

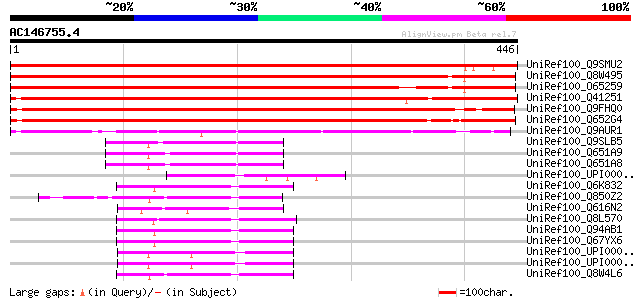
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146755.4 + phase: 0
(446 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SMU2 Putative calmodulin-binding heat-shock protein ... 702 0.0
UniRef100_Q8W495 Hypothetical protein At4g00500; F6N23.21 [Arabi... 600 e-170
UniRef100_O65259 F6N23.21 protein [Arabidopsis thaliana] 575 e-162
UniRef100_Q41251 Calmodulin-binding heat-shock protein [Nicotian... 534 e-150
UniRef100_Q9FHQ0 Calmodulin-binding heat-shock protein [Arabidop... 518 e-145
UniRef100_Q652G4 Putative calmodulin-binding heat-shock protein ... 505 e-141
UniRef100_Q9AUR1 Putative calmodulin-binding heat-shock protein ... 347 3e-94
UniRef100_Q9SLB5 Hypothetical protein At2g42450 [Arabidopsis tha... 76 2e-12
UniRef100_Q651A9 Hypothetical protein B1331F11.10-1 [Oryza sativa] 73 2e-11
UniRef100_Q651A8 Hypothetical protein B1331F11.10-2 [Oryza sativa] 73 2e-11
UniRef100_UPI0000498875 UPI0000498875 UniRef100 entry 70 1e-10
UniRef100_Q6K832 Lipase class 3 protein-like [Oryza sativa] 69 3e-10
UniRef100_Q850Z2 Putative heat-shock protein [Oryza sativa] 67 8e-10
UniRef100_Q616N2 Hypothetical protein CBG15206 [Caenorhabditis b... 66 2e-09
UniRef100_Q8L570 Lipase class 3-like protein [Oryza sativa] 65 4e-09
UniRef100_Q94AB1 AT3g14070/MAG2_2 [Arabidopsis thaliana] 65 5e-09
UniRef100_Q67YX6 Hypothetical protein At3g14075 [Arabidopsis tha... 65 5e-09
UniRef100_UPI000027CA7E UPI000027CA7E UniRef100 entry 64 7e-09
UniRef100_UPI0000018805 UPI0000018805 UniRef100 entry 64 1e-08
UniRef100_Q8W4L6 Hypothetical protein Z97340.15 [Arabidopsis tha... 64 1e-08
>UniRef100_Q9SMU2 Putative calmodulin-binding heat-shock protein [Arabidopsis
thaliana]
Length = 477
Score = 702 bits (1813), Expect = 0.0
Identities = 339/472 (71%), Positives = 397/472 (83%), Gaps = 26/472 (5%)
Query: 1 MSIICGL-PLVECVYCLACVRWAWKRCLHTAGHDSETWGFAATQEFEPVPRLCRYILAVY 59
MSI+CG PL+ECVYCL C RW +KRCL+TAGHDSE WG A T EFEPVPR CRYILAVY
Sbjct: 1 MSILCGCCPLLECVYCLGCARWGYKRCLYTAGHDSEDWGLATTDEFEPVPRFCRYILAVY 60
Query: 60 EDDLRNPLWAPPGGYGINPDWLLLRKTYKDTRGRAPPYILYLDHDHADIVLAIRGLNLAR 119
EDD+RNPLW PP GYGINPDWLLL+KTY+DT+GRAP YILYLDH H DIV+AIRGLNLA+
Sbjct: 61 EDDIRNPLWEPPEGYGINPDWLLLKKTYEDTQGRAPAYILYLDHVHQDIVVAIRGLNLAK 120
Query: 120 ESDYAVLLDNKLGKRKFDGGYVHNGLLKAAGWVMDAECEILRELVEKYPNYTLTFAGHSL 179
ESDYA+LLDNKLG+RKFDGGYVHNGL+K+AG+V+D EC++L+ELV+KYP+YTLTFAGHSL
Sbjct: 121 ESDYAMLLDNKLGERKFDGGYVHNGLVKSAGYVLDEECKVLKELVKKYPSYTLTFAGHSL 180
Query: 180 GSGVAAALSMVVVQNRDRLGNIERKRVRCYAIAPARCMSLNLAVRYADVINSVVLQDDFL 239
GSGVA L+++VV++ +RLGNI+RKRVRC+AIAPARCMSLNLAVRYADVINSV+LQDDFL
Sbjct: 181 GSGVATMLALLVVRHPERLGNIDRKRVRCFAIAPARCMSLNLAVRYADVINSVILQDDFL 240
Query: 240 PRTATPLEDIFKSLFCLPCLLCLRCMRDTCIPEEKMLKDPRRLYAPGRLYHIVERKPFRL 299
PRTATPLEDIFKS+FCLPCLLC+RCM+DTC+PE+KMLKDPRRLYAPGR+YHIVERKP RL
Sbjct: 241 PRTATPLEDIFKSVFCLPCLLCIRCMKDTCVPEQKMLKDPRRLYAPGRMYHIVERKPCRL 300
Query: 300 GRFPPVVRTAVPVDGRFEHIVLSCNATSDHAIIWIEKEAQRALDLMMEKDNTMEVPAKQI 359
GR+PPVV+TAVPVDGRFEHIVLSCNATSDHAIIWIE+EAQRAL+LMME + ME+P KQ
Sbjct: 301 GRYPPVVKTAVPVDGRFEHIVLSCNATSDHAIIWIEREAQRALNLMMENEKKMEIPEKQR 360
