

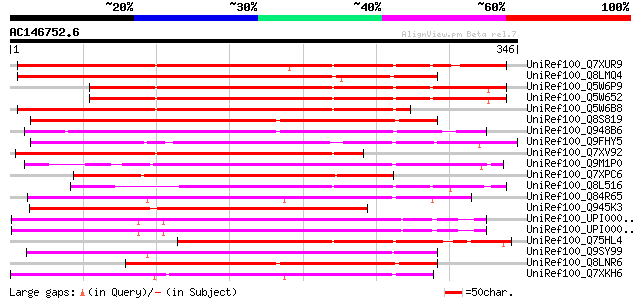
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.6 + phase: 0
(346 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XUR9 OSJNBa0084K11.21 protein [Oryza sativa] 272 1e-71
UniRef100_Q8LMQ4 Hypothetical protein OSJNBa0011L14.7 [Oryza sat... 263 5e-69
UniRef100_Q5W6P9 Putative polyprotein [Oryza sativa] 257 4e-67
UniRef100_Q5W652 Hypothetical protein OSJNBb0052F16.10 [Oryza sa... 257 4e-67
UniRef100_Q5W6B8 Putative polyprotein [Oryza sativa] 253 4e-66
UniRef100_Q8S819 Putative transposase [Oryza sativa] 234 2e-60
UniRef100_Q948B6 Putative transposase [Oryza sativa] 234 3e-60
UniRef100_Q9FHY5 Emb|CAB72466.1 [Arabidopsis thaliana] 228 2e-58
UniRef100_Q7XV92 OSJNBb0012E08.4 protein [Oryza sativa] 225 2e-57
UniRef100_Q9M1P0 Hypothetical protein T18B22.40 [Arabidopsis tha... 223 5e-57
UniRef100_Q7XPC6 OSJNBa0042N22.14 protein [Oryza sativa] 223 8e-57
UniRef100_Q8L516 Putative transposase [Oryza sativa] 212 1e-53
UniRef100_Q84R65 Hypothetical protein OSJNBb0113I20.11 [Oryza sa... 203 5e-51
UniRef100_Q945K3 Putative transposase [Zea mays] 201 2e-50
UniRef100_UPI000043028E UPI000043028E UniRef100 entry 201 3e-50
UniRef100_UPI000042EF69 UPI000042EF69 UniRef100 entry 201 3e-50
UniRef100_Q75HL4 Expressed protein [Oryza sativa] 199 9e-50
UniRef100_Q9SY99 T25B24.14 protein [Arabidopsis thaliana] 194 3e-48
UniRef100_Q8LNR6 Hypothetical protein OSJNBa0042E19.20 [Oryza sa... 193 5e-48
UniRef100_Q7XKH6 OSJNBb0033P05.11 protein [Oryza sativa] 181 3e-44
>UniRef100_Q7XUR9 OSJNBa0084K11.21 protein [Oryza sativa]
Length = 377
Score = 272 bits (695), Expect = 1e-71
Identities = 151/339 (44%), Positives = 214/339 (62%), Gaps = 15/339 (4%)
Query: 6 NSETSRNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFW 65
N T N++R+ +F + C + G L T +EEQVA L+ + HN +NR V
Sbjct: 42 NDTTCINMLRLRRASFFRFCKLFRDRGLLEDTIHMCIEEQVAMFLHTVGHNLRNRLVRTN 101
Query: 66 FRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTH 125
F RS ET+SR+ ++VL AI EL ++ I P T P +I+ + R+ PYFKDC+GAIDGTH
Sbjct: 102 FDRSRETVSRYFNKVLHAIGELRDELIRPPSLDT-PTKIAGNPRWDPYFKDCIGAIDGTH 160
Query: 126 IRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDK 185
IR V +RGRK + TQNV+AA FDL+FTYVLAGWEG+A D+ ++++AL RE+
Sbjct: 161 IRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTAHDAVVLRDALERENG 220
Query: 186 LKIP-----EGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASL 240
L++P +GKYYLVDAG+ G + P+R VRYHL E+ NP N KELFNLRH+SL
Sbjct: 221 LRVPQGNRLQGKYYLVDAGYGAKQGFLPPFRAVRYHLNEW-GNNPVQNEKELFNLRHSSL 279
Query: 241 RNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDA 300
R +ERAFG LK+RF++L ++T P + + Q I+ ACCI+HN++++ + ++LIA D
Sbjct: 280 RVTVERAFGSLKRRFKVLDDAT-PFFPFQTQVDIVVACCIIHNWVVN-DGIDELIAPPDW 337
Query: 301 ELANQNVSHDNHEASRSDMDEFAQGGIIKNGAAHQMWSN 339
S D E+ ++ A + G A QMW++
Sbjct: 338 S------SEDIDESLTGQANDHALMVQFRQGLADQMWAD 370
>UniRef100_Q8LMQ4 Hypothetical protein OSJNBa0011L14.7 [Oryza sativa]
Length = 1202
Score = 263 bits (672), Expect = 5e-69
Identities = 145/295 (49%), Positives = 185/295 (62%), Gaps = 12/295 (4%)
Query: 6 NSETSRNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFW 65
+ T +RM F KLC L +G L T SVEEQVA L + + V F
Sbjct: 588 SDRTCTQQLRMSRAVFYKLCARLRNKGLLVDTFHVSVEEQVAMFLKKVGQHHSVSCVGFS 647
