNQKKNDGNVKKENDHQKKSD 576
Query: 207 NNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESS 266
N K E K + G + N N +S+SKR + K+ AE VA+ KRGRES
Sbjct: 577 GNVKKENSKVKPRELRSSTGKKKVEVENNNSKSSSKR---KQTKETAE-VATGKRGRES- 631
Query: 267 ASGGKDKRPKKRSKK 281
G DK+P+KRS++
Sbjct: 632 --GKDDKQPRKRSRR 644
>UniRef100_Q84YJ6 DNA-binding protein family-like [Oryza sativa]
Length = 660
Score = 69.3 bits (168), Expect = 1e-10
Identities = 68/249 (27%), Positives = 100/249 (39%), Gaps = 31/249 (12%)
Query: 35 KPPILVCRKRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPP 94
K PI+VCRKRSSI + K + DKKE+ S+ K K + + T
Sbjct: 441 KAPIIVCRKRSSIAKAAAAAAKGEKAEKAE--TDKKEKDGSEEKK--KAAAAATTATAAA 496
Query: 95 KAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKK 154
AK+K G R+ +S + + K + + A G KK
Sbjct: 497 TAKDKKARGMRTNKSRGPARNQKTAKLSETGEGTKKSDKKGGGGGGSSSAGAAAGGVAKK 556
Query: 155 KNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKG 214
+N A FLNR+ +N S R S + T +++ ++GKG+
Sbjct: 557 RN--AVDFLNRMNQNGSPSTERV-------SLLETLKLSAAATEQQKKSSSSSSGKGDGR 607
Query: 215 KERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAA--ETVASTKRGRESSASGGKD 272
KE GSG K+ + R +GRPPK+AA T +KR ++ D
Sbjct: 608 KE---------AGGSGSKKGAAASTPPGRRIGRPPKRAAAPPTPPPSKRAKD-------D 651
Query: 273 KRPKKRSKK 281
K +KR KK
Sbjct: 652 KPTRKRGKK 660
>UniRef100_Q9M2Q0 Hypothetical protein T10K17.190 [Arabidopsis thaliana]
Length = 632
Score = 63.5 bits (153), Expect = 6e-09
Identities = 66/250 (26%), Positives = 101/250 (40%), Gaps = 29/250 (11%)
Query: 37 PILVCRKRSSIPVKPS---TTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEP 93
PI+ CRKRSS+ V+ T T K P +++K+ + +P+ D++E
Sbjct: 407 PIIACRKRSSLAVRSPASVTETLKKKTRVVPTVDEKQVSEEEEGRPS--------DKDEK 458
Query: 94 PKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGV--S 151
P +K GA + K S N K + G S N + K + +G+ S
Sbjct: 459 PIVSKKMARGAAPSTAKKVGSRNV--KTSLNAGISNRGRSPNGSSVLKKSVQQKKGINTS 516
Query: 152 DKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKG 211
K AA FL R+K S E + + K G EQ+K +N K
Sbjct: 517 GGSKKQSAASFLKRMKGVSSSETVVE----------TVKAESSNGKRGAEQRK--SNSKS 564
Query: 212 EKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGK 271
EK A + G +G + S +K+N G K+ ++ + + S G
Sbjct: 565 EKVD--AVKLPAGQKRLTGKRPTIEKGSPTKKNSGVASKRGTASLMAKRDSETSEKETGS 622
Query: 272 DKRPKKRSKK 281
RPKKRSK+
Sbjct: 623 STRPKKRSKR 632
>UniRef100_Q5R5X0 Hypothetical protein DKFZp459I0427 [Pongo pygmaeus]
Length = 719
Score = 63.2 bits (152), Expect = 7e-09
Identities = 63/281 (22%), Positives = 118/281 (41%), Gaps = 39/281 (13%)
Query: 14 QKSDSLPPNTPLSRPDSLSKHKP---------PILVCRKRS-----SIPVKPSTTTFSHK 59
++ +S P P+++P +H+P P+L + S S+ V T + +
Sbjct: 462 EEKESEPQPEPVAQPQPQPQHQPQLQLQSQSQPVLQSQPPSQPENLSLAVLQPTPQVTQE 521
Query: 60 GDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASN 119
H ++ ++KE P VK T P K+K K + +R R NK S + +
Sbjct: 522 QGH--LLPERKEFPVESVKLTEVPVEPVLTVHPESKSKTKTRSRSRGRARNKTSKSRSRS 579
Query: 120 KKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGS 179
S+S+ T SS+ + + ++ S ++ +G +R S S
Sbjct: 580 SSSSSSSSSSTSSSSGSSSSSGSSSSR----SSSSSSSSTSGSSSR----------DSSS 625
Query: 180 GGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGE-KGKERASRYNDGGGS-GSGDKRNKNI 237
S S+ G G ++ ++ V+ +G+ G ER+ + + GG S + ++KN
Sbjct: 626 STSSSSESRSRSRGRGHNRDRKHRRSVDRKRGDTSGLERSHKSSKGGSSRDTKGSKDKNS 685
Query: 238 ESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKR 278
S+ KR++ +E+ S KR S D++ K+R
Sbjct: 686 RSDRKRSI-------SESSRSGKRSSRSERDRKSDRKDKRR 719
>UniRef100_UPI0000369C76 UPI0000369C76 UniRef100 entry
Length = 455
Score = 62.8 bits (151), Expect = 1e-08
Identities = 62/280 (22%), Positives = 115/280 (40%), Gaps = 37/280 (13%)
Query: 14 QKSDSLPPNTPLSRPDSLSKHKP---------PILVCRKRS-----SIPVKPSTTTFSHK 59
++ +S P P+++P +H+P P+L + S S+ V T + +
Sbjct: 198 EEKESEPQPEPVAQPQPQPQHQPQLQLQSQSQPVLQSQPPSQPEDLSLAVLQPTPQLTQE 257
Query: 60 GDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASN 119
H ++ ++K+ P VK T P K+K K + +R R NK S + +
Sbjct: 258 QGH--LLPERKDFPVESVKLTEVPVEPVLTVHPESKSKTKTRSRSRGRARNKTSKSRSRS 315
Query: 120 KKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGS 179
S+S+ T SS+ + + G S + ++ ++ + +S S S
Sbjct: 316 SSSSSSSSSSTSSSSG--------SSSSSGSSSSRSSSSSSSSTSGSSSRDS-----SSS 362
Query: 180 GGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGS-GSGDKRNKNIE 238
S S G G + K ++ V + G ER+ + + GG S + ++KN
Sbjct: 363 TSSSSESRSRSRGRGHNRDRKHRRSVDRKRRDTSGLERSHKSSKGGSSRDTKGSKDKNSR 422
Query: 239 SNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKR 278
S+ KR++ +E+ S KR S D++ K+R
Sbjct: 423 SDRKRSI-------SESSRSGKRSSRSERDRKSDRKDKRR 455
>UniRef100_Q7PKP4 ENSANGP00000009670 [Anopheles gambiae str. PEST]
Length = 1198
Score = 62.4 bits (150), Expect = 1e-08
Identities = 54/210 (25%), Positives = 84/210 (39%), Gaps = 15/210 (7%)
Query: 79 PTLKPSYSETDEEEPPKAKEK---PVTGARSRRSNKNLSSNASNKKPPS--NSTPRTGSS 133
