

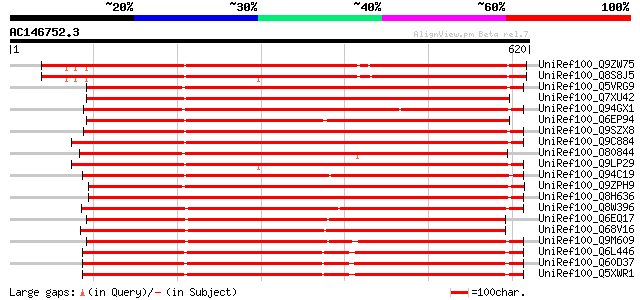
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.3 - phase: 0 /pseudo
(620 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ZW75 Hypothetical protein At2g43200 [Arabidopsis tha... 652 0.0
UniRef100_Q8S8J5 Hypothetical protein At2g43143 [Arabidopsis tha... 645 0.0
UniRef100_Q5VRG9 Hypothetical protein [Oryza sativa] 628 e-178
UniRef100_Q7XU42 OSJNBa0088I22.11 protein [Oryza sativa] 622 e-176
UniRef100_Q94GX1 Hypothetical protein OSJNBa0005K07.2 [Oryza sat... 617 e-175
UniRef100_Q6EP94 Dehydration-responsive family protein-like [Ory... 614 e-174
UniRef100_Q9SZX8 Hypothetical protein F7L13.20 [Arabidopsis thal... 608 e-172
UniRef100_Q9C884 Hypothetical protein T16O9.7 [Arabidopsis thali... 607 e-172
UniRef100_O80844 Hypothetical protein At2g45750 [Arabidopsis tha... 602 e-170
UniRef100_Q9LP29 T9L6.6 protein [Arabidopsis thaliana] 596 e-169
UniRef100_Q94C19 At1g26850/T2P11_4 [Arabidopsis thaliana] 593 e-168
UniRef100_Q9ZPH9 F15P23.1 protein [Arabidopsis thaliana] 591 e-167
UniRef100_Q8H636 Dehydration-responsive protein-like [Oryza sativa] 588 e-166
UniRef100_Q8W396 Hypothetical protein OSJNBa0013O08.23 [Oryza sa... 578 e-163
UniRef100_Q6EQ17 Dehydration-responsive protein-like [Oryza sativa] 578 e-163
UniRef100_Q68V16 Ankyrin protein kinase-like [Poa pratensis] 575 e-162
UniRef100_Q9M609 Hypothetical protein [Malus domestica] 570 e-161
UniRef100_Q6L446 Putative methyltransferase [Solanum demissum] 560 e-158
UniRef100_Q60D37 Putative methyltransferase [Solanum tuberosum] 560 e-158
UniRef100_Q5XWR1 Hypothetical protein [Solanum tuberosum] 560 e-158
>UniRef100_Q9ZW75 Hypothetical protein At2g43200 [Arabidopsis thaliana]
Length = 611
Score = 652 bits (1682), Expect = 0.0
Identities = 320/604 (52%), Positives = 416/604 (67%), Gaps = 33/604 (5%)
Query: 39 KRLILLHYHFNMFSKYLNIFFITTFFIF---------LYIITSSFFTS------------ 77
KRL L F +FS Y + I T I L++ + S + S
Sbjct: 14 KRLFLFFTPFLLFSLYYILTTIKTITISSQDRHHPPQLHVPSISHYYSLPETSENRSSPP 73
Query: 78 PLTLHSPITTKIS---HFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQ-NNERL 133
PL L P ++ S +F C N+TNY PC DP +++ + ++R+ERHCP E+
Sbjct: 74 PLLLPPPPSSSSSLSSYFPLCPKNFTNYLPCHDPSTARQYSIERHYRRERHCPDIAQEKF 133
Query: 134 TCLIPKPIGYKNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFP 193
CL+PKP GYK PFPWP+S+ AWF NVPF +L E KK+QNW+ L GDRFVFPGGGTSFP
Sbjct: 134 RCLVPKPTGYKTPFPWPESRKYAWFRNVPFKRLAELKKTQNWVRLEGDRFVFPGGGTSFP 193
Query: 194 DGVKGYVDDLKKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQV 253
GVK YVD + +LP L SG IRTVLD+GCGVASFGA L++Y ILTMSIAP D H+AQV
Sbjct: 194 GGVKDYVDVILSVLP--LASGSIRTVLDIGCGVASFGAFLLNYKILTMSIAPRDIHEAQV 251
Query: 254 MFALERGLPAMLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFW 313
FALERGLPAMLGV ST++L +PS+SFD+ HCSRCLV W + DGLYL E+DR+LRP G+W
Sbjct: 252 QFALERGLPAMLGVLSTYKLPYPSRSFDMVHCSRCLVNWTSYDGLYLMEVDRVLRPEGYW 311
Query: 314 VLSGPPINWRVNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKC 373
VLSGPP+ RV +K + + L+ + L ++ ++CWEK+AE + IW+KP NH++C
Sbjct: 312 VLSGPPVASRVKFKNQKRDSKELQNQMEKLNDVFRRLCWEKIAESYPVVIWRKPSNHLQC 371
Query: 374 MQKLNTLSSPKFCNSSDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPR 433