Query: 66 FRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTH 125
F RSGET+SR+ VL+A+ E+ + I +T S +FYPYFKDC+GA+DGTH
Sbjct: 648 FWRSGETVSRYFRIVLRAMCEIARELIYIRSTNTHSKITSKKNKFYPYFKDCIGALDGTH 707
Query: 126 IRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDK 185
IR V AK R+RGRK YPTQNVLAA FDL+F Y+LAGWEGSA DS ++++AL+R +
Sbjct: 708 IRASVPAKKVDRFRGRKPYPTQNVLAAVDFDLRFIYILAGWEGSAHDSLVLQDALSRPNG 767
Query: 186 LKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNP--------PLNYKELFNLRH 237
LKI EGK++L DAG+ G++ YR VRYHLKEF R P P KELFN RH
Sbjct: 768 LKILEGKFFLADAGYAARPGILPSYRRVRYHLKEF--RGPQGPHGPQGPKCPKELFNYRH 825
Query: 238 ASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNE 292
+SLR IERAFG LK RF+IL N +P +KAQ +++ ACC LHN+++ +E
Sbjct: 826 SSLRTTIERAFGALKNRFKILMN--KPFIPLKAQSMVVIACCALHNWILENGSDE 878
>UniRef100_Q5W6P9 Putative polyprotein [Oryza sativa]
Length = 1067
Score = 257 bits (656), Expect = 4e-67
Identities = 138/298 (46%), Positives = 193/298 (64%), Gaps = 17/298 (5%)
Query: 55 HNAKNREVNFWFRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYF 114
HN +NR V F RSGET+SR+ ++VL AI EL ++ I P T P +I+ + R+ PYF
Sbjct: 20 HNLRNRLVRTNFDRSGETVSRYFNKVLHAIGELRDELIRPPSLDT-PTKIAGNPRWDPYF 78
Query: 115 KDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSR 174
KDC+GAIDGTHIR V +RGRK + TQNV+AA FDL+FTYVLAGWEG+ D+
Sbjct: 79 KDCIGAIDGTHIRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTTHDAV 138
Query: 175 IIKNALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFN 234
++++AL RE+ L++P+GKYYLVDAG+ G + P+R VRYHL E+ NP N KELFN
Sbjct: 139 VLRDALERENSLRVPQGKYYLVDAGYGAKQGFLPPFRAVRYHLNEW-GNNPVQNEKELFN 197
Query: 235 LRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDL 294
LRH+SLR +ERAFG LK+RF++L ++T P + + Q I+ ACCI+HN++++ + ++L
Sbjct: 198 LRHSSLRVTVERAFGSLKRRFKVLDDAT-PFFPFRTQVDIVVACCIIHNWVVN-DGIDEL 255
Query: 295 IAEVDAELANQNVSHDNHEASRSDMDEFAQG-------------GIIKNGAAHQMWSN 339
IA D + + S + M +F QG GI K G+ H W++
Sbjct: 256 IAPPDWSSEDIDESLTGQANDHALMVQFRQGLADQMWADRNSHHGIGKGGSTHASWTS 313
>UniRef100_Q5W652 Hypothetical protein OSJNBb0052F16.10 [Oryza sativa]
Length = 1220
Score = 257 bits (656), Expect = 4e-67
Identities = 138/298 (46%), Positives = 193/298 (64%), Gaps = 17/298 (5%)
Query: 55 HNAKNREVNFWFRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYF 114
HN +NR V F RSGET+SR+ ++VL AI EL ++ I P T P +I+ + R+ PYF
Sbjct: 173 HNLRNRLVRTNFDRSGETVSRYFNKVLHAIGELRDELIRPPSLDT-PTKIAGNPRWDPYF 231
Query: 115 KDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSR 174
KDC+GAIDGTHIR V +RGRK + TQNV+AA FDL+FTYVLAGWEG+ D+
Sbjct: 232 KDCIGAIDGTHIRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTTHDAV 291
Query: 175 IIKNALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFN 234
++++AL RE+ L++P+GKYYLVDAG+ G + P+R VRYHL E+ NP N KELFN
Sbjct: 292 VLRDALERENSLRVPQGKYYLVDAGYGAKQGFLPPFRAVRYHLNEW-GNNPVQNEKELFN 350
Query: 235 LRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDL 294
LRH+SLR +ERAFG LK+RF++L ++T P + + Q I+ ACCI+HN++++ + ++L
Sbjct: 351 LRHSSLRVTVERAFGSLKRRFKVLDDAT-PFFPFRTQVDIVVACCIIHNWVVN-DGIDEL 408
Query: 295 IAEVDAELANQNVSHDNHEASRSDMDEFAQG-------------GIIKNGAAHQMWSN 339
IA D + + S + M +F QG GI K G+ H W++
Sbjct: 409 IAPPDWSSEDIDESLTGQANDHALMVQFRQGLADQMWADRNSHHGIGKGGSTHASWTS 466