P KP E + + P + EK P ++ + SS++S K S + P G S
Sbjct: 194 PPYKPISHEANSSDGPSSPEKEMEPTPATLQQQCGRISSSSSSGLKSSSRLSGEPSVGGS 253
Query: 134 ANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGG 193
++ + + +K S K++ +G + + S VL SG GGG G SG+S G
Sbjct: 254 SSTSSSSSSSSSKQRKSSSASKSSSGSGSGASLSSSSSSSVLASGGGGGSGGSGASGMSG 313
Query: 194 GGGTNTKEQKKV---VNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPK 250
+T V++ G G S + G GSG + +SK + G
Sbjct: 314 NSSMSTSSSSSSSSGVSSMSGSGGSGSVSNSSGSGIGGSGIAGSVGGSGSSKTSSG---- 369
Query: 251 KAAETVASTKRGRESSASGGKDKRPKKRSK 280
+ + G S+S KDK K R K
Sbjct: 370 ---SSGGTGGGGSSGSSSKEKDKYSKSRDK 396
Score = 53.5 bits (127), Expect = 6e-06
Identities = 44/196 (22%), Positives = 75/196 (37%), Gaps = 6/196 (3%)
Query: 80 TLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSN---ASNKKPPSNSTPRTGSSANK 136
T S S + ++ + K +G+ S S + SS+ AS S + +G S N
Sbjct: 256 TSSSSSSSSSKQRKSSSASKSSSGSGSGASLSSSSSSSVLASGGGGGSGGSGASGMSGNS 315
Query: 137 PAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGS---SGSSKGGG 193
T + + GVS + G+ N + GG G S SGSS G G
Sbjct: 316 SMSTSSSSSSSSGVSSMSGSGGSGSVSNSSGSGIGGSGIAGSVGGSGSSKTSSGSSGGTG 375
Query: 194 GGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAA 253
GGG++ K+ K ++S+ + + G N N +++ + G ++
Sbjct: 376 GGGSSGSSSKEKDKYSKSRDKSSKSSKSSSSSSTSGGSGSNFNNSTSTSSSAGGSSNQSK 435
Query: 254 ETVASTKRGRESSASG 269
+ G S + G
Sbjct: 436 DVDMGGTAGSSSGSMG 451
Score = 41.2 bits (95), Expect = 0.030
Identities = 55/253 (21%), Positives = 95/253 (36%), Gaps = 67/253 (26%)
Query: 86 SETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKN 145
S ++++ K+++K S +S+K+ SS++++ SN T +S++ + + K+
Sbjct: 383 SSKEKDKYSKSRDK------SSKSSKSSSSSSTSGGSGSNFNNSTSTSSSAGGSSNQSKD 436
Query: 146 KAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKV 205
G ++G+ G L ++S S GGG SG GGGG ++
Sbjct: 437 VDMG-GTAGSSSGSMGALGANASSQSSSQSGYSSTAGGGGSGPGGSGGGGSSSQSGGPLS 495
Query: 206 VNNGKGE--------------------------KGKERASR------------------- 220
N+G G GKE S+
Sbjct: 496 TNSGVGSGRAASLSPAAIPTTLIIKPPQDHTGGSGKEPLSKEAIAKMSTSSNFTETIVVN 555
Query: 221 ----YNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVAS----------TKRGRESS 266
YN GSGSG+ N ++ + K +G + + S + G SS
Sbjct: 556 SESVYNH-AGSGSGNAGNTSVIESGKLAMGGSGAGSGGSGGSYGGSTGPTGGSSTGSSSS 614
Query: 267 ASGGKDKRPKKRS 279
+SGGK ++ RS
Sbjct: 615 SSGGKKRKADARS 627
Score = 39.3 bits (90), Expect = 0.12
Identities = 41/182 (22%), Positives = 64/182 (34%), Gaps = 15/182 (8%)
Query: 114 SSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVE 173
S ++ S T GSS + E K + S K++ ++ N +
Sbjct: 361 SGSSKTSSGSSGGTGGGGSSGSSSKEKDKYSKSRDKSSKSSKSSSSSSTSGGSGSNFNNS 420
Query: 174 VLRSGSGGG----------GGSSGSSKGGGGG-GTNTKEQKKVVNNGKGEKGKERASRYN 222
S S GG GG++GSS G G G N Q +G S
Sbjct: 421 TSTSSSAGGSSNQSKDVDMGGTAGSSSGSMGALGANASSQSS-SQSGYSSTAGGGGSGPG 479
Query: 223 DGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVAST---KRGRESSASGGKDKRPKKRS 279
GG GS + + +NS GR + + +T K ++ + GK+ K+
Sbjct: 480 GSGGGGSSSQSGGPLSTNSGVGSGRAASLSPAAIPTTLIIKPPQDHTGGSGKEPLSKEAI 539
Query: 280 KK 281
K
Sbjct: 540 AK 541
>UniRef100_Q6FX23 Similarities with tr|Q08294 Saccharomyces cerevisiae YOL155c
[Candida glabrata]
Length = 1034
Score = 62.0 bits (149), Expect = 2e-08
Identities = 45/181 (24%), Positives = 74/181 (40%), Gaps = 5/181 (2%)
Query: 100 PVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGA 159
P +G RS S+ + S ++SN +P SN P +S +KP+ P + G ++ N G+
Sbjct: 755 PSSGPRSISSSISSSISSSNSRPSSN--PTGSNSGSKPSSNP---GGSTGNTNGGSNGGS 809
Query: 160 AGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERAS 219
N + S +GS G S +S G + + +NG + G S
Sbjct: 810 NSGTNSGSNSGSNAGSNAGSNAGSNSGANSGSNAGSNSGSNPGSNSGSNGGTKPGSNSGS 869
Query: 220 RYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRS 279
GS SG N SNS N G P + + + + G + ++ G + +
Sbjct: 870 NSGSNAGSNSGTNAGSNAGSNSGSNGGTKPGSNSGSNSGSNAGSNAGSNAGSNSGSNSGT 929
Query: 280 K 280
K
Sbjct: 930 K 930
Score = 55.5 bits (132), Expect = 2e-06
Identities = 55/206 (26%), Positives = 72/206 (34%), Gaps = 12/206 (5%)
Query: 82 KPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETP 141
+PS + T K P + N SN S SNS GS+A A +
Sbjct: 776 RPSSNPTGSNSGSKPSSNPGGSTGNTNGGSNGGSN-SGTNSGSNSGSNAGSNAGSNAGSN 834
Query: 142 KLKNKAEGV-SDKKKNNGAAGFLNRIKKNESVEVLRSGSGGG-------GGSSGSSKGGG 193
N S+ N G+ N K S SGS G G ++GS+ G
Sbjct: 835 SGANSGSNAGSNSGSNPGSNSGSNGGTKPGSNSGSNSGSNAGSNSGTNAGSNAGSNSGSN 894
Query: 194 GGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAA 253
GG TK +N G S GS SG K N SNS N G A
Sbjct: 895 GG---TKPGSNSGSNSGSNAGSNAGSNAGSNSGSNSGTKPGSNSGSNSGSNSGSNAGSNA 951
Query: 254 ETVASTKRGRESSASGGKDKRPKKRS 279
+ + + G S ++ G + K S
Sbjct: 952 ASNSGSNSGSNSGSNSGSNSGTKPSS 977
Score = 52.0 bits (123), Expect = 2e-05
Identities = 68/284 (23%), Positives = 96/284 (32%), Gaps = 31/284 (10%)
Query: 3 TFGQAQSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKGDH 62