++L L P C+SSD DA WY +M CI PLP+V D ++ VL+ WP RLN PR
Sbjct: 372 RKRLKALKFPGLCSSSDPDAAWYKEMEPCITPLPDVNDTNKT---VLKNWPERLN-HVPR 427
Query: 434 LRKENHDVFSLKTYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKY 493
++ + ++ + D +W++RV YY+ K LS+GKYRNV+DMNAG GGFAAAL+KY
Sbjct: 428 MKTGSIQGTTIAGFKADTNLWQRRVLYYDTKFKFLSNGKYRNVIDMNAGLGGFAAALIKY 487
Query: 494 PVWVMNVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCD 553
P+WVMNVVPFD K N LG++Y+RGLIGTYM+WCE STYPRTYDLIHA +FS+Y+DKCD
Sbjct: 488 PMWVMNVVPFDLKPNTLGVVYDRGLIGTYMNWCEALSTYPRTYDLIHANGVFSLYLDKCD 547
Query: 554 ITDIVIEMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIM 613
I DI++EM RILRPEG VIIRD DV++KVK IT++MRW G + +D + H +++
Sbjct: 548 IVDILLEMQRILRPEGAVIIRDRFDVLVKVKAITNQMRWNG--TMYPEDNSVFDHGTILI 605
Query: 614 VLNN 617
V N+
Sbjct: 606 VDNS 609
>UniRef100_Q8S8J5 Hypothetical protein At2g43143 [Arabidopsis thaliana]
Length = 617
Score = 645 bits (1665), Expect = 0.0
Identities = 320/610 (52%), Positives = 416/610 (67%), Gaps = 39/610 (6%)
Query: 39 KRLILLHYHFNMFSKYLNIFFITTFFIF---------LYIITSSFFTS------------ 77
KRL L F +FS Y + I T I L++ + S + S
Sbjct: 14 KRLFLFFTPFLLFSLYYILTTIKTITISSQDRHHPPQLHVPSISHYYSLPETSENRSSPP 73
Query: 78 PLTLHSPITTKIS---HFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQ-NNERL 133
PL L P ++ S +F C N+TNY PC DP +++ + ++R+ERHCP E+
Sbjct: 74 PLLLPPPPSSSSSLSSYFPLCPKNFTNYLPCHDPSTARQYSIERHYRRERHCPDIAQEKF 133
Query: 134 TCLIPKPIGYKNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFP 193
CL+PKP GYK PFPWP+S+ AWF NVPF +L E KK+QNW+ L GDRFVFPGGGTSFP
Sbjct: 134 RCLVPKPTGYKTPFPWPESRKYAWFRNVPFKRLAELKKTQNWVRLEGDRFVFPGGGTSFP 193
Query: 194 DGVKGYVDDLKKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQV 253
GVK YVD + +LP L SG IRTVLD+GCGVASFGA L++Y ILTMSIAP D H+AQV
Sbjct: 194 GGVKDYVDVILSVLP--LASGSIRTVLDIGCGVASFGAFLLNYKILTMSIAPRDIHEAQV 251
Query: 254 MFALERGLPAMLGVFSTHRLTFPSKSFDVAHCSRCLVPWIAN------DGLYLREIDRIL 307
FALERGLPAMLGV ST++L +PS+SFD+ HCSRCLV W + DGLYL E+DR+L
Sbjct: 252 QFALERGLPAMLGVLSTYKLPYPSRSFDMVHCSRCLVNWTSYERTFYPDGLYLMEVDRVL 311
Query: 308 RPGGFWVLSGPPINWRVNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKP 367
RP G+WVLSGPP+ RV +K + + L+ + L ++ ++CWEK+AE + IW+KP
Sbjct: 312 RPEGYWVLSGPPVASRVKFKNQKRDSKELQNQMEKLNDVFRRLCWEKIAESYPVVIWRKP 371
Query: 368 INHIKCMQKLNTLSSPKFCNSSDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRL 427
NH++C ++L L P C+SSD DA WY +M CI PLP+V D ++ VL+ WP RL
Sbjct: 372 SNHLQCRKRLKALKFPGLCSSSDPDAAWYKEMEPCITPLPDVNDTNKT---VLKNWPERL 428
Query: 428 NDSPPRLRKENHDVFSLKTYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFA 487
N P R++ + ++ + D +W++RV YY+ K LS+GKYRNV+DMNAG GGFA
Sbjct: 429 NHVP-RMKTGSIQGTTIAGFKADTNLWQRRVLYYDTKFKFLSNGKYRNVIDMNAGLGGFA 487
Query: 488 AALVKYPVWVMNVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSM 547
AAL+KYP+WVMNVVPFD K N LG++Y+RGLIGTYM+WCE STYPRTYDLIHA +FS+
Sbjct: 488 AALIKYPMWVMNVVPFDLKPNTLGVVYDRGLIGTYMNWCEALSTYPRTYDLIHANGVFSL 547
Query: 548 YIDKCDITDIVIEMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESS 607
Y+DKCDI DI++EM RILRPEG VIIRD DV++KVK IT++MRW G + +D +
Sbjct: 548 YLDKCDIVDILLEMQRILRPEGAVIIRDRFDVLVKVKAITNQMRWNG--TMYPEDNSVFD 605