>UniRef100_Q5W6B8 Putative polyprotein [Oryza sativa]
Length = 561
Score = 253 bits (647), Expect = 4e-66
Identities = 130/268 (48%), Positives = 182/268 (67%), Gaps = 3/268 (1%)
Query: 6 NSETSRNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFW 65
N T +++R+ +F + C + G L T +EEQVA L+ + HN +NR V
Sbjct: 19 NDTTCIDMLRLRKGSFFRFCKLFRDRGLLEDTIHMCIEEQVAMFLHTVGHNLRNRLVRTN 78
Query: 66 FRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTH 125
+ RSGET+SR+ ++VL AI EL ++ I P T P +I+ + R+ PYFKDC+GAIDGTH
Sbjct: 79 YSRSGETVSRYFNKVLHAIGELRDELIRPPSLDT-PTKIAGNPRWDPYFKDCIGAIDGTH 137
Query: 126 IRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDK 185
IR V +RGRK + TQNV+AA FDL+FTYVLAGWEG+A D+ ++++AL RE+
Sbjct: 138 IRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTAHDAVVLRDALERENG 197
Query: 186 LKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIE 245
L +P+GK+YLVDAG+ G + P+R VRYHLKE+ NP N KELFNLRH+SLR +E
Sbjct: 198 LHVPQGKFYLVDAGYGAKQGFLPPFRAVRYHLKEW-GNNPVQNEKELFNLRHSSLRITVE 256
Query: 246 RAFGVLKKRFEILSNSTEPAYGIKAQKL 273
RAFG LK+RF++L ++T P + + Q++
Sbjct: 257 RAFGSLKRRFKVLDDAT-PFFPFRTQEM 283
>UniRef100_Q8S819 Putative transposase [Oryza sativa]
Length = 1003
Score = 234 bits (598), Expect = 2e-60
Identities = 127/279 (45%), Positives = 169/279 (60%), Gaps = 5/279 (1%)
Query: 15 RMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGETIS 74
RM P F + L E LR TR +VEE++ LY+++HNA ++ F SGETI
Sbjct: 329 RMEPNIFRAIVTYLRTEHLLRDTRGITVEEKLGHFLYMISHNASYEDLQHEFHHSGETIH 388
Query: 75 RHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVKVSAKD 134
RH+ V K I L +FI Q +IS+ F+PYF++C+GAIDGTH+ + +S
Sbjct: 389 RHIKAVFKVIPSLTYRFIKQTTRVETHWKISTDQLFFPYFQNCLGAIDGTHVPITISQDL 448
Query: 135 APRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDKLKIPEGKYY 194
YR RK +QNV+ C FDL F ++ +GWEGSA+D+R++++A+ + +P+GKYY
Sbjct: 449 QAPYRNRKGTLSQNVMLVCDFDLNFLFIPSGWEGSATDARVLRSAMLK--GFNVPQGKYY 506
Query: 195 LVDAGFMLTSGLITPYRGVRYHLKEFS-ARNPPLNYKELFNLRHASLRNAIERAFGVLKK 253
LVD G+ T + PYRGVRYHLKEF + P NYKELFN RHA LRN IERA GVLKK
Sbjct: 507 LVDGGYANTPSFLAPYRGVRYHLKEFGRGQQRPRNYKELFNHRHAILRNHIERAIGVLKK 566
Query: 254 RFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNE 292
RF IL T + IK Q I A + HN + +E
Sbjct: 567 RFPILKVGTH--HRIKNQVKIPVATVVFHNLIRMLNGDE 603
>UniRef100_Q948B6 Putative transposase [Oryza sativa]
Length = 435
Score = 234 bits (597), Expect = 3e-60
Identities = 129/317 (40%), Positives = 191/317 (59%), Gaps = 16/317 (5%)
Query: 11 RNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSG 70
R RM P+ F+ L + L E + TR VEE++A LY+L+HNA ++ F SG
Sbjct: 65 RVAFRMEPEIFISLANYLRTEKLVDDTR-IKVEEKLAFFLYMLSHNASFEDLQEKFGHSG 123
Query: 71 ETISRHLHQVLKAIL-ELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVK 129
++ RH+ +++ L ++F+ P+ + + +I RFYPYFK+C+GAIDGTHI +
Sbjct: 124 DSFHRHVKHFFNSVVPSLSKRFLKPPNPNEVHWKIEKDPRFYPYFKNCLGAIDGTHIPIS 183
Query: 130 VSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDKLKIP 189
+S+ A +R RK +QNV+ AC FDLK T++ GWEGSA+D+R++ +A+ + ++P
Sbjct: 184 ISSDKAAPFRNRKNTLSQNVMIACDFDLKITFMSTGWEGSATDARVLTSAVNK--GFQVP 241
Query: 190 EGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSA-RNPPLNYKELFNLRHASLRNAIERAF 248
GK+YLVD G+ T+ + PYR VRYHLKE+ A R P NYKELFN RHA LRN +ER