T G S+ + S P + P S P S P + SSI S + + G +
Sbjct: 726 TGGSGVSSGPSSAPSSGPSSGPSSGPSSGPSSGPRSISSSISSSISSSNSRPSSNPTGSN 785
Query: 63 KPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKP 122
S KP+ P S + T + S S N SNA +
Sbjct: 786 ------------SGSKPSSNPGGSTGNTNGGSNGGSNSGTNSGSN-SGSNAGSNAGSNAG 832
Query: 123 PSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEV-----LRS 177
SNS +GS+A + + N K +N + + N S
Sbjct: 833 -SNSGANSGSNAGSNSGSNPGSNSGSNGGTKPGSNSGSNSGSNAGSNSGTNAGSNAGSNS 891
Query: 178 GSGGG---GGSSGSSKGGGGG-------GTNTKEQK--KVVNNGKGEKGKERASRYNDGG 225
GS GG G +SGS+ G G G+N+ K +N G S
Sbjct: 892 GSNGGTKPGSNSGSNSGSNAGSNAGSNAGSNSGSNSGTKPGSNSGSNSGSNSGSNAGSNA 951
Query: 226 GSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASG 269
S SG N SNS N G P ++ + ++ R S SG
Sbjct: 952 ASNSGSNSGSNSGSNSGSNSGTKPSSSSNSGSNASAVRGGSGSG 995
>UniRef100_UPI000042E61B UPI000042E61B UniRef100 entry
Length = 967
Score = 61.2 bits (147), Expect = 3e-08
Identities = 72/285 (25%), Positives = 99/285 (34%), Gaps = 46/285 (16%)
Query: 14 QKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKGDHKPIINDKKERP 73
Q S S P++ P S S P SS P + + G P
Sbjct: 240 QSSTSRGPSSASRGPSSASSSTPS-----GPSSTPSRAPSGIGGRGGASSSAPRGPSSTP 294
Query: 74 SSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSR--------RSNKNLSSNASNKKPPSN 125
SS T + S T P +A + G R R + S S S
Sbjct: 295 SSTPSSTPSSTPSSTPSSTPSRAPSRSPGGLGGRGGASSSSPRGPSSTPSGPSGTLSESR 354
Query: 126 STPR--------------TGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNES 171
STP TG S++ P+ P G+S ++GA G
Sbjct: 355 STPSGTGSAPTGAGKVATTGGSSSSPS-APSDTGSLGGISGGATSSGAGG--------SG 405
Query: 172 VEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGD 231
SGSG GGGSSG GGGGG +E + E+G E +D G+G
Sbjct: 406 SGAGGSGSGAGGGSSGGR--GGGGGRRGREDGR-------EEGNEDEDTDDDESGNGEDS 456
Query: 232 KRNKNIESNSKRNVGRPPKKAAETVAS-TKRGRESSASGGKDKRP 275
+ N + + + + KK ET A +R +ES K +P
Sbjct: 457 DTSSNSSGSRREEILKKLKKIKETAAKMNQRTKESREKRNKSHKP 501
Score = 43.9 bits (102), Expect = 0.005
Identities = 50/225 (22%), Positives = 88/225 (38%), Gaps = 17/225 (7%)
Query: 68 DKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGAR-SRRSNKNLSSNASNKKPPSNS 126
D E + + T S EE K K+ T A+ ++R+ ++ + KP S
Sbjct: 446 DDDESGNGEDSDTSSNSSGSRREEILKKLKKIKETAAKMNQRTKESREKRNKSHKPGGFS 505
Query: 127 TPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNR-IKKNESVEVLRSGSGGG--- 182
+ + SS++ + L + +S+ + ++ + +E SG+GGG
Sbjct: 506 SSSSLSSSSGSLFSSDLSDSHRDLSELLLSASSSSTSREALAAQGPLETSESGTGGGAAA 565
Query: 183 ------GGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKN 236
GG+SG + GG GG + V G G++G A R G SG
Sbjct: 566 SDGGASGGASGGASGGASGGASGGASGGVGALGGGKRGAGGAGRGAVRGASGGASGGASG 625
Query: 237 IESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRSKK 281
S+ R +++ S +R R S S + + K R+ +
Sbjct: 626 GASSGTR------ARSSLVTKSMQRSRSRSRSRSRSRSSKSRTSR 664
Score = 42.0 bits (97), Expect = 0.018
Identities = 40/158 (25%), Positives = 57/158 (35%), Gaps = 21/158 (13%)
Query: 124 SNSTPRTGSSANKPAETPKL-KNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGG 182
S+ST R +A P ET + SD + GA+G G
Sbjct: 538 SSSTSREALAAQGPLETSESGTGGGAAASDGGASGGASG------------------GAS 579
Query: 183 GGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSK 242
GG+SG + GG GG K G G AS GG SG + S
Sbjct: 580 GGASGGASGGASGGVGALGGGKRGAGGAGRGAVRGASGGASGGASGGASSGTRARSSLVT 639
Query: 243 RNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRSK 280
+++ R ++ S R +S S + R + RS+
Sbjct: 640 KSMQR--SRSRSRSRSRSRSSKSRTSRSRTSRSRSRSR 675
Score = 35.4 bits (80), Expect = 1.7
Identities = 41/168 (24%), Positives = 62/168 (36%), Gaps = 8/168 (4%)
Query: 42 RKRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPV 101
R RSS+ K + S + K S + + S S EE A
Sbjct: 632 RARSSLVTKSMQRSRSRSRSRSRSRSSKSRTSRS--RTSRSRSRSRAMEESSCAASSSRS 689
Query: 102 TGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNN---- 157
+ +R+S+ + SS + P + + SS++ KL + G+ D K
Sbjct: 690 SLLSTRQSSVSASSVFLSISAPVSLSVSVSSSSSGLVVL-KLSDSEVGIVDPSKTRTTSR 748
Query: 158 -GAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKK 204
G+ G + GSGG GG G +GGGG G K+ K
Sbjct: 749 GGSEGGERGGRGGRGGRGGEGGSGGRGGGGGGGRGGGGSGDGGKKAPK 796
Score = 33.5 bits (75), Expect = 6.3
Identities = 27/96 (28%), Positives = 41/96 (42%), Gaps = 9/96 (9%)
Query: 176 RSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRY-NDGGGSGSGDKRN 234
R GS G G + G+S GG + + KV GKG+ +++ R + GG GS
Sbjct: 188 RKGSAGSGAACGTS----GGPSKADKTGKVGTAGKGKGTRDKQGRTPSRPGGRGSA---- 239
Query: 235 KNIESNSKRNVGRPPKKAAETVASTKRGRESSASGG 270
++ S + R P A+ + S S A G
Sbjct: 240 QSSTSRGPSSASRGPSSASSSTPSGPSSTPSRAPSG 275
>UniRef100_UPI000035F990 UPI000035F990 UniRef100 entry
Length = 520
Score = 60.8 bits (146), Expect = 4e-08
Identities = 57/279 (20%), Positives = 103/279 (36%), Gaps = 12/279 (4%)