Query: 608 HPEMIMVLNN 617
H +++V N+
Sbjct: 606 HGTILIVDNS 615
>UniRef100_Q5VRG9 Hypothetical protein [Oryza sativa]
Length = 631
Score = 628 bits (1619), Expect = e-178
Identities = 282/523 (53%), Positives = 373/523 (70%), Gaps = 5/523 (0%)
Query: 92 FQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPK 151
F C Y+ Y PC+DP+R +KFPK +ERHCP+ E CLIP P YKNPF WP+
Sbjct: 100 FPPCQLKYSEYTPCQDPRRARKFPKTMMQYRERHCPRKEELFRCLIPAPPKYKNPFKWPQ 159
Query: 152 SKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNL 211
+D AW+ N+P +L K QNWI + G RF FPGGGT FP G Y+DD+ L ++L
Sbjct: 160 CRDFAWYDNIPHRELSIEKAVQNWIQVEGKRFRFPGGGTMFPHGADAYIDDINAL--ISL 217
Query: 212 DSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTH 271
G IRT LD GCGVAS+GA L+ +I+TMS AP D H+AQV FALERG+PAM+GV ST
Sbjct: 218 TDGNIRTALDTGCGVASWGAYLIKRNIITMSFAPRDSHEAQVQFALERGVPAMIGVISTE 277
Query: 272 RLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQT 331
R+ +P++SFD+AHCSRCL+PW DG+YL E+DR++RPGG+W+LSGPPI+W+ +K W+
Sbjct: 278 RIPYPARSFDMAHCSRCLIPWNKFDGIYLIEVDRVIRPGGYWILSGPPIHWKKYFKGWER 337
Query: 332 EPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSSDS 391
L++EQ+ +E+LA ++CW+KV E +AIWQKPINHI+C+ +P+ C S+D
Sbjct: 338 TEEDLKQEQDEIEDLAKRLCWKKVVEKDDLAIWQKPINHIECVNSRKIYETPQICKSNDV 397
Query: 392 DAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTYSEDN 451
D+ WY KM CI PLP+V DE+AGG LEKWP R PPR+ + + + + + EDN
Sbjct: 398 DSAWYKKMETCISPLPDVNSEDEVAGGALEKWPKRAFAVPPRISRGSVSGLTTEKFQEDN 457
Query: 452 MIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSNNLG 511
+W +R YY+ ++ L+ G+YRNVMDMNAG GGFAAAL+KYP+WVMNVVP + + LG
Sbjct: 458 KVWAERADYYKKLIPPLTKGRYRNVMDMNAGMGGFAAALMKYPLWVMNVVPSGSAHDTLG 517
Query: 512 IIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPEGTV 571
IIYERG IGTY DWCE FSTYPRTYD IHA +FS Y D+CD+T I++EM RILRPEGTV
Sbjct: 518 IIYERGFIGTYQDWCEAFSTYPRTYDFIHADKIFSFYQDRCDVTYILLEMDRILRPEGTV 577
Query: 572 IIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
I RD+ +V++K++ IT+ MRW+ + D ++ +PE I+V
Sbjct: 578 IFRDTVEVLVKIQSITEGMRWKS---QIMDHESGPFNPEKILV 617
>UniRef100_Q7XU42 OSJNBa0088I22.11 protein [Oryza sativa]
Length = 646
Score = 622 bits (1603), Expect = e-176
Identities = 285/509 (55%), Positives = 365/509 (70%), Gaps = 5/509 (0%)
Query: 92 FQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPK 151
++ C Y+ Y PCED +R +FP+ +ERHCP ERL CL+P P GY+NPFPWP
Sbjct: 113 YEACPAKYSEYTPCEDVERSLRFPRDRLVYRERHCPSEGERLRCLVPAPQGYRNPFPWPT 172
Query: 152 SKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNL 211
S+D AWF+NVP +L K QNWI + G++F FPGGGT FP G Y+DD+ K++P L
Sbjct: 173 SRDVAWFANVPHKELTVEKAVQNWIRVEGEKFRFPGGGTMFPHGAGAYIDDIGKIIP--L 230
Query: 212 DSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTH 271
G IRT LD GCGVAS+GA L+ +IL MS AP D H+AQV FALERG+PAM+GV S++
Sbjct: 231 HDGSIRTALDTGCGVASWGAYLLSRNILAMSFAPRDSHEAQVQFALERGVPAMIGVLSSN 290
Query: 272 RLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQT 331
RLT+P+++FD+AHCSRCL+PW DGLYL E+DRILRPGG+W+LSGPPINW+ ++K WQ
Sbjct: 291 RLTYPARAFDMAHCSRCLIPWQLYDGLYLAEVDRILRPGGYWILSGPPINWKKHWKGWQR 350
Query: 332 EPTVLEKEQNNLEELAMQMCWEKVA--EGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSS 389