Sbjct: 242 PGKFYLVDGGYANTNSFLAPYRKVRYHLKEYGAGRRRPQNYKELFNHRHAVLRNHVERTL 301
Query: 249 GVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDAELANQNVS 308
GV+KKRF IL +T + IK Q I A + HN ++S +E + N
Sbjct: 302 GVVKKRFPILKVAT--FHKIKNQVKIPVAAAVFHNIILSLNGDEQWL---------NNQP 350
Query: 309 HDNHEASRSDMDEFAQG 325
H+ H ++ D+ + +G
Sbjct: 351 HNIHPSNYVDLPDGDEG 367
>UniRef100_Q9FHY5 Emb|CAB72466.1 [Arabidopsis thaliana]
Length = 374
Score = 228 bits (581), Expect = 2e-58
Identities = 134/344 (38%), Positives = 187/344 (53%), Gaps = 29/344 (8%)
Query: 15 RMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGETIS 74
RM F KLCD+L+ G LR T +E Q+A L+I+ HN + R V F SGETIS
Sbjct: 47 RMDKPVFYKLCDLLQTRGLLRHTNRIKIEAQLAIFLFIIGHNLRTRAVQELFCYSGETIS 106
Query: 75 RHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVKVSAKD 134
RH + VL A++ + + F QP+ ++ LE PYFKDCVG +D HI V V +
Sbjct: 107 RHFNNVLNAVIAISKDFF-QPNSNSDTLENDD-----PYFKDCVGVVDSFHIPVMVGVDE 160
Query: 135 APRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDKLKIPEGKYY 194
+R TQNVLAA +FDL+F YVLAGWEGSASD +++ ALTR +KL++P+GKYY
Sbjct: 161 QGPFRNGNGLLTQNVLAASSFDLRFNYVLAGWEGSASDQQVLNAALTRRNKLQVPQGKYY 220
Query: 195 LVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGVLKKR 254
+VD + G I PY GV + +E KE+FN RH L AI R FG LK+R
Sbjct: 221 IVDNKYPNLPGFIAPYHGVSTNSRE--------EAKEMFNERHKLLHRAIHRTFGALKER 272
Query: 255 FEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDAELANQNVSHDNHEA 314
F IL ++ P Y ++ Q ++ A C LHNY+ +P +DL+ + E D A
Sbjct: 273 FPILLSA--PPYPLQTQVKLVIAACALHNYVRLEKP-DDLVFRMFEEETLAEAGEDREVA 329
Query: 315 SRSDM------------DEFAQGGIIKNGAAHQMWSNYKNNGQT 346
+ +E +++ A ++W++Y N T
Sbjct: 330 LEEEQVEIVGQEHGFRPEEVEDSLRLRDEIASELWNHYVQNMST 373
>UniRef100_Q7XV92 OSJNBb0012E08.4 protein [Oryza sativa]
Length = 261
Score = 225 bits (573), Expect = 2e-57
Identities = 115/237 (48%), Positives = 159/237 (66%), Gaps = 2/237 (0%)
Query: 5 KNSETSRNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNF 64
+N T +++R+ +F + C + G L T +EEQVA L+ + HN +NR V
Sbjct: 18 RNDTTCIDMLRLRRGSFFRFCKLFRDCGLLEDTIHMCIEEQVAMFLHTVGHNLRNRLVRT 77
Query: 65 WFRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGT 124
+ RSGET+SR+ ++VL AI EL ++ I P T P +I+ + R+ PYFKDC+GAIDGT
Sbjct: 78 NYDRSGETVSRYFNKVLHAIGELRDELIRPPSLDT-PTKIAGNPRWDPYFKDCIGAIDGT 136
Query: 125 HIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTRED 184
HIR + +RGRK + TQNV+AA FDL+FTYVLAGWEG+ D+ ++++AL RE+
Sbjct: 137 HIRASIRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTTHDAVVLRDALEREN 196
Query: 185 KLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLR 241
L +P+GK+YLVDAG+ G + P+R VRYHLKE+ NP N KELFNLRH+SLR
Sbjct: 197 GLHVPQGKFYLVDAGYGAKQGFLPPFRAVRYHLKEW-GNNPVQNEKELFNLRHSSLR 252
>UniRef100_Q9M1P0 Hypothetical protein T18B22.40 [Arabidopsis thaliana]
Length = 377
Score = 223 bits (569), Expect = 5e-57
Identities = 126/336 (37%), Positives = 184/336 (54%), Gaps = 45/336 (13%)
Query: 11 RNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSG 70
R + RM P+ F KLCD+ IL +N E+ +++
Sbjct: 72 RQLYRMEPEVFHKLCDV------------------------ILLYNGHISEIKLFYKHK- 106
Query: 71 ETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVKV 130
H+ LKA+ + ++ +P+ +P +I ST FYPYFKDC+GAIDGTHI V