Query: 1 MKTFGQAQSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKG 60
+ + + S+ T S S +TP S P S P SS PS + S
Sbjct: 5 LSSSSSSSSSPSTSSSSSSSSSTPPSPPSPSSSSSSPSPSSSSDSSSSTPPSPPSSSSPS 64
Query: 61 DHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNK 120
++ + SS + K S S T +K + + S + + SS +S+
Sbjct: 65 SSS--LSSSPAQSSSSSTNSSKSS-SSTSRSSSSSSKSSSSSSSSSSSKSSSNSSRSSSS 121
Query: 121 KPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSG 180
K SNS+ + SS++K + + + + S K ++ ++ ++ N S S
Sbjct: 122 KSSSNSSCSSSSSSSKSSSSTSRSSSSS--SSKSSSSSSSSSSSKRSSNSSRSSSSKSSS 179
Query: 181 GGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESN 240
SS SS T+ ++ +S + G S ++K+ S+
Sbjct: 180 NSSCSSSSSSSKSSSSTSRSSSSSSSSSSSSSSSSSSSSSSSSSSGRSSSSSKSKSSSSS 239
Query: 241 SKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRS 279
S N+ + + +ST R S +S + RS
Sbjct: 240 SSSNI-------SSSSSSTSSSRSSCSSSSSSSSSRSRS 271
Score = 59.7 bits (143), Expect = 8e-08
Identities = 52/245 (21%), Positives = 96/245 (38%), Gaps = 5/245 (2%)
Query: 37 PILVCRKRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKA 96
P + SS PST++ S P SS P+ S S++ PP
Sbjct: 1 PSSLLSSSSSSSSSPSTSSSSSSSSSTPPSPPSPSSSSSSPSPS---SSSDSSSSTPPSP 57
Query: 97 KEKPVTGARSRRSNKNLSSNAS-NKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKK 155
+ S S+ SS++S N S+ST R+ SS++K + + + ++ S+ +
Sbjct: 58 PSSSSPSSSSLSSSPAQSSSSSTNSSKSSSSTSRSSSSSSKSSSSSSSSSSSKSSSNSSR 117
Query: 156 NNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKV-VNNGKGEKG 214
++ + N + S S S SS SSK +++ ++ + K
Sbjct: 118 SSSSKSSSNSSCSSSSSSSKSSSSTSRSSSSSSSKSSSSSSSSSSSKRSSNSSRSSSSKS 177
Query: 215 KERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKR 274
+S + S S +++ S+S + ++ + +S+ GR SS+S K
Sbjct: 178 SSNSSCSSSSSSSKSSSSTSRSSSSSSSSSSSSSSSSSSSSSSSSSSGRSSSSSKSKSSS 237
Query: 275 PKKRS 279
S
Sbjct: 238 SSSSS 242
Score = 53.9 bits (128), Expect = 5e-06
Identities = 50/278 (17%), Positives = 104/278 (36%), Gaps = 3/278 (1%)
Query: 2 KTFGQAQSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKGD 61
++ ++S++ S+S ++ S S S + SS S+++ S
Sbjct: 165 RSSNSSRSSSSKSSSNSSCSSSSSSSKSSSSTSRSSSSSSSSSSSSSSSSSSSSSSSSSS 224
Query: 62 HKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKK 121
+ + K + SS + S S T + + +RSR SN SS++S+
Sbjct: 225 GRSSSSSKSKSSSSSSSSNISSSSSSTSSSRSSCSSSSSSSSSRSR-SNSCSSSSSSSSS 283
Query: 122 PPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGG 181
SNS+ R SS++K + + + S K ++ ++ ++ + S S
Sbjct: 284 SSSNSSGR--SSSSKSSSSSSSSKSSSSSSSKSSSSSSSSSSSKSSSSSSSSSSSKSSSS 341
Query: 182 GGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNS 241
SS S +++ K ++ + +S + S S + + S+S
Sbjct: 342 SSSSSSKSSSSSSSSSSSSGSSKSSSSSSSSSSSKSSSTSSSSSSSKSSSSSSSSSSSSS 401
Query: 242 KRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRS 279
K + ++ + +S+ SS+S S
Sbjct: 402 KSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 439
>UniRef100_Q9UQR9 MEMA [Homo sapiens]
Length = 586
Score = 60.8 bits (146), Expect = 4e-08
Identities = 62/278 (22%), Positives = 113/278 (40%), Gaps = 33/278 (11%)
Query: 14 QKSDSLPPNTPLSRPD-------SLSKHKPPILVCRKRS-----SIPVKPSTTTFSHKGD 61
++ +S P P+++P L P+L + S S+ V T + +
Sbjct: 329 EEKESEPQPEPVAQPQPQSQPQLQLQSQSQPVLQSQPPSQPEDLSLAVLQPTPQVTQEQG 388
Query: 62 HKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKK 121
H ++ ++K+ P VK T P K+K K + +R R NK S + +
Sbjct: 389 H--LLPERKDFPVESVKLTEVPVEPVLTVHPESKSKTKTRSRSRGRARNKTSKSRSRSSS 446
Query: 122 PPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGG 181
S+S+ T SS+ + + G S + ++ ++ + +S S S
Sbjct: 447 SSSSSSSSTSSSSG--------SSSSSGSSSSRSSSSSSSSTSGSSSRDSSS---STSTS 495
Query: 182 GGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGS-GSGDKRNKNIESN 240
S S G G + K ++ V + G ER+ + + GGGS + ++KN S+
Sbjct: 496 SSSESRSRSRGRGHNRDRKHRRSVDRKRRDTSGLERSHKSSKGGGSRDTKGSKDKNSRSD 555
Query: 241 SKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKR 278
KR++ +E+ S KR S D++ K+R
Sbjct: 556 RKRSI-------SESSRSGKRSSRSERDRKSDRKDKRR 586
>UniRef100_Q627N5 Hypothetical protein CBG00596 [Caenorhabditis briggsae]
Length = 279
Score = 59.3 bits (142), Expect = 1e-07
Identities = 61/197 (30%), Positives = 80/197 (39%), Gaps = 29/197 (14%)
Query: 76 DVKPTLKPSYSETDEEEPPKAKEK--PVTGARSRRSNK-----NLSSNASNKKPPSNST- 127
DVKP + + D PP EK V G R +K +L + +KPP NS
Sbjct: 83 DVKPGTRDNRYPEDVTFPPSDIEKVYDVMGDEDARKSKTMTLVSLDEGSLEEKPPKNSES 142
Query: 128 ----PRTGSSANKPAETPKLKNKAEGV------SDKKKNNGAAGFLNRIKKNESVEVLRS 177
P S+A P P K + + S K N G +K SVE L
Sbjct: 143 DKRKPVIPSAARTPTNNPFQKQLSREILLTPKQSLKTANEGT-------RKTASVEQLVV 195
Query: 178 GSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASR--YNDGGGSGSGDKRNK 235
+ SS G GGG +K+ + +K KE SR + GGG G+ DKR
Sbjct: 196 AKKSREPVTPSSGGSSGGGRRSKKNWDEMGAFDEKKSKELPSREQISGGGGGGTADKRTP 255
Query: 236 NIESNSKRNVGR--PPK 250
+ ESNSK + PPK
Sbjct: 256 SRESNSKSKSKKALPPK 272
Score = 36.6 bits (83), Expect = 0.75