Query: 360 MQRQKTMTR-HGQEYKAALQRAKTLDIPHAFTPPSEYGTFD--------------EEGEE 404
M+RQ+++ R H EY+AAL+RA TLD+PHA + EYGTFD EE EE
Sbjct: 361 MERQESLAREHNLEYRAALRRAVTLDVPHAESMAYEYGTFDKTQEDETEEEEVETEEEEE 420
Query: 405 SS--------RSEAESSVSSTNRSTVNE--SWDVLIERLFDKDEHGHMVLKR 446
+ S + SSV T R N SWD LIE LF++DE G++ ++
Sbjct: 421 DTDSIAPMVGESSSSSSVKPTYRIRRNRRVSWDELIEHLFERDESGNLTFEK 472
>UniRef100_Q8W495 Hypothetical protein At4g00500; F6N23.21 [Arabidopsis thaliana]
Length = 460
Score = 600 bits (1546), Expect = e-170
Identities = 286/449 (63%), Positives = 355/449 (78%), Gaps = 6/449 (1%)
Query: 1 MSIICGLPLVECVYCLACVRWAWKRCLHTAGHDSETWGFAATQEFEPVPRLCRYILAVYE 60
MSI+C +P++ECVYCL C W WK+CL++AGH+SE WG A + EFEP+PR+CR ILAVYE
Sbjct: 1 MSILCCVPVLECVYCLGCTHWLWKKCLYSAGHESENWGLATSDEFEPIPRICRLILAVYE 60
Query: 61 DDLRNPLWAPPGGYGINPDWLLLRKTYKDTRGRAPPYILYLDHDHADIVLAIRGLNLARE 120
++L +P+WAPP GYGI+P+ ++L+K Y T GR PY++YLDH++ D+VLAIRGLNLA+E
Sbjct: 61 ENLHDPMWAPPDGYGIDPNHVILKKDYDQTEGRVTPYMIYLDHENGDVVLAIRGLNLAKE 120
Query: 121 SDYAVLLDNKLGKRKFDGGYVHNGLLKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLG 180
DYAVLLDNKLG+ KFDGGYVHNGLLKAA WV + E +LREL+E P+Y+LTF GHSLG
Sbjct: 121 CDYAVLLDNKLGQTKFDGGYVHNGLLKAAMWVFEEEHVVLRELLEANPSYSLTFVGHSLG 180
Query: 181 SGVAAALSMVVVQNRDRLGNIERKRVRCYAIAPARCMSLNLAVRYADVINSVVLQDDFLP 240
+GV + L + V+QNR RLGNIERKR+RC+AIAP RCMSL+LAV YADVINSVVLQDDFLP
Sbjct: 181 AGVVSLLVLFVIQNRVRLGNIERKRIRCFAIAPPRCMSLHLAVTYADVINSVVLQDDFLP 240
Query: 241 RTATPLEDIFKSLFCLPCLLCLRCMRDTCIPEEKMLKDPRRLYAPGRLYHIVERKPFRLG 300
RT T LE++FKS+ CLPCLLCL C++DT EE+ LKD RRLYAPGRLYHIV RKP RLG
Sbjct: 241 RTTTALENVFKSIICLPCLLCLTCLKDTFTFEERKLKDARRLYAPGRLYHIVVRKPLRLG 300
Query: 301 RFPPVVRTAVPVDGRFEHIVLSCNATSDHAIIWIEKEAQRALDLMMEKDNTMEVPAKQIM 360
R+PPVVRTAVPVDGRFE IVLSCNAT+DHAIIWIE+E+QRALDLM+E+D M++P +Q +
Sbjct: 301 RYPPVVRTAVPVDGRFEQIVLSCNATADHAIIWIERESQRALDLMVEEDQVMQIPVEQKI 360
Query: 361 QRQKTMTR-HGQEYKAALQRAKTLDIPHAFTPPSEYGTF--DEEGEESSRSEAESSVSST 417
RQK++ H +EY+AA+ +A +L+IP +P YGTF EEGE S+ S E S S
Sbjct: 361 VRQKSIVEDHDEEYRAAIMKAASLNIP--MSPSPSYGTFHDTEEGESSAGSGMEGSPSGW 418
Query: 418 NRSTVNESWDVLIERLFD-KDEHGHMVLK 445
+ + WD I+ F D HM+ K
Sbjct: 419 SFKGMRRKWDQFIDCHFPVNDNSEHMIFK 447
>UniRef100_O65259 F6N23.21 protein [Arabidopsis thaliana]
Length = 445
Score = 575 bits (1481), Expect = e-162
Identities = 277/448 (61%), Positives = 344/448 (75%), Gaps = 19/448 (4%)
Query: 1 MSIICGLPLVECVYCLACVRWAWKRCLHTAGHDSETWGFAATQEFEPVPRLCRYILAVYE 60
MSI+C +P++ECVYCL C W WK+CL++AGH+SE WG A + EFEP+PR+CR ILAVYE
Sbjct: 1 MSILCCVPVLECVYCLGCTHWLWKKCLYSAGHESENWGLATSDEFEPIPRICRLILAVYE 60
Query: 61 DDLRNPLWAPPGGYGINPDWLLLRKTYKDTRGRAPPYILYLDHDHADIVLAIRGLNLARE 120
++L +P+WAPP GYGI+P+ ++L+K Y T GR PY++YLDH++ D+VLAIRGLNLA+E
Sbjct: 61 ENLHDPMWAPPDGYGIDPNHVILKKDYDQTEGRVTPYMIYLDHENGDVVLAIRGLNLAKE 120
Query: 121 SDYAVLLDNKLGKRKFDGGYVHNGLLKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLG 180
DYAVLLDNKLG+ KFDGGYVHNGLLKAA WV + E +LREL+E P+Y+LTF GHSLG
Sbjct: 121 CDYAVLLDNKLGQTKFDGGYVHNGLLKAAMWVFEEEHVVLRELLEANPSYSLTFVGHSLG 180