Sbjct: 107 ------FHKALKALAFVAPNWMAKPE-VRVPAKIRESTSFYPYFKDCIGAIDGTHIFAMV 159
Query: 131 SAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTRE-DKLKIP 189
DA +R RK + +QNVLAAC FDL+F Y+L+GW+GSA DS+++ +ALTR ++L++P
Sbjct: 160 PTCDAASFRNRKGFISQNVLAACNFDLQFIYILSGWKGSAHDSKVLNDALTRNTNRLQVP 219
Query: 190 EGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSAR-NPPLNYKELFNLRHASLRNAIERAF 248
EGK+YLVD G+ + P+R RYHL++F + P ELFNLRHASLRN IER F
Sbjct: 220 EGKFYLVDCGYANRRKFLAPFRRTRYHLQDFRGQGRDPKTQNELFNLRHASLRNVIERIF 279
Query: 249 GVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDAELANQNVS 308
G+ K RF I ++ P Y K Q ++ AC LHN+L +++ E N + +
Sbjct: 280 GIFKSRFLIFKSA--PPYPFKTQTELVLACAGLHNFLHQECRSDEFPPETSINEENSDDN 337
Query: 309 HDNHEASRSDMD-------EFAQGGIIKNGAAHQMW 337
+N+ D+ E+A ++ +H MW
Sbjct: 338 EENNYEENGDVGILESQQREYANNW--RDTISHNMW 371
>UniRef100_Q7XPC6 OSJNBa0042N22.14 protein [Oryza sativa]
Length = 401
Score = 223 bits (567), Expect = 8e-57
Identities = 112/219 (51%), Positives = 157/219 (71%), Gaps = 2/219 (0%)
Query: 44 EQVAKSLYILTHNAKNREVNFWFRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIPLE 103
EQVA L+ HN +NR V F RSGETISR+ + VL A+ EL ++ +++P T P +
Sbjct: 45 EQVAMFLHTFGHNVRNRVVATNFYRSGETISRYFNLVLHAVGELRKE-LIRPPSITTPSK 103
Query: 104 ISSSTRFYPYFKDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVL 163
I + + PYFKDC+GAIDGTH+RV V+ +RGRK++ TQNV+AA FDLKFTYVL
Sbjct: 104 ILGNPMWDPYFKDCIGAIDGTHVRVSVTKDMELSFRGRKDHATQNVMAAIDFDLKFTYVL 163
Query: 164 AGWEGSASDSRIIKNALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSAR 223
AGWE +A D+ ++++++ R D L++P+GK+YLVD G+ G + P+RGVRYHL E+ +
Sbjct: 164 AGWETTAHDAVVLRDSIERTDGLRVPQGKFYLVDVGYGAKPGFLPPFRGVRYHLNEWGS- 222
Query: 224 NPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNST 262
NP N KELFNLRH+SLR + RAF LK+R++IL +++
Sbjct: 223 NPVQNEKELFNLRHSSLRVTVARAFRSLKRRWKILDDAS 261
>UniRef100_Q8L516 Putative transposase [Oryza sativa]
Length = 535
Score = 212 bits (539), Expect = 1e-53
Identities = 120/300 (40%), Positives = 176/300 (58%), Gaps = 52/300 (17%)
Query: 42 VEEQVAKSLYILTHNAKNREVNFWFRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIP 101
+EEQVA L+ + HN +NR V + RSGET
Sbjct: 24 IEEQVAMFLHTVGHNLRNRLVRTNYGRSGET----------------------------- 54
Query: 102 LEISSSTRFYPYFKDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTY 161
DC+GAIDGTHIR V +RGRK + TQNV+AA FDL+FTY
Sbjct: 55 --------------DCIGAIDGTHIRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTY 100
Query: 162 VLAGWEGSASDSRIIKNALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFS 221
VLAGWEG+A D+ ++++AL RE+ L +P+GK+YLVD G+ G + P+R VRYHLKE+
Sbjct: 101 VLAGWEGTAHDAVVLRDALERENGLHVPQGKFYLVDGGYGAKQGFLPPFRAVRYHLKEW- 159
Query: 222 ARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCIL 281
NP N KELFNLRH+SLR +ERAFG LK+RF++L ++T P + + Q I+ ACCI+
Sbjct: 160 GNNPVQNEKELFNLRHSSLRITVERAFGSLKRRFKVLDDAT-PFFPFRTQVDIVVACCII 218
Query: 282 HNYLMSAEPNEDLIAEVD--AELANQNVSHDNHEASRSDMDEFAQGGIIKNGAAHQMWSN 339
HN++++ + ++L+A D +E +++ + ++ + +MD + K G+ H W++
Sbjct: 219 HNWVIN-DGIDELVAPSDWSSEDIDESPTWQANDHALMEMDNNSG----KGGSTHASWTS 273
>UniRef100_Q84R65 Hypothetical protein OSJNBb0113I20.11 [Oryza sativa]
Length = 411
Score = 203 bits (517), Expect = 5e-51
Identities = 118/317 (37%), Positives = 169/317 (53%), Gaps = 16/317 (5%)