Identities = 42/174 (24%), Positives = 61/174 (34%), Gaps = 22/174 (12%)
Query: 33 KHKPPILVCRKRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEE 92
K K LV S+ KP + S K KP+I PS+ PT P + E
Sbjct: 118 KSKTMTLVSLDEGSLEEKPPKNSESDK--RKPVI------PSAARTPTNNPFQKQLSREI 169
Query: 93 PPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSD 152
K+ T R ++ KK TP +G S+ G
Sbjct: 170 LLTPKQSLKTANEGTRKTASVEQLVVAKKSREPVTPSSGGSSG-------------GGRR 216
Query: 153 KKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVV 206
KKN G + KK++ + SGGGGG + + + + KK +
Sbjct: 217 SKKNWDEMGAFDE-KKSKELPSREQISGGGGGGTADKRTPSRESNSKSKSKKAL 269
>UniRef100_Q9H307 Pinin [Homo sapiens]
Length = 717
Score = 59.3 bits (142), Expect = 1e-07
Identities = 61/278 (21%), Positives = 112/278 (39%), Gaps = 35/278 (12%)
Query: 14 QKSDSLPPNTPLSRPD-------SLSKHKPPILVCRKRS-----SIPVKPSTTTFSHKGD 61
++ +S P P+++P L P+L + S S+ V T + +
Sbjct: 462 EEKESEPQPEPVAQPQPQSQPQLQLQSQSQPVLQSQPPSQPEDLSLAVLQPTPQVTQEQG 521
Query: 62 HKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKK 121
H ++ ++K+ P VK T P K+K K + +R R NK S + +
Sbjct: 522 H--LLPERKDFPVESVKLTEVPVEPVLTVHPESKSKTKTRSRSRGRARNKTSKSRSRSSS 579
Query: 122 PPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGG 181
S+S+ T SS+ + + G S + ++ ++ + +S S S
Sbjct: 580 SSSSSSSSTSSSSG--------SSSSSGSSSSRSSSSSSSSTSGSSSRDS-----SSSTS 626
Query: 182 GGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGS-GSGDKRNKNIESN 240
S S G G + K ++ V + G ER+ + + GG S + ++KN S+
Sbjct: 627 SSSESRSRSRGRGHNRDRKHRRSVDRKRRDTSGLERSPKSSKGGSSRDTKGSKDKNSRSD 686
Query: 241 SKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKR 278
KR++ +E+ S KR S D++ K+R
Sbjct: 687 RKRSI-------SESSRSGKRSSRSERDRKSDRKDKRR 717
>UniRef100_Q6P5X4 Pinin, desmosome associated protein [Homo sapiens]
Length = 717
Score = 59.3 bits (142), Expect = 1e-07
Identities = 61/278 (21%), Positives = 112/278 (39%), Gaps = 35/278 (12%)
Query: 14 QKSDSLPPNTPLSRPD-------SLSKHKPPILVCRKRS-----SIPVKPSTTTFSHKGD 61
++ +S P P+++P L P+L + S S+ V T + +
Sbjct: 462 EEKESEPQPEPVAQPQPQSQPQLQLQSQSQPVLQSQPPSQPEDLSLAVLQPTPQVTQEQG 521
Query: 62 HKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKK 121
H ++ ++K+ P VK T P K+K K + +R R NK S + +
Sbjct: 522 H--LLPERKDFPVESVKLTEVPVEPVLTVHPESKSKTKTRSRSRGRARNKTSKSRSRSSS 579
Query: 122 PPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGG 181
S+S+ T SS+ + + G S + ++ ++ + +S S S
Sbjct: 580 SSSSSSSSTSSSSG--------SSSSSGSSSSRSSSSSSSSTSGSSSRDS-----SSSTS 626
Query: 182 GGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGS-GSGDKRNKNIESN 240
S S G G + K ++ V + G ER+ + + GG S + ++KN S+
Sbjct: 627 SSSESRSRSRGRGHNRDRKHRRSVDRKRRDTSGLERSHKSSKGGSSRDTKGSKDKNSRSD 686
Query: 241 SKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKR 278
KR++ +E+ S KR S D++ K+R
Sbjct: 687 RKRSI-------SESSRSGKRSSRSERDRKSDRKDKRR 717
>UniRef100_P40631 Micronuclear linker histone polyprotein (MIC LH) [Contains:
Micronuclear linker histone-alpha; Micronuclear linker
histone-beta; Micronuclear linker histone-delta;
Micronuclear linker histone-gamma] [Tetrahymena
thermophila]
Length = 633
Score = 59.3 bits (142), Expect = 1e-07
Identities = 71/278 (25%), Positives = 120/278 (42%), Gaps = 32/278 (11%)
Query: 4 FGQAQSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKGDHK 63
+GQA T++ +S T SR S SK K + R +S+ + + ++ S
Sbjct: 167 YGQAAQKKQTKRKNS----TSKSRRSS-SKGKSSVSKGRTKSTSSKRRADSSASQGRSQS 221
Query: 64 PIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPP 123
N +K S D K T S ++ + G ++ SNK SS +S+K+
Sbjct: 222 SSSNRRKASSSKDQKGTRSSSRKASNSK-----------GRKNSTSNKRNSS-SSSKRSS 269
Query: 124 SNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGG 183
S+ ++ SS NK + + K + S + A+ NR K+ + +S S G
Sbjct: 270 SSKNKKSSSSKNKKSSSSKGRK-----SSSSRGRKASSSKNR--KSSKSKDRKSSSSKGR 322
Query: 184 GSSGSSKGGGGGGTNTKEQKKVVNNG-KGEKGKERASRYNDGGG--SGSGDKRNKNIES- 239
SS SSK ++++ +K + G K K +ER + + D G KR ++ S
Sbjct: 323 KSSSSSKSNKRKASSSRGRKSSSSKGRKSSKSQERKNSHADTSKQMEDEGQKRRQSSSSA 382
Query: 240 ----NSKRNVGRPPKKAAETVASTKRGRESSASGGKDK 273
+SK++ K+A A+ K ++S SG K K
Sbjct: 383 KRDESSKKSRRNSMKEARTKKANNKSASKASKSGSKSK 420
Score = 53.9 bits (128), Expect = 5e-06
Identities = 55/244 (22%), Positives = 92/244 (37%), Gaps = 47/244 (19%)
Query: 50 KPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRS 109
+ ++T+ S + K + K R S S + + + + + ++ ++
Sbjct: 178 RKNSTSKSRRSSSKGKSSVSKGRTKSTSSKRRADSSASQGRSQSSSSNRRKASSSKDQKG 237
Query: 110 NKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKN 169
++ S ASN K NST +S++ + KNK S KK++ + G
Sbjct: 238 TRSSSRKASNSKGRKNSTSNKRNSSSSSKRSSSSKNKKSSSSKNKKSSSSKG-------- 289
Query: 170 ESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGS 229
R S G + SSK N K K K+R S + G S S
Sbjct: 290 ------RKSSSSRGRKASSSK-----------------NRKSSKSKDRKSSSSKGRKSSS 326
Query: 230 GDKRNKNIESNSK-----RNVGRPPKKAAETVAS-----------TKRGRESSASGGKDK 273
K NK S+S+ + GR K+ E S ++ R+SS+S +D+