Query: 181 SGVAAALSMVVVQNRDRLGNIERKRVRCYAIAPARCMSLNLAVRYADVINSVVLQDDFLP 240
+GV + L + V+QNR RLGNIERKR+RC+AIAP RCMSL+LAV YADVINSVVLQDDFLP
Sbjct: 181 AGVVSLLVLFVIQNRVRLGNIERKRIRCFAIAPPRCMSLHLAVTYADVINSVVLQDDFLP 240
Query: 241 RTATPLEDIFKSLFCLPCLLCLRCMRDTCIPEEKMLKDPRRLYAPGRLYHIVERKPFRLG 300
RT T LE++FKS+ CLPCLLCL C++DT EE+ LKD RRLYAPGRLYHIV RKP RLG
Sbjct: 241 RTTTALENVFKSIICLPCLLCLTCLKDTFTFEERKLKDARRLYAPGRLYHIVVRKPLRLG 300
Query: 301 RFPPVVRTAVPVDGRFEHIVLSCNATSDHAIIWIEKEAQRALDLMMEKDNTMEVPAKQIM 360
R+PPVVRTAVPVDGRFE IVLSCNAT+DHAIIWIE+E+QRALD +I+
Sbjct: 301 RYPPVVRTAVPVDGRFEQIVLSCNATADHAIIWIERESQRALD--------------KIV 346
Query: 361 QRQKTMTRHGQEYKAALQRAKTLDIPHAFTPPSEYGTF--DEEGEESSRSEAESSVSSTN 418
+++ + H +EY+AA+ +A +L+IP +P YGTF EEGE S+ S E S S +
Sbjct: 347 RQKSIVEDHDEEYRAAIMKAASLNIP--MSPSPSYGTFHDTEEGESSAGSGMEGSPSGWS 404
Query: 419 RSTVNESWDVLIERLFD-KDEHGHMVLK 445
+ WD I+ F D HM+ K
Sbjct: 405 FKGMRRKWDQFIDCHFPVNDNSEHMIFK 432
>UniRef100_Q41251 Calmodulin-binding heat-shock protein [Nicotiana tabacum]
Length = 449
Score = 534 bits (1375), Expect = e-150
Identities = 262/449 (58%), Positives = 334/449 (74%), Gaps = 9/449 (2%)
Query: 1 MSIICGLPLVECVYCLACVRWAWKRCLHTAGHDSETWGFAATQEFEPVPRLCRYILAVYE 60
MS+ CG ECV L C+RWAWKRC +T DS TW A +EFEPVPR+CR ILAVYE
Sbjct: 1 MSVACG---AECVLVLGCLRWAWKRCTYTGNDDSATWPTATYEEFEPVPRICRTILAVYE 57
Query: 61 DDLRNPLWAPPGGYGINPDWLLLRKTYKDTRGRAPPYILYLDHDHADIVLAIRGLNLARE 120
+LR+P + P GGY +NPDW++ R TY+ T G APPY++Y DH+H +IV+AIRGLNL E
Sbjct: 58 PNLRSPKYPPKGGYRLNPDWVIKRVTYEQTSGNAPPYLIYCDHEHQEIVVAIRGLNLLNE 117
Query: 121 SDYAVLLDNKLGKRKFDGGYVHNGLLKAAGWVMDAECEILREL-VEKYPNYTLTFAGHSL 179
SDY VLLDN+LGK+ FDGGYVH+GLLK+A WV++ E E L++L +E +Y + FAGHSL
Sbjct: 118 SDYKVLLDNRLGKQMFDGGYVHHGLLKSAVWVLNNESETLKKLWIENGRSYKMIFAGHSL 177
Query: 180 GSGVAAALSMVVVQNRDRLGNIERKRVRCYAIAPARCMSLNLAVRYADVINSVVLQDDFL 239
GSGVA+ L+++V ++DRLG I R +RCYA+APARCMSLNLAV+YAD+I+SVVLQDDFL
Sbjct: 178 GSGVASLLTVIVANHKDRLGGIPRSLLRCYAVAPARCMSLNLAVKYADIIHSVVLQDDFL 237
Query: 240 PRTATPLEDIFKSLFCLPCLLCLRCMRDTCIPEEKMLKDPRRLYAPGRLYHIVERKPFRL 299
PRTATPLEDIFKS+FCLPCL+ L C+RDT IPE + L+DPRRLYAPGR+YHIVER+ R
Sbjct: 238 PRTATPLEDIFKSIFCLPCLIFLVCLRDTFIPEGRKLRDPRRLYAPGRMYHIVERRFCRC 297
Query: 300 GRFPPVVRTAVPVDGRFEHIVLSCNATSDHAIIWIEKEAQRALDLMME--KDNTMEVPAK 357
GRF P VRTA+PVDGRFEHIVLS NAT DH IIWIE+E+++ L + E + T P
Sbjct: 298 GRFTPDVRTAIPVDGRFEHIVLSRNATVDHGIIWIERESEKVLARLKEASAETTTTPPKV 357
Query: 358 QIMQRQKTMTRHGQEYKAALQRAKTLDIPHAFTPPSEYGTFDEEGEESSRSEAESSVSST 417
Q ++R KT+ + E+K AL+RA +L+IPHA E T E S + E ++ S
Sbjct: 358 QKIERLKTLEK---EHKDALERAVSLNIPHAVDADEEESTESITEESSQKQEEDAMTSKA 414
Query: 418 NRSTVNESWDVLIERLFDKDEHGHMVLKR 446
S +W+ ++E+LF++DE G + LKR
Sbjct: 415 QCSDARTNWNEVVEKLFNRDESGKLRLKR 443
>UniRef100_Q9FHQ0 Calmodulin-binding heat-shock protein [Arabidopsis thaliana]
Length = 449
Score = 518 bits (1334), Expect = e-145
Identities = 259/446 (58%), Positives = 328/446 (73%), Gaps = 17/446 (3%)
Query: 1 MSIICGLPLVECVYCLACVRWAWKRCLHTAGHDSETWGFAATQEFEPVPRLCRYILAVYE 60
MS+ CGL ECV+C+ RWAWKRC H DS TW A +EFEP+PR+ R ILAVYE
Sbjct: 1 MSVACGL---ECVFCVGFSRWAWKRCTHVGSDDSATWTSATPEEFEPIPRISRVILAVYE 57