Query: 13 IIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGET 72
++RM TFL L D+L + L PT E +A LYIL + N+ F+ SGET
Sbjct: 75 MLRMNGNTFLALHDLLVDKYKLAPTVHMHTMEALAIFLYILGDGSSNQRAQNCFKHSGET 134
Query: 73 ISRHLHQVLKAILELEEKFI--VQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVKV 130
ISR + +VL A++EL + P+ S + I R +P+FKDC+GA+DGTHI V
Sbjct: 135 ISRKIEEVLFAVVELGRDIVRPKDPNFSNVHERIRKDRRMWPHFKDCIGAVDGTHILAVV 194
Query: 131 SAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDKL--KI 188
D RY GR + TQNV+A C D++FTY G GS D+ ++ NAL + +
Sbjct: 195 PDDDKIRYIGRSKSTTQNVMAICDHDMRFTYASIGQPGSMHDTTVLFNALRTDIDIFPHP 254
Query: 189 PEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAF 248
P+GKYYLVDAG+ G + PY+G RYH+ EF + P KE FN H S+R IER F
Sbjct: 255 PQGKYYLVDAGYPNRPGYLAPYKGQRYHVPEFRRGSAPSGVKEKFNFLHCSVRTIIERCF 314
Query: 249 GVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMS----------AEPNEDLIAEV 298
GV K ++ +L P++ + QK+++ A LHN++ E N D + +
Sbjct: 315 GVWKMKWRVLLKM--PSFPLWKQKMVVAATMALHNFIRDHNAPDRHFHRFERNPDYVPTI 372
Query: 299 DAELANQNVSHDNHEAS 315
+ +S D + S
Sbjct: 373 PSRFRRYVISQDASDTS 389
>UniRef100_Q945K3 Putative transposase [Zea mays]
Length = 298
Score = 201 bits (512), Expect = 2e-50
Identities = 107/232 (46%), Positives = 147/232 (63%), Gaps = 5/232 (2%)
Query: 14 IRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGETI 73
+R+ ++F LC +L + T SVEE+VA L ++ H K R + + S E I
Sbjct: 69 LRLTKRSFSDLCTILRERCDMCDTLNVSVEEKVAIFLLVVGHGTKMRMIRSSYGWSLEPI 128
Query: 74 SRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVKVSAK 133
SR+ ++VL+ +L L +FI PD PL + + +F+DC+GA+DGTHI V V
Sbjct: 129 SRYFNEVLRGVLSLCHEFIKLPD----PLAVQPEDSKWRWFEDCLGALDGTHIDVFVPLA 184
Query: 134 DAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDKLKIPEGKY 193
D RYR RK+ T NVL C +KF YVLAGWEGSASDSR++++A++R+D IP GKY
Sbjct: 185 DQGRYRNRKQQITTNVLGVCDRHMKFVYVLAGWEGSASDSRVLRDAMSRDDAFAIPSGKY 244
Query: 194 YLVDAGFMLTSGLITPYRGVRYHLKEFSAR-NPPLNYKELFNLRHASLRNAI 244
YLVDAG+ G + PYR RYHL E++A+ N P N KELFNLRH++ RN +
Sbjct: 245 YLVDAGYTNGPGFLAPYRSTRYHLNEWAAQGNNPSNAKELFNLRHSTARNYV 296
>UniRef100_UPI000043028E UPI000043028E UniRef100 entry
Length = 427
Score = 201 bits (510), Expect = 3e-50
Identities = 123/329 (37%), Positives = 178/329 (53%), Gaps = 19/329 (5%)
Query: 2 YRLKNSETSRNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNRE 61
Y + R+ + +G + F+ L E G + ++ E +A +L L
Sbjct: 74 YLEHRGRSMRHHLGVGLEVFMLLLREFEEHGIQLASHDATAAEHLAIALAYLRKKTSVNW 133
Query: 62 VNFWFRRSGETISRHLHQVLKAILE--LEEKFIVQPDGSTIPLE--ISSSTRFYPYFKDC 117
+ F++S E IS +H V+ A+ + ++++ P G P Y +F
Sbjct: 134 IGEEFKKSNEFISFCIHDVVNALTSKVIYDRWVKYPTGREPPNAKVADDPLNRYRHFHGA 193
Query: 118 VGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIK 177
VG+IDGTHI VKVS + P YR RK + NVLAAC FDLKFTY++AGWEGSA+DS
Sbjct: 194 VGSIDGTHIAVKVSPRMQPAYRNRKGVVSINVLAACDFDLKFTYIIAGWEGSANDSFTFN 253
Query: 178 NALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEF-SARNPPLNYKELFNLR 236
A+ R + ++P +Y+L DAGF + L+TPYRG RYHL +F +R P + KE+FNL
Sbjct: 254 QAVHRYNFPQLPSNRYFLADAGFPICDQLLTPYRGTRYHLADFHPSRGAPQSEKEVFNLA 313
Query: 237 HASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIA 296
HA LRN++ER FG+ K+R+ +LS E G Q II ACC+LHN +S +D +