Sbjct: 327 SSKSNKRKASSSRGRKSSSSKGRKSSKSQERKNSHADTSKQMEDEGQKRRQSSSSAKRDE 386
Query: 274 RPKK 277
KK
Sbjct: 387 SSKK 390
Score = 51.2 bits (121), Expect = 3e-05
Identities = 46/189 (24%), Positives = 75/189 (39%), Gaps = 7/189 (3%)
Query: 95 KAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKN--KAEGVSD 152
+ K K T R S+K SS + + ++S R SSA++ N KA D
Sbjct: 175 QTKRKNSTSKSRRSSSKGKSSVSKGRTKSTSSKRRADSSASQGRSQSSSSNRRKASSSKD 234
Query: 153 KKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGE 212
+K ++ + K ++ + S S SSK + K+ ++ KG
Sbjct: 235 QKGTRSSSRKASNSKGRKNSTSNKRNSSSSSKRSSSSKNKKSSSSKNKKS----SSSKGR 290
Query: 213 KGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKD 272
K R + K S+SK K+ + AS+ RGR+SS+S G+
Sbjct: 291 KSSSSRGRKASSSKNRKSSKSKDRKSSSSKGRKSSSSSKSNKRKASSSRGRKSSSSKGR- 349
Query: 273 KRPKKRSKK 281
K K + +K
Sbjct: 350 KSSKSQERK 358
Score = 51.2 bits (121), Expect = 3e-05
Identities = 67/284 (23%), Positives = 111/284 (38%), Gaps = 45/284 (15%)
Query: 26 SRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSY 85
SR S SK + ++ SS K S+++ + K + K + SS K S
Sbjct: 242 SRKASNSKGRKNSTSNKRNSSSSSKRSSSSKNKKS------SSSKNKKSSSSKGRKSSSS 295
Query: 86 -----SETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSAN--KPA 138
S + + K+K++ + ++ R+S+ SS+ SNK+ S+S R SS+ K +
Sbjct: 296 RGRKASSSKNRKSSKSKDRKSSSSKGRKSS---SSSKSNKRKASSSRGRKSSSSKGRKSS 352
Query: 139 ETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTN 198
++ + KN S + ++ G K+ +S + S K N
Sbjct: 353 KSQERKNSHADTSKQMEDEGQ-------KRRQSSSSAKRDESSKKSRRNSMKEARTKKAN 405
Query: 199 TKEQKKVVNNG---KGE-----KGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPK 250
K K +G KG+ KGK + N S S K N SN+ +
Sbjct: 406 NKSASKASKSGSKSKGKSASKSKGKSSSKGKNSKSRSASKPKSNAAQNSNNTHQTADSSE 465
Query: 251 KAAETVASTKRGRE--------------SSASGGKDKRPKKRSK 280
A+ T + RGR+ SS+ G K+ + RSK
Sbjct: 466 NASSTTQTRTRGRQREQKDMVNEKSNSKSSSKGKKNSKSNTRSK 509
Score = 49.3 bits (116), Expect = 1e-04
Identities = 36/177 (20%), Positives = 70/177 (39%), Gaps = 7/177 (3%)
Query: 104 ARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFL 163
A ++ K +S + +++ S G S+ T +K S + +
Sbjct: 170 AAQKKQTKRKNSTSKSRRSSSK-----GKSSVSKGRTKSTSSKRRADSSASQGRSQSSSS 224
Query: 164 NRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYND 223
NR K + S + + G+ + +SKG +N + K K+ +S N
Sbjct: 225 NRRKASSSKD--QKGTRSSSRKASNSKGRKNSTSNKRNSSSSSKRSSSSKNKKSSSSKNK 282
Query: 224 GGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRSK 280
S G K + + + + R K+ + +S+ +GR+SS+S +KR S+
Sbjct: 283 KSSSSKGRKSSSSRGRKASSSKNRKSSKSKDRKSSSSKGRKSSSSSKSNKRKASSSR 339
Score = 45.1 bits (105), Expect = 0.002
Identities = 47/226 (20%), Positives = 76/226 (32%), Gaps = 24/226 (10%)
Query: 59 KGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSR---RSNKNLSS 115
KG + K R +S K + + T + T R+R R K++ +
Sbjct: 428 KGKSSSKGKNSKSRSASKPKSNAAQNSNNTHQTADSSENASSTTQTRTRGRQREQKDMVN 487
Query: 116 NASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVL 175
SN K S + S+ +++ + S KK+ G R K
Sbjct: 488 EKSNSKSSSKGKKNSKSNTRSKSKSKSASKSRKNASKSKKDTTNHGRQTRSK-------- 539
Query: 176 RSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNK 235
S SK + +V+ K E + + DK++K
Sbjct: 540 ---------SRSESKSKSEAPNKPSNKMEVIEQPKEESSDRKRRESRSQSAKKTSDKKSK 590
Query: 236 NIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRSKK 281
N S+SK+ PKK A +G++ S G KK +KK
Sbjct: 591 N-RSDSKKMTAEDPKK---NNAEDSKGKKKSKEGKTGAYGKKANKK 632
Score = 43.9 bits (102), Expect = 0.005
Identities = 46/193 (23%), Positives = 82/193 (41%), Gaps = 9/193 (4%)
Query: 96 AKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNK----AEGVS 151
AK K +T A+ + +N K+ A E K K A+
Sbjct: 116 AKHKDLTNAKILKIMSEDFNNLPQKEVKVYEDAYQKEYAQYLVEFKKWNEKYGQAAQKKQ 175
Query: 152 DKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKG 211
K+KN+ + + K SV R+ S + SS G +++ ++K ++ K
Sbjct: 176 TKRKNSTSKSRRSSSKGKSSVSKGRTKSTSSKRRADSSASQGRSQSSSSNRRKA-SSSKD 234
Query: 212 EKGKERASRY-NDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAET---VASTKRGRESSA 267
+KG +SR ++ G + +N S+SKR+ KK++ + +S+ +GR+SS+
Sbjct: 235 QKGTRSSSRKASNSKGRKNSTSNKRNSSSSSKRSSSSKNKKSSSSKNKKSSSSKGRKSSS 294
Query: 268 SGGKDKRPKKRSK 280
S G+ K K
Sbjct: 295 SRGRKASSSKNRK 307
>UniRef100_Q9UHZ9 Nuclear protein SDK3 [Homo sapiens]
Length = 717
Score = 58.9 bits (141), Expect = 1e-07
Identities = 62/278 (22%), Positives = 113/278 (40%), Gaps = 35/278 (12%)
Query: 14 QKSDSLPPNTPLSRPD-------SLSKHKPPILVCRKRS-----SIPVKPSTTTFSHKGD 61
++ +S P P+++P L P+L + S S+ V T + +
Sbjct: 462 EEKESEPQPEPVAQPQPQSQPQLQLQSQSQPVLQSQPPSQPEDLSLAVLQPTPQVTQEQG 521
Query: 62 HKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKK 121
H ++ ++K+ P VK T P K+K K + +R R NK S + +
Sbjct: 522 H--LLPERKDFPVESVKLTEVPVEPVLTVHPESKSKTKTRSRSRGRARNKTSKSRSRSSS 579
Query: 122 PPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGG 181
S+S+ T SS+ + + +++ S +G++G +S S S