Query: 61 DDLRNPLWAPP-GGYGINPDWLLLRKTYKDTRGRAPPYILYLDHDHADIVLAIRGLNLAR 119
DLRNP +P G + +NP+W++ R T++ T+GR+PPYI+Y+DHDH +IVLAIRGLNLA+
Sbjct: 58 PDLRNPKISPSLGTFDLNPEWVIKRVTHEKTQGRSPPYIIYIDHDHREIVLAIRGLNLAK 117
Query: 120 ESDYAVLLDNKLGKRKFDGGYVHNGLLKAAGWVMDAECEIL-RELVEKYPNYTLTFAGHS 178
ESDY +LLDNKLG++ GGYVH GLLK+A WV++ E E L R E Y L FAGHS
Sbjct: 118 ESDYKILLDNKLGQKMLGGGYVHRGLLKSAAWVLNQESETLWRVWEENGREYDLVFAGHS 177
Query: 179 LGSGVAAALSMVVVQNRDRLGNIERKRVRCYAIAPARCMSLNLAVRYADVINSVVLQDDF 238
LGSGVAA ++++VV +G+I R +VRC+A+APARCMSLNLAV+YADVI+SV+LQDDF
Sbjct: 178 LGSGVAALMAVLVVNTPAMIGDIPRNKVRCFALAPARCMSLNLAVKYADVISSVILQDDF 237
Query: 239 LPRTATPLEDIFKSLFCLPCLLCLRCMRDTCIPEEKMLKDPRRLYAPGRLYHIVERKPFR 298
LPRTATPLEDIFKS+FCLPCLL L C+RDT IPE + L+DPRRLYAPGR+YHIVERK R
Sbjct: 238 LPRTATPLEDIFKSVFCLPCLLFLVCLRDTFIPEGRKLRDPRRLYAPGRIYHIVERKFCR 297
Query: 299 LGRFPPVVRTAVPVDGRFEHIVLSCNATSDHAIIWIEKEAQRALDLMMEKDNTMEVPAKQ 358
GRFPP VRTA+PVDGRFEHIVLS NATSDHAI+WIE+EA++AL ++ EK + V
Sbjct: 298 CGRFPPEVRTAIPVDGRFEHIVLSSNATSDHAILWIEREAEKALQILREKSSETVVTMAP 357
Query: 359 IMQRQKTMTRHGQEYKAALQRAKTLDIPHAFTPPSEYGTFDEEGEESSRSEAESSVSSTN 418
+R + ++ +E+K AL+RA +L+IPHA + EE EE + EA S
Sbjct: 358 KEKRMERLSTLEKEHKDALERAVSLNIPHAVSTA-------EEEEECNNGEA-----SAE 405
Query: 419 RSTVNESWDVLIERLFDKDEHGHMVL 444
T ++WD ++++LF + G VL
Sbjct: 406 LKTKKKNWDEVVDKLFHRSNSGEFVL 431
>UniRef100_Q652G4 Putative calmodulin-binding heat-shock protein [Oryza sativa]
Length = 453
Score = 505 bits (1300), Expect = e-141
Identities = 259/450 (57%), Positives = 331/450 (73%), Gaps = 13/450 (2%)
Query: 1 MSIICGLPLVECVYCLACVRWAWKRCLHTAGHDSETWGFAATQEFEPVPRLCRYILAVYE 60
MS+ CGL E V CL C RWAWKR + +DSE W AA EFEPVPR+CR ILA+YE
Sbjct: 1 MSVSCGL---EWVVCLGCTRWAWKRLTYIGAYDSEAWPAAAPGEFEPVPRICRVILAIYE 57
Query: 61 DDLRNPL-WAPPG-GY-GINPDWLLLRKTYKDTRGRAPPYILYLDHDHADIVLAIRGLNL 117
DDL NP +APPG GY G++ ++ R TY+ PPYI+Y+DH H ++VLAIRGLNL
Sbjct: 58 DDLSNPTKFAPPGRGYAGVDLAGVVKRATYEHVGNTCPPYIVYVDHRHKEVVLAIRGLNL 117
Query: 118 ARESDY-AVLLDNKLGKRKFDGGYVHNGLLKAAGWVMDAECEILRELVEKY-PNYTLTFA 175
R +DY VL+DNKLG + FDGGYVH+GLLKAA ++++ E + L+EL+++ P+Y L FA
Sbjct: 118 TRNADYKVVLMDNKLGMQMFDGGYVHHGLLKAAQFILERETKTLQELLQQNGPDYKLIFA 177
Query: 176 GHSLGSGVAAALSMVVVQNRDRLGNIERKRVRCYAIAPARCMSLNLAVRYADVINSVVLQ 235
GHSLGSG+AA ++++VV NR GNI R ++RCYA+APARCMSLNLAV+YADVINSVVLQ
Sbjct: 178 GHSLGSGIAALMTVLVVNNRKMFGNIPRSQIRCYALAPARCMSLNLAVKYADVINSVVLQ 237
Query: 236 DDFLPRTATPLEDIFKSLFCLPCLLCLRCMRDTCIPEEKMLKDPRRLYAPGRLYHIVERK 295
DDFLPRT TPLE IF S+FCLPCLL + C+RDT +++ KDPRRLYAPGR+YHIVERK
Sbjct: 238 DDFLPRTPTPLEYIFGSIFCLPCLLFIMCLRDTFKQDKRKFKDPRRLYAPGRMYHIVERK 297
Query: 296 PFRLGRFPPVVRTAVPVDGRFEHIVLSCNATSDHAIIWIEKEAQRALDLMMEKDNTMEVP 355
R GRFPP VRTA+PV+GRFEHIVLSC+ TSDHAI+WIE+E+++AL+LM + P
Sbjct: 298 FCRCGRFPPEVRTAIPVEGRFEHIVLSCSTTSDHAIVWIERESEKALELMKGNEKPTTPP 357
Query: 356 AKQIMQRQKTMTRHGQEYKAALQRAKTLDIPHAFTPPSEYGTFDEEGEESSRSEAESSVS 415
A+Q M+R ++ +E+K AL+RAKTLD+PHA SE E + S+ S +
Sbjct: 358 AQQKMERLQSFE---EEHKNALERAKTLDVPHA-VDLSEV-EIQEGSSPTPPSDTHSEAT 412
Query: 416 STNRSTVNESWDVLIERLFDKDEHGHMVLK 445
S +S SWD L+ +LF +DE G +V+K