Sbjct: 314 HAQLRNSVERIFGITKQRWGVLSGGLEKFKG-GMQSQIILACCVLHNMCISI---KDRVP 369
Query: 297 EVDAELANQNVSHDNHEASRSDMDEFAQG 325
E D A++ DM+E +G
Sbjct: 370 ERDVGEADEG----------GDMEEEVEG 388
>UniRef100_UPI000042EF69 UPI000042EF69 UniRef100 entry
Length = 427
Score = 201 bits (510), Expect = 3e-50
Identities = 123/329 (37%), Positives = 178/329 (53%), Gaps = 19/329 (5%)
Query: 2 YRLKNSETSRNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNRE 61
Y + R+ + +G + F+ L E G + ++ E +A +L L
Sbjct: 74 YLEHRGRSMRHHLGVGLEVFMLLLREFEEHGIQLASHDATAAEHLAIALAYLRKKTSVNW 133
Query: 62 VNFWFRRSGETISRHLHQVLKAILE--LEEKFIVQPDGSTIPLE--ISSSTRFYPYFKDC 117
+ F++S E IS +H V+ A+ + ++++ P G P Y +F
Sbjct: 134 IGEEFKKSNEFISFCIHDVVNALTSKVIYDRWVKYPTGREPPNAKVADDPLNRYRHFHGA 193
Query: 118 VGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIK 177
VG+IDGTHI VKVS + P YR RK + NVLAAC FDLKFTY++AGWEGSA+DS
Sbjct: 194 VGSIDGTHIAVKVSPRMQPAYRNRKGVVSINVLAACDFDLKFTYIIAGWEGSANDSFTFN 253
Query: 178 NALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEF-SARNPPLNYKELFNLR 236
A+ R + ++P +Y+L DAGF + L+TPYRG RYHL +F +R P + KE+FNL
Sbjct: 254 QAVHRYNFPQLPSNRYFLADAGFPICDQLLTPYRGTRYHLADFHPSRGAPQSEKEVFNLA 313
Query: 237 HASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIA 296
HA LRN++ER FG+ K+R+ +LS E G Q II ACC+LHN +S +D +
Sbjct: 314 HAQLRNSVERIFGITKQRWGVLSGGLEKFKG-GMQSQIILACCVLHNMCISI---KDRVP 369
Query: 297 EVDAELANQNVSHDNHEASRSDMDEFAQG 325
E D A++ DM+E +G
Sbjct: 370 ERDVGEADEG----------GDMEEEVEG 388
>UniRef100_Q75HL4 Expressed protein [Oryza sativa]
Length = 608
Score = 199 bits (506), Expect = 9e-50
Identities = 104/231 (45%), Positives = 151/231 (65%), Gaps = 13/231 (5%)
Query: 115 KDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSR 174
+DC+GAIDGTH+R V +RGRK + TQNV+AA FDL+FTYVLAGWEG+A D
Sbjct: 138 RDCIGAIDGTHVRASVPKNMESSFRGRKNHATQNVMAAVDFDLRFTYVLAGWEGAAHDVV 197
Query: 175 IIKNALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFN 234
++++AL RE+ L +P+GK+YLVD G+ G + P+R RYHL E+ NP N KELFN
Sbjct: 198 VLRDALERENGLHVPQGKFYLVDVGYGAKPGFLPPFRSTRYHLNEW-GNNPVQNEKELFN 256
Query: 235 LRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDL 294
LRH+SLR +ERAFG LK+RF+IL ++T P + + Q I+ ACCI+HN++++ + +E +
Sbjct: 257 LRHSSLRITVERAFGSLKRRFKILDDAT-PFFPFQTQVNIVVACCIIHNWVINDDIDELV 315
Query: 295 IAEVDAELANQNVSHDNHEASRSDMDEFAQGGIIKNGAAHQ---MWSNYKN 342
+ N+++ DN + +DMD G++ + + M SN N
Sbjct: 316 MG------PNESIIQDND--TSTDMDTNNGKGVVASWTSSMSSLMLSNLAN 358
>UniRef100_Q9SY99 T25B24.14 protein [Arabidopsis thaliana]
Length = 404
Score = 194 bits (493), Expect = 3e-48
Identities = 107/284 (37%), Positives = 159/284 (55%), Gaps = 4/284 (1%)
Query: 12 NIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGE 71
+I+RM TF LC +L + L + +EE VA + ++ + R + ++ S E
Sbjct: 70 DILRMHQSTFRTLCKILSEQYKLEESCNIYLEESVAMFIEMVAQDLTVRVIAERYQHSLE 129
Query: 72 TISRHLHQVLKAILELEEKFI--VQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVK 129
T+ R L +VL A+L+L + + D + I + + R+ PYF DC+GA+DGTH+ V+
Sbjct: 130 TVKRKLDEVLSALLKLAADIVKPTRDDVTGISPFLVNDKRYMPYFIDCIGALDGTHVSVR 189
Query: 130 VSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDKLKIP 189
+ D RYRGRK T N+LA C F +KF G G A D++++ T E P
Sbjct: 190 PPSGDVERYRGRKSEATMNILAVCNFSMKFNIAYVGVPGRAHDTKVLTYCATHEASFPHP 249