Sbjct: 580 SSSSSSSSTSSSSGSSSSSGSSSSRSSS-SSSSSTSGSSG-------RDS-----SSSTS 626
Query: 182 GGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGS-GSGDKRNKNIESN 240
S S G G + K ++ V + G ER+ + + GG S + ++KN S+
Sbjct: 627 SSSESRSRSRGRGHNRDRKHRRSVDRKRRDTSGLERSHKSSKGGSSRDTKGSKDKNSRSD 686
Query: 241 SKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKR 278
KR++ +E+ S KR S D++ K+R
Sbjct: 687 RKRSI-------SESSRSGKRSSRSERDRKSDRKDKRR 717
>UniRef100_Q6FX21 Similarities with tr|Q08294 Saccharomyces cerevisiae YOL155c
[Candida glabrata]
Length = 1036
Score = 58.9 bits (141), Expect = 1e-07
Identities = 54/231 (23%), Positives = 82/231 (35%), Gaps = 20/231 (8%)
Query: 51 PSTTTFSHKGDHKPIINDKKERPSSDV-------KPTLKPSYSETDEEEPPKAKEKPVTG 103
PS+ S I+ RPSS+ KP+ P S + + T
Sbjct: 754 PSSAPSSAPRSISSSISSSNSRPSSNPTGSNSSSKPSSNPGGSNGNTNGGSNSGSNSGTN 813
Query: 104 ARSRRSNKNLSSNASNK--KPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAG 161
+ S + + S++ SN KP SNS +GS++ A + N +N +
Sbjct: 814 SGSNSGSNSASNSGSNSGTKPGSNSGSNSGSNSGSNAGSNAGSNAGSNAGSNAGSNAGSN 873
Query: 162 FLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRY 221
+ N S SG GS+ S G GTN+ +N G S
Sbjct: 874 SGTKPGSNSG-----SNSGSNAGSNSGSNSGTNSGTNSG------SNSGSNSGSNAGSNA 922
Query: 222 NDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKD 272
GS SG N SN+ N G + T + T G + ++ G +
Sbjct: 923 GSNSGSNSGTNSGTNSGSNAGSNAGSNSGSNSGTNSGTNSGSNAGSNSGSN 973
Score = 53.1 bits (126), Expect = 8e-06
Identities = 43/180 (23%), Positives = 72/180 (39%), Gaps = 7/180 (3%)
Query: 100 PVTGARSRRSN--KNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKN-KAEGVSDKKKN 156
P +G S S+ +++SS+ S+ +S P +S++KP+ P N G S+ N
Sbjct: 750 PSSGPSSAPSSAPRSISSSISSSNSRPSSNPTGSNSSSKPSSNPGGSNGNTNGGSNSGSN 809
Query: 157 NGAAGFLNRIKKNESVEVLRSGS--GGGGGSSGSSKGGGGGGTNTKEQ--KKVVNNGKGE 212
+G N + S SG+ G GS+ S G G+N +N
Sbjct: 810 SGTNSGSNSGSNSASNSGSNSGTKPGSNSGSNSGSNSGSNAGSNAGSNAGSNAGSNAGSN 869
Query: 213 KGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKD 272
G ++ GS SG N SNS N G + + + + G + ++ G +
Sbjct: 870 AGSNSGTKPGSNSGSNSGSNAGSNSGSNSGTNSGTNSGSNSGSNSGSNAGSNAGSNSGSN 929
Score = 47.4 bits (111), Expect = 4e-04
Identities = 56/250 (22%), Positives = 89/250 (35%), Gaps = 15/250 (6%)
Query: 32 SKHKPPILVCRKRSSIPVKPS-------TTTFSHKGDH-KPIINDKKERPSSDVKPTLKP 83
S KP +V ++IP+KPS T+ F G I+ S L
Sbjct: 650 SNGKPTTIV----TTIPLKPSDGGEADYTSVFVSDGHTITKIVTHVTTTDSLGHTVVLSS 705
Query: 84 SYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKL 143
+Y D + P S S +S +S P +G S+ P+ P+
Sbjct: 706 TYLTYDRVDDGAVFYSPSVSVSSNVSRSVDTSMVPTGGSGVSSGPSSGPSS-APSSAPRS 764
Query: 144 KNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQK 203
+ + S+ + ++ G N K S +G+ GG +SGS+ G G +
Sbjct: 765 ISSSISSSNSRPSSNPTGS-NSSSKPSSNPGGSNGNTNGGSNSGSNSGTNSGSNSGSNSA 823
Query: 204 KVVNNGKGEK-GKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRG 262
+ G K G S GS +G N SN+ N G + T + G
Sbjct: 824 SNSGSNSGTKPGSNSGSNSGSNSGSNAGSNAGSNAGSNAGSNAGSNAGSNSGTKPGSNSG 883
Query: 263 RESSASGGKD 272
S ++ G +
Sbjct: 884 SNSGSNAGSN 893
Score = 45.1 bits (105), Expect = 0.002
Identities = 60/262 (22%), Positives = 91/262 (33%), Gaps = 7/262 (2%)
Query: 3 TFGQAQSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKGDH 62
T G S+ + S P + P S S+S S+ KPS+ G+
Sbjct: 741 TGGSGVSSGPSSGPSSAPSSAPRSISSSISSSNSRPSSNPTGSNSSSKPSSNPGGSNGNT 800
Query: 63 KPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKP 122
N ++ + S S + K + + S S N SNA +
Sbjct: 801 NGGSNSGSNSGTNSGSNSGSNSASNSGSNSGTKPGSNSGSNSGSN-SGSNAGSNAGSNAG 859
Query: 123 PSNSTPRTGSSANKPAET-PKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGG 181
SN+ GS+A + T P + + S+ N+G+ N + S SGS
Sbjct: 860 -SNAGSNAGSNAGSNSGTKPGSNSGSNSGSNAGSNSGSNSGTNSGTNSGSNSGSNSGSNA 918
Query: 182 GG--GSSGSSKGGGGGGTNTKEQ--KKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNI 237
G GS+ S G GTN+ +N G + GS SG N
Sbjct: 919 GSNAGSNSGSNSGTNSGTNSGSNAGSNAGSNSGSNSGTNSGTNSGSNAGSNSGSNSGSNS 978
Query: 238 ESNSKRNVGRPPKKAAETVAST 259
SNS N + + + A+T
Sbjct: 979 GSNSGSNSASAVRGGSGSGAAT 1000
Score = 43.5 bits (101), Expect = 0.006
Identities = 39/152 (25%), Positives = 53/152 (34%), Gaps = 14/152 (9%)
Query: 109 SNKNLSSNA---SNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNR 165
+ N SNA S KP SNS +GS+A + + N +N + +
Sbjct: 862 AGSNAGSNAGSNSGTKPGSNSGSNSGSNAGSNSGSNSGTNSGTNSGSNSGSNSGSNAGSN 921
Query: 166 IKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGG 225
N S SG G++ S G G+N+ +N G S
Sbjct: 922 AGSNSG-----SNSGTNSGTNSGSNAGSNAGSNSG------SNSGTNSGTNSGSNAGSNS 970
Query: 226 GSGSGDKRNKNIESNSKRNVGRPPKKAAETVA 257
GS SG N SNS V A TV+
Sbjct: 971 GSNSGSNSGSNSGSNSASAVRGGSGSGAATVS 1002
>UniRef100_UPI000049934A UPI000049934A UniRef100 entry
Length = 472
Score = 58.2 bits (139), Expect = 2e-07
Identities = 54/211 (25%), Positives = 89/211 (41%), Gaps = 23/211 (10%)