Sbjct: 413 SEAKSAGRTSWDELMHKLFTRDEGGKLVVK 442
>UniRef100_Q9AUR1 Putative calmodulin-binding heat-shock protein [Oryza sativa]
Length = 426
Score = 347 bits (891), Expect = 3e-94
Identities = 212/444 (47%), Positives = 265/444 (58%), Gaps = 39/444 (8%)
Query: 1 MSIICGLPLVECVYCLACVRWAWKRCLHTAGHDSETWGFAATQEFEPVPRLCRYILAVYE 60
M++ C EC LAC RWA +R + DS +W A+ F PVPR CR LA ++
Sbjct: 1 MALSCA---AECALSLACARWALRRLSLSGADDSASWPAASPSSFAPVPRACRSALAAWQ 57
Query: 61 DDLRNPLWAPPGGYGINPDWLLLRKTYKDTRGRAPPYILYLDHDHADIVLAIRGLNLARE 120
AP P L PPY L D ++VLA+RGL LAR
Sbjct: 58 QGHDEAEQAPTPA----PSRL------------CPPYRLSHDRARGEVVLAVRGLGLARL 101
Query: 121 SDYAVLLDNKLGKRKFDGGYVHNGLLKAAGWVMDAECEILRELVEKY--PNYTLTFAGHS 178
DY VLLD G F GG+ H GLL+AA W++D E +R +V + L F GHS
Sbjct: 102 EDYRVLLDAG-GPEPFAGGHAHRGLLRAAVWLLDREGPAIRRMVAEAGPAGCRLVFVGHS 160
Query: 179 LGSGVAAALSMVVVQN-RDRLGNIERKRVRCYAIAPARCMSLNLAVRYADVINSVVLQDD 237
LG+GVAA ++V V+ +RLG + R VRCYA+AP RCMSL LAV YADV++SVVLQDD
Sbjct: 161 LGAGVAALAAVVAVRCWLERLG-LRRGDVRCYAMAPPRCMSLGLAVEYADVVHSVVLQDD 219
Query: 238 FLPRTATPLEDIFKSLFCLPCLLCLRCMRDTCIPEEKMLKDPRRLYAPGRLYHIVERKPF 297
FLPRT PL+ IF S+FCLPCLLC CMRDT + EEK LKD +LYAPGR++HIVER+
Sbjct: 220 FLPRTPAPLQHIFGSIFCLPCLLCFICMRDTFVSEEK-LKDASKLYAPGRVFHIVERENC 278
Query: 298 RLGRFPPVVRTAVPVDGRFEHIVLSCNATSDHAIIWIEKEAQRALDLMMEKDNTMEVPAK 357
R GR PP VRTAVP +GRFEH+VLSCNATSDH IIWIEKEAQ+ALDLM +++ T+ P++
Sbjct: 279 RCGRLPPQVRTAVPAEGRFEHVVLSCNATSDHGIIWIEKEAQKALDLMEQEELTLP-PSQ 337
Query: 358 QIMQRQKTMTRHGQEYKAALQRAKTLDIPHAFTPPSEYGTFDEEGEESSRSEAESSVSST 417
Q M R K K + + D + +P S S R+ SS+ S
Sbjct: 338 QKMLRVKETESLADHQKLSAGNPQEDDTLSSSSPFS-----------SPRTSTTSSLRSE 386
Query: 418 NRSTVNESWDVLIE-RLFDKDEHG 440
+ ST +E WD L+E L D +E G
Sbjct: 387 SSSTRSE-WDELVEIFLSDHEEDG 409
>UniRef100_Q9SLB5 Hypothetical protein At2g42450 [Arabidopsis thaliana]
Length = 508
Score = 75.9 bits (185), Expect = 2e-12
Identities = 46/160 (28%), Positives = 82/160 (50%), Gaps = 7/160 (4%)
Query: 85 KTYKDTRGRAPPYILYLDHDHADIVLAIRGLNLARE--SDYAVLLDNKLGKRKFDGGYVH 142
K KD+ P Y + +DH +V IRG + + +D D ++ F+G H
Sbjct: 181 KFVKDSSVMRPGYYIGVDHRRKLVVFGIRGTHTIYDLITDIVSSSDEEV---TFEGYSTH 237
Query: 143 NGLLKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLGSGVAAALSMVVVQN-RDRLGNI 201
G +AA W ++ E + +R + KY Y L GHSLG +A+ +++++ + R+ LG
Sbjct: 238 FGTAEAARWFLNHELQTIRRCLAKYEGYKLRLVGHSLGGAIASLMAIMLKKMPREELG-F 296
Query: 202 ERKRVRCYAIAPARCMSLNLAVRYADVINSVVLQDDFLPR 241
+ + + A C+S LA ++ + ++V+QDD +PR
Sbjct: 297 DAEIISAVGYATPPCVSKELAENCSEFVTTIVMQDDIIPR 336
>UniRef100_Q651A9 Hypothetical protein B1331F11.10-1 [Oryza sativa]
Length = 518
Score = 72.8 bits (177), Expect = 2e-11
Identities = 49/160 (30%), Positives = 80/160 (49%), Gaps = 7/160 (4%)
Query: 85 KTYKDTRGRAPPYILYLDHDHADIVLAIRGLNLARE--SDYAVLLDNKLGKRKFDGGYVH 142
K KD+ P Y + +D ++L IRG + + +D L D K+ + F H
Sbjct: 199 KFVKDSSILRPGYYIAIDPRTKLVILGIRGTHTVYDLVTDLIALSDKKVSPKGFS---TH 255
Query: 143 NGLLKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLGSGVAAALS-MVVVQNRDRLGNI 201
G +AA W + E ++R+ +EK+ +Y L GHSLG AA L+ M+ ++++ LG