L EQ +E +A +CW+K+ E G IAIWQKP NHI C + SP FC++
Sbjct: 351 TKEDLNAEQQAIEAVAKSLCWKKITLKEVGDIAIWQKPTNHIHCKASRKVVKSPPFCSNK 410
Query: 390 DSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTYSE 449
+ DA WY KM ACI PLPEV DI EIAGG L+KWP RL PPR+ + + + + + E
Sbjct: 411 NPDAAWYDKMEACITPLPEVSDIKEIAGGQLKKWPERLTAVPPRIASGSIEGVTDEMFVE 470
Query: 450 DNMIWKKRVSYYEVMLKSL-SSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSN 508
D +W+KRV +Y+ ++ G+YRN++DMNA FGGFAAALV PVWVMN+VP S
Sbjct: 471 DTKLWQKRVGHYKSVISQFGQKGRYRNLLDMNARFGGFAAALVDDPVWVMNMVPTVGNST 530
Query: 509 NLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPE 568
LG+IYERGLIG+Y DWCE STYPRTYDLIHA ++F++Y D+C + +I++EM RILRPE
Sbjct: 531 TLGVIYERGLIGSYQDWCEGMSTYPRTYDLIHADSVFTLYKDRCQMDNILLEMDRILRPE 590
Query: 569 GTVIIRDSRDVILKVKEITDKMRWEGGTV 597
GTVIIRD D+++K+K ITD MRW V
Sbjct: 591 GTVIIRDDVDMLVKIKSITDGMRWNSQIV 619
>UniRef100_Q94GX1 Hypothetical protein OSJNBa0005K07.2 [Oryza sativa]
Length = 634
Score = 617 bits (1590), Expect = e-175
Identities = 283/526 (53%), Positives = 371/526 (69%), Gaps = 6/526 (1%)
Query: 89 ISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFP 148
+ F C N++ Y PCED KR ++F + +ERHCP +E + CLIP P Y+ PF
Sbjct: 101 VQPFPACPLNFSEYTPCEDRKRGRRFERAMLVYRERHCPGKDEEIRCLIPAPPKYRTPFK 160
Query: 149 WPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLP 208
WP+S+D AWF+N+P +L K QNWI + G RF FPGGGT FP G Y+DD+ KL
Sbjct: 161 WPQSRDFAWFNNIPHKELSIEKAVQNWIQVDGQRFRFPGGGTMFPRGADAYIDDIGKL-- 218
Query: 209 VNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVF 268
++L G+IRT +D GCGVAS+GA L+ +IL MS AP D H+AQV FALERG+PA++GV
Sbjct: 219 ISLTDGKIRTAIDTGCGVASWGAYLLKRNILAMSFAPRDTHEAQVQFALERGVPAIIGVM 278
Query: 269 STHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKA 328
RL +PS+SFD+AHCSRCL+PW DG+YL E+DRILRPGG+W+LSGPPINW+ +YK
Sbjct: 279 GKQRLPYPSRSFDMAHCSRCLIPWHEFDGIYLAEVDRILRPGGYWILSGPPINWKTHYKG 338
Query: 329 WQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNS 388
W+ L++EQ+N+E++A +CW KV E G ++IWQKP NH++C +P C S
Sbjct: 339 WERTKEDLKEEQDNIEDVARSLCWNKVVEKGDLSIWQKPKNHLECANIKKKYKTPHICKS 398
Query: 389 SDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTYS 448
+ DA WY +M AC+ PLPEV + EIAGG LE+WP R PPR+++ +
Sbjct: 399 DNPDAAWYKQMEACVTPLPEVSNQGEIAGGALERWPQRAFAVPPRVKRGMIPGIDASKFE 458
Query: 449 EDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSN 508
ED +W+KRV+YY+ L ++ G+YRNVMDMNA GGFAA+LVKYPVWVMNVVP ++ +
Sbjct: 459 EDKKLWEKRVAYYKRTL-PIADGRYRNVMDMNANLGGFAASLVKYPVWVMNVVPVNSDRD 517
Query: 509 NLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPE 568
LG IYERG IGTY DWCE FSTYPRTYDL+HA LFS+Y D+CDIT+I++EM RILRPE
Sbjct: 518 TLGAIYERGFIGTYQDWCEAFSTYPRTYDLLHADNLFSIYQDRCDITNILLEMDRILRPE 577
Query: 569 GTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
GT IIRD+ DV+ KV+ I +MRWE + D ++ +PE ++V
Sbjct: 578 GTAIIRDTVDVLTKVQAIAKRMRWESR---ILDHEDGPFNPEKVLV 620
>UniRef100_Q6EP94 Dehydration-responsive family protein-like [Oryza sativa]
Length = 646
Score = 614 bits (1583), Expect = e-174
Identities = 275/507 (54%), Positives = 361/507 (70%), Gaps = 6/507 (1%)
Query: 92 FQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPK 151
+Q C Y+ Y PCED KR ++P++ +ERHCP ERL CL+P P GY+NPFPWP