Query: 79 PTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPA 138
P KPS + T+ E K+K GA++ + KN S +KK + +T NK
Sbjct: 116 PQEKPSVTATNTHEEEKSKGNTEGGAQNEATTKN-SETGESKKQEGSGNGQTSEGENKAI 174
Query: 139 ETPKLKNKAEGVSDKKKN------------NGAAGFLNRIKKNESVEVLRSGSGGGGGSS 186
+ ++ + K + +G+A K +ES + SG+ G GGS
Sbjct: 175 KGTEVSGGGSSSNQKVSSGENSAETGAPSASGSATTGKGGKGSESEHTVESGTSGQGGSE 234
Query: 187 GSSKGGG-GGGTNTKEQKK-----VVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESN 240
+S+G T+ K Q+ V G+G+K D G SG+ + + E N
Sbjct: 235 TTSEGSKVSEKTDVKNQESGGSQTVGQEGQGQKENGETGGKTDKGSSGTSSSQTQ--EQN 292
Query: 241 SKRNVGRPPKKAAETVASTKRGRESSASGGK 271
S+ G+ ++ +S K G+E SGG+
Sbjct: 293 SQNINGKGEVGGSD--SSGKSGKEGQPSGGE 321
Score = 47.8 bits (112), Expect = 3e-04
Identities = 47/186 (25%), Positives = 69/186 (36%), Gaps = 24/186 (12%)
Query: 90 EEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEG 149
EE P +EKP S A+N S T A A T KN G
Sbjct: 110 EEAKPTPQEKP-------------SVTATNTHEEEKSKGNTEGGAQNEATT---KNSETG 153
Query: 150 VSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGS--SKGGGGGGTNTKEQKKVVN 207
S K++ +G + + E+ + + GGG SS S G T
Sbjct: 154 ESKKQEGSGNG----QTSEGENKAIKGTEVSGGGSSSNQKVSSGENSAETGAPSASGSAT 209
Query: 208 NGKGEKGKERASRYNDG-GGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRG-RES 265
GKG KG E G G G + ++ + + K +V ++TV +G +E+
Sbjct: 210 TGKGGKGSESEHTVESGTSGQGGSETTSEGSKVSEKTDVKNQESGGSQTVGQEGQGQKEN 269
Query: 266 SASGGK 271
+GGK
Sbjct: 270 GETGGK 275
Score = 44.7 bits (104), Expect = 0.003
Identities = 48/222 (21%), Positives = 80/222 (35%), Gaps = 37/222 (16%)
Query: 90 EEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEG 149
E P A TG + S + + + T GS K +E +KN+ G
Sbjct: 198 ETGAPSASGSATTGKGGKGSESEHTVESGTSGQGGSETTSEGS---KVSEKTDVKNQESG 254
Query: 150 VSDK-------KKNNGAAG----------FLNRIKKNESVEVLRSGSGGGGGSSGSS--- 189
S +K NG G ++ ++ S + G GG SSG S
Sbjct: 255 GSQTVGQEGQGQKENGETGGKTDKGSSGTSSSQTQEQNSQNINGKGEVGGSDSSGKSGKE 314
Query: 190 -------KGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKN---IES 239
KG G +++++ N G +GK+ + D G +G G K N+N ++
Sbjct: 315 GQPSGGEKGDSGQEAGKEQKEQAGNEGATGEGKKES----DSGVAGEGQKENENKGSVDD 370
Query: 240 NSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRSKK 281
N + G +K + VA + G D + ++
Sbjct: 371 NKSDSKGGDGEKGNDGVAGNQAKDNKETEGQTDNNEQNNQQQ 412
>UniRef100_Q6HW01 Hypothetical protein [Bacillus anthracis]
Length = 441
Score = 58.2 bits (139), Expect = 2e-07
Identities = 45/212 (21%), Positives = 85/212 (39%), Gaps = 18/212 (8%)
Query: 64 PIINDKKERPSSDVKPTLKPS--------YSETDEEEPPKAKEKPVTGARSRRSNKNLSS 115
P+ N+++ + D P + P Y + + +E K +E+P + +
Sbjct: 229 PVENEEERQSQPDSSPDVVPDLSSVKDKKYEKPEYKEQKKIEEQPTKQIKENNGRGSQQE 288
Query: 116 NASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVL 175
N N++ + + G++ N+ + + +++ NNG N + E+
Sbjct: 289 NRGNQQENNGRESQQGNNGNQQGNNGRESQQGNN-GNQQGNNGRGSQGNNGHQQEN---N 344
Query: 176 RSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNK 235
GS G G+ + G G G N +Q+ NNG+G +G + ++G GS G+ N+
Sbjct: 345 GRGSQGNNGNQQGNNGRGSQGNNGHQQE---NNGRGSQGNNGNQQGDNGRGSQQGNNGNQ 401
Query: 236 ---NIESNSKRNVGRPPKKAAETVASTKRGRE 264
N + K NVG RG +
Sbjct: 402 QGDNGRGSQKENVGNEQGNNGRGSQQENRGHQ 433
Score = 41.6 bits (96), Expect = 0.023
Identities = 43/230 (18%), Positives = 88/230 (37%), Gaps = 16/230 (6%)
Query: 20 PPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKGDHKPIINDKK---ERPSSD 76
PP TP + ++ + + + SS V P ++ K KP ++K E+P+
Sbjct: 222 PPATPSNPVENEEERQS-----QPDSSPDVVPDLSSVKDKKYEKPEYKEQKKIEEQPTKQ 276
Query: 77 VKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNK-NLSSNASNKKPPSNSTPRTGSSAN 135
+K + ++ + ++ G S++ N N N + N+ + G++
Sbjct: 277 IKEN-----NGRGSQQENRGNQQENNGRESQQGNNGNQQGNNGRESQQGNNGNQQGNNGR 331
Query: 136 KPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGG 195
+ + G + N G R + + + G G++G+ +G G
Sbjct: 332 GSQGNNGHQQENNGRGSQGNNGNQQGNNGRGSQGNNGHQQENNGRGSQGNNGNQQGDNGR 391
Query: 196 GTNT-KEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRN 244
G+ + +NG+G + KE G GS + + + N K+N
Sbjct: 392 GSQQGNNGNQQGDNGRGSQ-KENVGNEQGNNGRGSQQENRGHQQGNEKKN 440
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.299 0.122 0.330
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 547,619,165
Number of Sequences: 2790947
Number of extensions: 28467401
Number of successful extensions: 369853
Number of sequences better than 10.0: 9509
Number of HSP's better than 10.0 without gapping: 2198
Number of HSP's successfully gapped in prelim test: 7723
Number of HSP's that attempted gapping in prelim test: 216395
Number of HSP's gapped (non-prelim): 53057
length of query: 281
length of database: 848,049,833
effective HSP length: 126
effective length of query: 155
effective length of database: 496,390,511
effective search space: 76940529205
effective search space used: 76940529205
T: 11
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146756.6