Sbjct: 256 FGTYEAARWYLRHELGLIRKCLEKHKDYKLRLVGHSLGGASAALLAIMLRKKSKEELG-F 314
Query: 202 ERKRVRCYAIAPARCMSLNLAVRYADVINSVVLQDDFLPR 241
+ C+S +A A +++VVLQDD +PR
Sbjct: 315 SPDVISAVGYGTPPCVSREIAQSCASYVSTVVLQDDIIPR 354
>UniRef100_Q651A8 Hypothetical protein B1331F11.10-2 [Oryza sativa]
Length = 328
Score = 72.8 bits (177), Expect = 2e-11
Identities = 49/160 (30%), Positives = 80/160 (49%), Gaps = 7/160 (4%)
Query: 85 KTYKDTRGRAPPYILYLDHDHADIVLAIRGLNLARE--SDYAVLLDNKLGKRKFDGGYVH 142
K KD+ P Y + +D ++L IRG + + +D L D K+ + F H
Sbjct: 9 KFVKDSSILRPGYYIAIDPRTKLVILGIRGTHTVYDLVTDLIALSDKKVSPKGFS---TH 65
Query: 143 NGLLKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLGSGVAAALS-MVVVQNRDRLGNI 201
G +AA W + E ++R+ +EK+ +Y L GHSLG AA L+ M+ ++++ LG
Sbjct: 66 FGTYEAARWYLRHELGLIRKCLEKHKDYKLRLVGHSLGGASAALLAIMLRKKSKEELG-F 124
Query: 202 ERKRVRCYAIAPARCMSLNLAVRYADVINSVVLQDDFLPR 241
+ C+S +A A +++VVLQDD +PR
Sbjct: 125 SPDVISAVGYGTPPCVSREIAQSCASYVSTVVLQDDIIPR 164
>UniRef100_UPI0000498875 UPI0000498875 UniRef100 entry
Length = 432
Score = 70.1 bits (170), Expect = 1e-10
Identities = 58/175 (33%), Positives = 85/175 (48%), Gaps = 25/175 (14%)
Query: 139 GYVHNGLLKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLGSGVAAALSMVVVQNR-DR 197
G+ H G+ KAA L+ L KYP++ +T AGHSLG GVA L++ + +N D
Sbjct: 216 GFTHAGIYKAALNKYQQIIPTLKMLRLKYPSFQITIAGHSLGGGVAQLLTLEINKNHPDW 275
Query: 198 LGNIERKRVRCYAIAPARCMSLNLAVR--YADVINSVVLQDDFLPRTA-------TPLED 248
L V Y +APA +SLN+A +I+SVV ++D +PR + PL +
Sbjct: 276 L-------VHGYCLAPALVLSLNIASSPLVRSLIDSVVSKNDIVPRLSFDSIKNIQPLIN 328
Query: 249 IFKSLFCLPCLLCLRCMRDT--------CIPEEKMLKDPRRLYAPGRLYHIVERK 295
F+S++ L+ L T E D L PGR++HI +RK
Sbjct: 329 EFRSIYNNTSLISLNSKETTEQYQQAFNRFYESTNTIDSSVLVPPGRVFHIQKRK 383
>UniRef100_Q6K832 Lipase class 3 protein-like [Oryza sativa]
Length = 657
Score = 68.9 bits (167), Expect = 3e-10
Identities = 47/165 (28%), Positives = 77/165 (46%), Gaps = 16/165 (9%)
Query: 95 PPYILYLDHDHADIVLAIRGLNLARESDYAVL---------LDNKLGKRKFDGGYVHNGL 145
P + + LD D I+L IRG + R++ A + + G GY H G+
Sbjct: 174 PAFTIILDRDKQCILLLIRGTHSIRDTLTAATGAVVPFHHTIVQEGGVSDLVLGYAHFGM 233
Query: 146 LKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLGSGVAAALSMVVVQNRDRLGNIERKR 205
+ AA W+ L + + +P+Y + GHSLG G AA L+ V+ + + E
Sbjct: 234 VAAARWIAKLAAPCLAQALHTHPDYKIKIVGHSLGGGTAALLTYVLREQQ------EFAS 287
Query: 206 VRCYAIAPARCMSLNLAVRYADVINSVVLQDDFLPR-TATPLEDI 249
C + APA CM+ +LA I +V+ D +P +A ++D+
Sbjct: 288 TTCVSFAPAACMTWDLAESGVHFITTVINGADLVPTFSAASVDDL 332
>UniRef100_Q850Z2 Putative heat-shock protein [Oryza sativa]
Length = 594
Score = 67.4 bits (163), Expect = 8e-10
Identities = 57/227 (25%), Positives = 98/227 (43%), Gaps = 34/227 (14%)
Query: 26 CLHTAGHDSETWGFAATQEFEPVPRLCRYI-LAVYEDDLRNPLWAPPGGYGINPDWLLLR 84
CL GHD+ T V L ++ + ++ + GGY N + +L+
Sbjct: 125 CLQLKGHDAHT----------EVAYLLEHLKICMFYSKKTFSAFLQFGGY--NQEDILIH 172
Query: 85 KTYKDTRGRAPPYILYLDHDHADIVLAIRGLNLARES-----------DYAVLLDNKLGK 133
K R P + L D +L IRG +E + VL + ++
Sbjct: 173 KAR--ARLMQPSFALVCDKKSKCFLLFIRGAISTKERLTAATAAEVPFHHIVLSEGQISN 230
Query: 134 RKFDGGYVHNGLLKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLGSGVAAALSMVVVQ 193
GY H G+L AA W+ + L + V+++P+Y + GHS+G+G+ A L+ ++ +
Sbjct: 231 VVL--GYAHCGMLAAARWIANLAKPHLHKAVQEFPDYQIKVIGHSMGAGIGAILTYILHE 288
Query: 194 NRDRLGNIERKRVRCYAIAPARCMSLNLAVRYADVINSVVLQDDFLP 240
+ E C A AP CMS LA + + S++ ++D +P
Sbjct: 289 HH------EFSSCTCLAFAPPACMSWELAESGKEFVTSLINRNDVVP 329
>UniRef100_Q616N2 Hypothetical protein CBG15206 [Caenorhabditis briggsae]
Length = 651
Score = 66.2 bits (160), Expect = 2e-09
Identities = 51/166 (30%), Positives = 75/166 (44%), Gaps = 30/166 (18%)
Query: 96 PYILYLDHDHADIVLAIRG-----------------LNLARESDYAVLLDNKLGKRKFDG 138
P+ + DHD IV+ IRG + + + D + D + +R
Sbjct: 333 PFAVIADHDRKSIVITIRGSCSLIDLVTDLSLEDELMTVDVDQDATLSQDENIDRR--GD 390
Query: 139 GYVHNGLLKAAGWVMDA--ECEILRELVEKYPNYTLTFAGHSLGSGVAAALSMVVVQNRD 196
VH G+L++A +V D + +IL +L PNY L GHSLG+GV + L+M++ Q
Sbjct: 391 VRVHRGMLRSARYVFDTLNKNKILNDLFISNPNYQLVVCGHSLGAGVGSLLTMLLKQ--- 447
Query: 197 RLGNIERKRVRCYAIAPARCMSLNLAV-RYADVINSVVLQDDFLPR 241
E RV CYA AP C+ + SVV DD + R
Sbjct: 448 -----EYPRVICYAFAPPGCVISEYGQDEMEKYVMSVVSGDDIVSR 488
>UniRef100_Q8L570 Lipase class 3-like protein [Oryza sativa]
Length = 635
Score = 65.1 bits (157), Expect = 4e-09
Identities = 47/168 (27%), Positives = 76/168 (44%), Gaps = 17/168 (10%)
Query: 95 PPYILYLDHDHADIVLAIRGLNLARESDYAV----------LLDNKLGKRKFDGGYVHNG 144
P + + D ++ IRG + +++ A LLD G K GY H G
Sbjct: 166 PAHTILRDECTKSFLVLIRGTHSMKDTLTAATGAVVPFHHSLLDEG-GVSKLVLGYAHCG 224
Query: 145 LLKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLGSGVAAALSMVVVQNRDRLGNIERK 204
++ AA W+ + L + V + P+Y + GHSLG G AA L+ ++ +++ E
Sbjct: 225 MVAAARWIARSITPCLCQAVSQCPDYQIRVVGHSLGGGTAALLTYILREHQ------ELS 278
Query: 205 RVRCYAIAPARCMSLNLAVRYADVINSVVLQDDFLPRTATPLEDIFKS 252
C A APA CM+ LA + ++V D +P +T D +S
Sbjct: 279 STTCVAFAPASCMTWELAESGKHFVRTIVNGADLVPTVSTSSIDDLRS 326
>UniRef100_Q94AB1 AT3g14070/MAG2_2 [Arabidopsis thaliana]
Length = 642
Score = 64.7 bits (156), Expect = 5e-09
Identities = 46/165 (27%), Positives = 77/165 (45%), Gaps = 16/165 (9%)
Query: 95 PPYILYLDHDHADIVLAIRGLNLARESDYAVL---------LDNKLGKRKFDGGYVHNGL 145
P + + +DH+ +L IRG + +++ A + N+ G GY H G+
Sbjct: 169 PAFTVLVDHNTKYFLLLIRGTHSIKDTLTAATGAIVPFHHTVVNERGVSNLVLGYAHCGM 228
Query: 146 LKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLGSGVAAALSMVVVQNRDRLGNIERKR 205
+ AA + L + +E+YP+Y + GHSLG G AA L+ ++ + +
Sbjct: 229 VAAARCIAKLATPCLLKGLEQYPDYKIKIVGHSLGGGTAALLTYIMREQK------MLST 282
Query: 206 VRCYAIAPARCMSLNLAVRYADVINSVVLQDDFLPR-TATPLEDI 249
C APA CM+ LA D I SV+ D +P +A ++D+
Sbjct: 283 ATCVTFAPAACMTWELADSGNDFIVSVINGADLVPTFSAAAVDDL 327
>UniRef100_Q67YX6 Hypothetical protein At3g14075 [Arabidopsis thaliana]
Length = 642
Score = 64.7 bits (156), Expect = 5e-09
Identities = 46/165 (27%), Positives = 77/165 (45%), Gaps = 16/165 (9%)
Query: 95 PPYILYLDHDHADIVLAIRGLNLARESDYAVL---------LDNKLGKRKFDGGYVHNGL 145
P + + +DH+ +L IRG + +++ A + N+ G GY H G+
Sbjct: 169 PAFTVLVDHNTKYFLLLIRGTHSIKDTLTAATGAIVPFHHTVVNERGVSNLVLGYAHCGM 228
Query: 146 LKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLGSGVAAALSMVVVQNRDRLGNIERKR 205