Query: 190 E-GKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAF 248
GKYYLVD+G+ SG + P+R RYHL+ F+ PP N +ELFN RH+SLR+ IER F
Sbjct: 250 PAGKYYLVDSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLRSVIERTF 309
Query: 249 GVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNE 292
GV K ++ IL P Y +K I+ + LHNY+ ++ +
Sbjct: 310 GVWKAKWRILDRK-HPKYEVKKWIKIVTSTMALHNYIRDSQQED 352
>UniRef100_Q8LNR6 Hypothetical protein OSJNBa0042E19.20 [Oryza sativa]
Length = 481
Score = 193 bits (491), Expect = 5e-48
Identities = 100/214 (46%), Positives = 135/214 (62%), Gaps = 5/214 (2%)
Query: 80 VLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVKVSAKDAPRYR 139
V I L +FI QP G +IS+ +F+PYF++C+GAIDGTH+ + +S YR
Sbjct: 3 VFNVIPSLTYRFIKQPTGVETHWKISTDQQFFPYFQNCLGAIDGTHVPITISQDLQAPYR 62
Query: 140 GRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDKLKIPEGKYYLVDAG 199
RK +QNV+ C FDL F+++ +GWEGSA+D+R++++A+ + +P+GKYYLVD G
Sbjct: 63 NRKGTLSQNVMLVCDFDLNFSFISSGWEGSATDARVLRSAMLK--GFNVPQGKYYLVDGG 120
Query: 200 FMLTSGLITPYRGVRYHLKEFS-ARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFEIL 258
+ T + PYRGVRYHLKEF + P NYKELFN RHA LRN IER GVLKKRF IL
Sbjct: 121 YANTPSFLAPYRGVRYHLKEFGCGQQRPRNYKELFNHRHAILRNHIERTIGVLKKRFPIL 180
Query: 259 SNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNE 292
T + I+ Q I A + HN + +E
Sbjct: 181 KVGTH--HSIENQVKIPVATVVFHNLIRMLNGDE 212
>UniRef100_Q7XKH6 OSJNBb0033P05.11 protein [Oryza sativa]
Length = 877
Score = 181 bits (458), Expect = 3e-44
Identities = 108/293 (36%), Positives = 161/293 (54%), Gaps = 7/293 (2%)
Query: 1 MYRLKNSETSRNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNR 60
M L N N+ RM F +L +L L+ T E + L+I+ R
Sbjct: 534 MRTLANRTACYNMFRMHRPVFERLHSVLVESYQLKSTNNMDSMECLGLFLWIVGAPQSVR 593
Query: 61 EVNFWFRRSGETISRHLHQVLKAILELEEKFIVQPDG--STIPLEISSSTRFYPYFKDCV 118
+ F RS +T+ + VL A+L+L + I D +T+ ++ S ++ PY +C+
Sbjct: 594 QAQDRFVRSLQTVHSKFNAVLTALLKLAQDIIRPKDPLFTTVHKKLLSP-QYTPYLDNCI 652
Query: 119 GAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKN 178
GAIDGTHI+V V A ++R R + +QNV+ C FD++FT+VLAGW GS D R+ +
Sbjct: 653 GAIDGTHIQVVVPNSAAVQHRNRHQEKSQNVMCVCDFDMRFTFVLAGWPGSVHDMRVFND 712
Query: 179 ALTREDKL--KIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLR 236
A TR K P GK+YLVD+G+ G + PY+G+ YH E++ P +E FN
Sbjct: 713 AQTRFSAKFPKPPLGKFYLVDSGYPNRPGYLAPYKGITYHFHEYNESTLPRGRREHFNYC 772
Query: 237 HASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAE 289
H+S RN IER+FGVLK ++ IL + P+Y + Q II AC LHN++ ++
Sbjct: 773 HSSCRNVIERSFGVLKNKWRILFSL--PSYSQEKQSRIIHACIALHNFIRDSQ 823
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 580,313,220
Number of Sequences: 2790947
Number of extensions: 23639808
Number of successful extensions: 52267
Number of sequences better than 10.0: 232
Number of HSP's better than 10.0 without gapping: 121
Number of HSP's successfully gapped in prelim test: 111
Number of HSP's that attempted gapping in prelim test: 51852
Number of HSP's gapped (non-prelim): 253
length of query: 346
length of database: 848,049,833
effective HSP length: 128
effective length of query: 218
effective length of database: 490,808,617
effective search space: 106996278506
effective search space used: 106996278506
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146752.6