Sbjct: 118 YQACPARYSEYTPCEDVKRSLRYPRERLVYRERHCPTGRERLRCLVPAPSGYRNPFPWPA 177
Query: 152 SKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNL 211
S+D AWF+NVP +L K QNWI + GD+F FPGGGT FP G Y+DD+ KL+P L
Sbjct: 178 SRDVAWFANVPHKELTVEKAVQNWIRVDGDKFRFPGGGTMFPHGADAYIDDIGKLIP--L 235
Query: 212 DSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTH 271
G +RT LD GCGVAS+GA L+ DIL MS AP D H+AQV FALERG+PAM+GV +++
Sbjct: 236 HDGSVRTALDTGCGVASWGAYLLSRDILAMSFAPRDSHEAQVQFALERGVPAMIGVLASN 295
Query: 272 RLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQT 331
RLT+P+++FD+AHCSRCL+PW DGLYL E+DR+LRPGG+W+LSGPPINW+ +K W+
Sbjct: 296 RLTYPARAFDMAHCSRCLIPWHLYDGLYLIEVDRVLRPGGYWILSGPPINWKKYWKGWER 355
Query: 332 EPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSSDS 391
L EQ +E +A +CW+K+ E G IA+WQKP NH C + SP FC+ +
Sbjct: 356 TKEDLNAEQQAIEAVARSLCWKKIKEAGDIAVWQKPANHASCKA---SRKSPPFCSHKNP 412
Query: 392 DAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTYSEDN 451
DA WY KM AC+ PLPEV D E+AGG L+KWP RL PPR+ + + + K + +D
Sbjct: 413 DAAWYDKMEACVTPLPEVSDASEVAGGALKKWPQRLTAVPPRISRGSIKGVTSKAFVQDT 472
Query: 452 MIWKKRVSYYEVMLKSL-SSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSNNL 510
+W+KR+ +Y+ ++ G+YRNV+DMNAG GGFAAAL P+WVMN+VP S+ L
Sbjct: 473 ELWRKRIQHYKGVINQFEQKGRYRNVLDMNAGLGGFAAALASDPLWVMNMVPTVGNSSTL 532
Query: 511 GIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPEGT 570
G++YERGLIG+Y DWCE STYPRTYDLIHA ++F++Y ++C++ I++EM RILRPEGT
Sbjct: 533 GVVYERGLIGSYQDWCEGMSTYPRTYDLIHADSVFTLYKNRCEMDIILLEMDRILRPEGT 592
Query: 571 VIIRDSRDVILKVKEITDKMRWEGGTV 597
VIIRD D+++KVK D MRW+ V
Sbjct: 593 VIIRDDVDMLVKVKSAADGMRWDSQIV 619
>UniRef100_Q9SZX8 Hypothetical protein F7L13.20 [Arabidopsis thaliana]
Length = 633
Score = 608 bits (1567), Expect = e-172
Identities = 270/526 (51%), Positives = 374/526 (70%), Gaps = 5/526 (0%)
Query: 89 ISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFP 148
I +F+ C + + Y PCED +R ++F + +ERHCP +E L CLIP P YK PF
Sbjct: 90 IKYFEPCELSLSEYTPCEDRQRGRRFDRNMMKYRERHCPVKDELLYCLIPPPPNYKIPFK 149
Query: 149 WPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLP 208
WP+S+D AW+ N+P +L K QNWI + GDRF FPGGGT FP G Y+DD+ +L+P
Sbjct: 150 WPQSRDYAWYDNIPHKELSVEKAVQNWIQVEGDRFRFPGGGTMFPRGADAYIDDIARLIP 209
Query: 209 VNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVF 268
L G IRT +D GCGVASFGA L+ DI+ +S AP D H+AQV FALERG+PA++G+
Sbjct: 210 --LTDGGIRTAIDTGCGVASFGAYLLKRDIMAVSFAPRDTHEAQVQFALERGVPAIIGIM 267
Query: 269 STHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKA 328
+ RL +P+++FD+AHCSRCL+PW NDGLYL E+DR+LRPGG+W+LSGPPINW+ ++
Sbjct: 268 GSRRLPYPARAFDLAHCSRCLIPWFKNDGLYLMEVDRVLRPGGYWILSGPPINWKQYWRG 327
Query: 329 WQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNS 388
W+ L+KEQ+++E++A +CW+KV E G ++IWQKP+NHI+C + SP C+S
Sbjct: 328 WERTEEDLKKEQDSIEDVAKSLCWKKVTEKGDLSIWQKPLNHIECKKLKQNNKSPPICSS 387
Query: 389 SDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTYS 448
++D+ WY + CI PLPE + D+ AGG LE WP R PPR+ + + + +
Sbjct: 388 DNADSAWYKDLETCITPLPETNNPDDSAGGALEDWPDRAFAVPPRIIRGTIPEMNAEKFR 447
Query: 449 EDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSN 508
EDN +WK+R+++Y+ ++ LS G++RN+MDMNA GGFAA+++KYP WVMNVVP DA+