+ AA + L + +E+YP+Y + GHSLG G AA L+ ++ + +
Sbjct: 229 VAAARCIAKLATPCLLKGLEQYPDYKIKIVGHSLGGGTAALLTYIMREQK------MLST 282
Query: 206 VRCYAIAPARCMSLNLAVRYADVINSVVLQDDFLPR-TATPLEDI 249
C APA CM+ LA D I SV+ D +P +A ++D+
Sbjct: 283 ATCVTFAPAACMTWELADSGNDFIVSVINGADLVPTFSAAAVDDL 327
>UniRef100_UPI000027CA7E UPI000027CA7E UniRef100 entry
Length = 597
Score = 64.3 bits (155), Expect = 7e-09
Identities = 48/160 (30%), Positives = 80/160 (50%), Gaps = 14/160 (8%)
Query: 96 PYILYLDHDHADIVLAIRGLNLARE--SDYAVLLDNKLGKRKFDGGYVHNGLLKAAGWVM 153
P+ + LDH +V+A+RG ++ +D + +N + Y H G+ +AAG++
Sbjct: 292 PFFVALDHKKEAVVVAVRGTLSLKDVLTDLSAECENLPLEGVPGACYAHKGISQAAGFIY 351
Query: 154 DAECE--ILRELVEKYPNYTLTFAGHSLGSGVAAALSMVVVQNRDRLGNIERKRVRCYAI 211
IL + + P Y L GHSLG+G A+ L++++ + L +CYA
Sbjct: 352 KKLVNDGILSQALSTVPEYKLVITGHSLGAGTASVLAVLLRSSFPTL--------QCYAF 403
Query: 212 A-PARCMSLNLAVRYADVINSVVLQDDFLPRTATP-LEDI 249
+ P +S LA D + SVVL D +PR + P +ED+
Sbjct: 404 SPPGGLLSKALADYSKDFVVSVVLGKDLVPRLSIPNMEDL 443
>UniRef100_UPI0000018805 UPI0000018805 UniRef100 entry
Length = 667
Score = 63.5 bits (153), Expect = 1e-08
Identities = 47/160 (29%), Positives = 80/160 (49%), Gaps = 14/160 (8%)
Query: 96 PYILYLDHDHADIVLAIRGLNLARE--SDYAVLLDNKLGKRKFDGGYVHNGLLKAAGWVM 153
P+ + LDH +++A+RG ++ +D + +N + Y H G+ +AAG++
Sbjct: 369 PFFVALDHKREAVLVAVRGTLSLKDVLTDLSAECENLPVEGVPGACYAHKGISQAAGYIY 428
Query: 154 DAECE--ILRELVEKYPNYTLTFAGHSLGSGVAAALSMVVVQNRDRLGNIERKRVRCYAI 211
IL + + P Y L GHSLG+G A+ L++++ + L +CYA
Sbjct: 429 KKLVNDGILNQALSIVPEYKLVITGHSLGAGTASVLAILLRTSFPTL--------QCYAF 480
Query: 212 A-PARCMSLNLAVRYADVINSVVLQDDFLPRTATP-LEDI 249
+ P +S LA D + SVVL D +PR + P +ED+
Sbjct: 481 SPPGGLLSKALADYSKDFVVSVVLGKDLVPRLSIPNMEDL 520
>UniRef100_Q8W4L6 Hypothetical protein Z97340.15 [Arabidopsis thaliana]
Length = 654
Score = 63.5 bits (153), Expect = 1e-08
Identities = 42/167 (25%), Positives = 80/167 (47%), Gaps = 20/167 (11%)
Query: 95 PPYILYLDHDHADIVLAIRGLNLARES-----------DYAVLLDNKLGKRKFDGGYVHN 143
P + + D + I+L IRG + +++ ++VL D L GY H
Sbjct: 167 PAFTIIRDTNSKCILLLIRGTHSIKDTLTAATGAVVPFHHSVLHDGGLSNLVL--GYAHC 224
Query: 144 GLLKAAGWVMDAECEILRELVEKYPNYTLTFAGHSLGSGVAAALSMVVVQNRDRLGNIER 203
G++ AA W+ L + +++ P++ + GHSLG G A+ L+ ++ + + E
Sbjct: 225 GMVAAARWIAKLSVPCLLKALDENPSFKVQIVGHSLGGGTASLLTYILREQK------EF 278
Query: 204 KRVRCYAIAPARCMSLNLAVRYADVINSVVLQDDFLPR-TATPLEDI 249
C+ APA CM+ +LA I +++ D +P +A+ ++D+
Sbjct: 279 ASATCFTFAPAACMTWDLAESGKHFITTIINGSDLVPTFSASSVDDL 325
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 773,050,422
Number of Sequences: 2790947
Number of extensions: 32326590
Number of successful extensions: 73888
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 87
Number of HSP's that attempted gapping in prelim test: 73685
Number of HSP's gapped (non-prelim): 182
length of query: 446
length of database: 848,049,833
effective HSP length: 131
effective length of query: 315
effective length of database: 482,435,776
effective search space: 151967269440
effective search space used: 151967269440
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146755.4