Sbjct: 448 EDNEVWKERIAHYKKIVPELSHGRFRNIMDMNAFLGGFAASMLKYPSWVMNVVPVDAEKQ 507
Query: 509 NLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPE 568
LG+IYERGLIGTY DWCE FSTYPRTYD+IHA LFS+Y +CD+T I++EM RILRPE
Sbjct: 508 TLGVIYERGLIGTYQDWCEGFSTYPRTYDMIHAGGLFSLYEHRCDLTLILLEMDRILRPE 567
Query: 569 GTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
GTV++RD+ + + KV++I M+W+ + D + +PE I+V
Sbjct: 568 GTVVLRDNVETLNKVEKIVKGMKWKS---QIVDHEKGPFNPEKILV 610
>UniRef100_Q9C884 Hypothetical protein T16O9.7 [Arabidopsis thaliana]
Length = 639
Score = 607 bits (1565), Expect = e-172
Identities = 276/542 (50%), Positives = 377/542 (68%), Gaps = 7/542 (1%)
Query: 75 FTSPLTLHSPITTK-ISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERL 133
F S L IT + + +F+ C + + Y PCED +R ++F + +ERHCP +E L
Sbjct: 92 FESHHKLELKITNQTVKYFEPCDMSLSEYTPCEDRERGRRFDRNMMKYRERHCPSKDELL 151
Query: 134 TCLIPKPIGYKNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFP 193
CLIP P YK PF WP+S+D AW+ N+P +L K QNWI + G+RF FPGGGT FP
Sbjct: 152 YCLIPPPPNYKIPFKWPQSRDYAWYDNIPHKELSIEKAIQNWIQVEGERFRFPGGGTMFP 211
Query: 194 DGVKGYVDDLKKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQV 253
G Y+DD+ +L+P L G IRT +D GCGVASFGA L+ DI+ MS AP D H+AQV
Sbjct: 212 RGADAYIDDIARLIP--LTDGAIRTAIDTGCGVASFGAYLLKRDIVAMSFAPRDTHEAQV 269
Query: 254 MFALERGLPAMLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFW 313
FALERG+PA++G+ + RL +P+++FD+AHCSRCL+PW NDGLYL E+DR+LRPGG+W
Sbjct: 270 QFALERGVPAIIGIMGSRRLPYPARAFDLAHCSRCLIPWFQNDGLYLTEVDRVLRPGGYW 329
Query: 314 VLSGPPINWRVNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKC 373
+LSGPPINW+ +K W+ L++EQ+++E+ A +CW+KV E G ++IWQKPINH++C
Sbjct: 330 ILSGPPINWKKYWKGWERSQEDLKQEQDSIEDAARSLCWKKVTEKGDLSIWQKPINHVEC 389
Query: 374 MQKLNTLSSPKFCNSSD-SDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPP 432
+ +P C+ SD D WY + +C+ PLPE DE AGG LE WP R PP
Sbjct: 390 NKLKRVHKTPPLCSKSDLPDFAWYKDLESCVTPLPEANSSDEFAGGALEDWPNRAFAVPP 449
Query: 433 RLRKENHDVFSLKTYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVK 492
R+ + + + EDN +WK+R+SYY+ ++ LS G++RN+MDMNA GGFAAA++K
Sbjct: 450 RIIGGTIPDINAEKFREDNEVWKERISYYKQIMPELSRGRFRNIMDMNAYLGGFAAAMMK 509
Query: 493 YPVWVMNVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKC 552
YP WVMNVVP DA+ LG+I+ERG IGTY DWCE FSTYPRTYDLIHA LFS+Y ++C
Sbjct: 510 YPSWVMNVVPVDAEKQTLGVIFERGFIGTYQDWCEGFSTYPRTYDLIHAGGLFSIYENRC 569
Query: 553 DITDIVIEMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMI 612
D+T I++EM RILRPEGTV+ RD+ +++ K++ IT+ MRW+ + D + +PE I
Sbjct: 570 DVTLILLEMDRILRPEGTVVFRDTVEMLTKIQSITNGMRWKSR---ILDHERGPFNPEKI 626
Query: 613 MV 614
++
Sbjct: 627 LL 628
>UniRef100_O80844 Hypothetical protein At2g45750 [Arabidopsis thaliana]
Length = 631
Score = 602 bits (1551), Expect = e-170
Identities = 277/516 (53%), Positives = 363/516 (69%), Gaps = 7/516 (1%)
Query: 84 PITTKISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGY 143
P+T F C+ + + PCED KR KF ++ ++RHCP+ E L C IP P GY
Sbjct: 79 PVTETAVSFPSCAAALSEHTPCEDAKRSLKFSRERLEYRQRHCPEREEILKCRIPAPYGY 138
Query: 144 KNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDL 203
K PF WP S+D AWF+NVP T+L KK+QNW+ DRF FPGGGT FP G Y+DD+
Sbjct: 139 KTPFRWPASRDVAWFANVPHTELTVEKKNQNWVRYENDRFWFPGGGTMFPRGADAYIDDI 198
Query: 204 KKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPA 263
+L ++L G IRT +D GCGVASFGA L+ +I TMS AP D H+AQV FALERG+PA
Sbjct: 199 GRL--IDLSDGSIRTAIDTGCGVASFGAYLLSRNITTMSFAPRDTHEAQVQFALERGVPA 256
Query: 264 MLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWR 323
M+G+ +T RL +PS++FD+AHCSRCL+PW NDG YL E+DR+LRPGG+W+LSGPPINW+
Sbjct: 257 MIGIMATIRLPYPSRAFDLAHCSRCLIPWGQNDGAYLMEVDRVLRPGGYWILSGPPINWQ 316
Query: 324 VNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSP 383
+K W+ L EQ +E++A +CW+KV + +AIWQKP NHI C + L +P
Sbjct: 317 KRWKGWERTMDDLNAEQTQIEQVARSLCWKKVVQRDDLAIWQKPFNHIDCKKTREVLKNP 376
Query: 384 KFC-NSSDSDAGWYTKMTACIFPLPEVKDIDE---IAGGVLEKWPIRLNDSPPRLRKENH 439
+FC + D D WYTKM +C+ PLPEV D ++ +AGG +EKWP RLN PPR+ K
Sbjct: 377 EFCRHDQDPDMAWYTKMDSCLTPLPEVDDAEDLKTVAGGKVEKWPARLNAIPPRVNKGAL 436
Query: 440 DVFSLKTYSEDNMIWKKRVSYYEVMLKSL-SSGKYRNVMDMNAGFGGFAAALVKYPVWVM 498
+ + + + E+ +WK+RVSYY+ + L +G+YRN++DMNA GGFAAAL PVWVM
Sbjct: 437 EEITPEAFLENTKLWKQRVSYYKKLDYQLGETGRYRNLVDMNAYLGGFAAALADDPVWVM 496
Query: 499 NVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIV 558
NVVP +AK N LG+IYERGLIGTY +WCE STYPRTYD IHA ++F++Y +C+ +I+
Sbjct: 497 NVVPVEAKLNTLGVIYERGLIGTYQNWCEAMSTYPRTYDFIHADSVFTLYQGQCEPEEIL 556
Query: 559 IEMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEG 594
+EM RILRP G VIIRD DV++KVKE+T + WEG
Sbjct: 557 LEMDRILRPGGGVIIRDDVDVLIKVKELTKGLEWEG 592
>UniRef100_Q9LP29 T9L6.6 protein [Arabidopsis thaliana]
Length = 656
Score = 596 bits (1537), Expect = e-169
Identities = 276/559 (49%), Positives = 377/559 (67%), Gaps = 24/559 (4%)
Query: 75 FTSPLTLHSPITTK-ISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERL 133
F S L IT + + +F+ C + + Y PCED +R ++F + +ERHCP +E L
Sbjct: 92 FESHHKLELKITNQTVKYFEPCDMSLSEYTPCEDRERGRRFDRNMMKYRERHCPSKDELL 151
Query: 134 TCLIPKPIGYKNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFP 193
CLIP P YK PF WP+S+D AW+ N+P +L K QNWI + G+RF FPGGGT FP
Sbjct: 152 YCLIPPPPNYKIPFKWPQSRDYAWYDNIPHKELSIEKAIQNWIQVEGERFRFPGGGTMFP 211
Query: 194 DGVKGYVDDLKKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQV 253
G Y+DD+ +L+P L G IRT +D GCGVASFGA L+ DI+ MS AP D H+AQV
Sbjct: 212 RGADAYIDDIARLIP--LTDGAIRTAIDTGCGVASFGAYLLKRDIVAMSFAPRDTHEAQV 269
Query: 254 MFALERGLPAMLGVFSTHRLTFPSKSFDVAHCSRCLVPWIAN-----------------D 296
FALERG+PA++G+ + RL +P+++FD+AHCSRCL+PW N D
Sbjct: 270 QFALERGVPAIIGIMGSRRLPYPARAFDLAHCSRCLIPWFQNGFLIGVANNQKKNWMCVD 329
Query: 297 GLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVA 356
GLYL E+DR+LRPGG+W+LSGPPINW+ +K W+ L++EQ+++E+ A +CW+KV
Sbjct: 330 GLYLTEVDRVLRPGGYWILSGPPINWKKYWKGWERSQEDLKQEQDSIEDAARSLCWKKVT 389
Query: 357 EGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSSD-SDAGWYTKMTACIFPLPEVKDIDEI 415
E G ++IWQKPINH++C + +P C+ SD D WY + +C+ PLPE DE
Sbjct: 390 EKGDLSIWQKPINHVECNKLKRVHKTPPLCSKSDLPDFAWYKDLESCVTPLPEANSSDEF 449
Query: 416 AGGVLEKWPIRLNDSPPRLRKENHDVFSLKTYSEDNMIWKKRVSYYEVMLKSLSSGKYRN 475
AGG LE WP R PPR+ + + + EDN +WK+R+SYY+ ++ LS G++RN
Sbjct: 450 AGGALEDWPNRAFAVPPRIIGGTIPDINAEKFREDNEVWKERISYYKQIMPELSRGRFRN 509
Query: 476 VMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRT 535
+MDMNA GGFAAA++KYP WVMNVVP DA+ LG+I+ERG IGTY DWCE FSTYPRT
Sbjct: 510 IMDMNAYLGGFAAAMMKYPSWVMNVVPVDAEKQTLGVIFERGFIGTYQDWCEGFSTYPRT 569
Query: 536 YDLIHAYALFSMYIDKCDITDIVIEMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEGG 595
YDLIHA LFS+Y ++CD+T I++EM RILRPEGTV+ RD+ +++ K++ IT+ MRW+
Sbjct: 570 YDLIHAGGLFSIYENRCDVTLILLEMDRILRPEGTVVFRDTVEMLTKIQSITNGMRWKSR 629
Query: 596 TVVVADDQNESSHPEMIMV 614
+ D + +PE I++
Sbjct: 630 ---ILDHERGPFNPEKILL 645
>UniRef100_Q94C19 At1g26850/T2P11_4 [Arabidopsis thaliana]
Length = 616
Score = 593 bits (1529), Expect = e-168
Identities = 278/527 (52%), Positives = 359/527 (67%), Gaps = 6/527 (1%)
Query: 88 KISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPF 147
K+ F+ C YT+Y PC+D +R FP+ + +ERHC NE+L CLIP P GY PF
Sbjct: 82 KVKAFEPCDGRYTDYTPCQDQRRAMTFPRDSMIYRERHCAPENEKLHCLIPAPKGYVTPF 141
Query: 148 PWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLL 207
WPKS+D ++N P+ L K QNWI GD F FPGGGT FP G Y+D L ++
Sbjct: 142 SWPKSRDYVPYANAPYKALTVEKAIQNWIQYEGDVFRFPGGGTQFPQGADKYIDQLASVI 201
Query: 208 PVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGV 267
P +++G +RT LD GCGVAS+GA L ++ MS AP D H+AQV FALERG+PA++GV
Sbjct: 202 P--MENGTVRTALDTGCGVASWGAYLWSRNVRAMSFAPRDSHEAQVQFALERGVPAVIGV 259
Query: 268 FSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYK 327
T +L +P+++FD+AHCSRCL+PW ANDG+YL E+DR+LRPGG+W+LSGPPINW+VNYK
Sbjct: 260 LGTIKLPYPTRAFDMAHCSRCLIPWGANDGMYLMEVDRVLRPGGYWILSGPPINWKVNYK 319
Query: 328 AWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCN 387
AWQ L++EQ +EE A +CWEK E G+IAIWQK +N C + + + FC
Sbjct: 320 AWQRPKEDLQEEQRKIEEAAKLLCWEKKYEHGEIAIWQKRVNDEACRSRQDDPRA-NFCK 378
Query: 388 SSDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTY 447
+ D+D WY KM ACI P PE DE+AGG L+ +P RLN PPR+ + ++ Y
Sbjct: 379 TDDTDDVWYKKMEACITPYPETSSSDEVAGGELQAFPDRLNAVPPRISSGSISGVTVDAY 438
Query: 448 SEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKS 507
+DN WKK V Y+ + L +G+YRN+MDMNAGFGGFAAAL +WVMNVVP A+
Sbjct: 439 EDDNRQWKKHVKAYKRINSLLDTGRYRNIMDMNAGFGGFAAALESQKLWVMNVVPTIAEK 498
Query: 508 NNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRP 567
N LG++YERGLIG Y DWCE FSTYPRTYDLIHA LFS+Y +KC+ DI++EM RILRP
Sbjct: 499 NRLGVVYERGLIGIYHDWCEAFSTYPRTYDLIHANHLFSLYKNKCNADDILLEMDRILRP 558
Query: 568 EGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
EG VIIRD D ++KVK I MRW+ V D ++ PE +++
Sbjct: 559 EGAVIIRDDVDTLIKVKRIIAGMRWDAKLV---DHEDGPLVPEKVLI 602
>UniRef100_Q9ZPH9 F15P23.1 protein [Arabidopsis thaliana]
Length = 633
Score = 591 bits (1524), Expect = e-167
Identities = 272/524 (51%), Positives = 367/524 (69%), Gaps = 8/524 (1%)
Query: 95 CSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPKSKD 154
C ++ Y PCE R FP++ +ERHCP+ +E + C IP P GY PF WP+S+D
Sbjct: 99 CGVEFSEYTPCEFVNRSLNFPRERLIYRERHCPEKHEIVRCRIPAPYGYSLPFRWPESRD 158
Query: 155 NAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNLDSG 214
AWF+NVP T+L KK+QNW+ DRF+FPGGGT FP G Y+D++ +L +NL G
Sbjct: 159 VAWFANVPHTELTVEKKNQNWVRYEKDRFLFPGGGTMFPRGADAYIDEIGRL--INLKDG 216
Query: 215 RIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTHRLT 274
IRT +D GCGVASFGA LM +I+TMS AP D H+AQV FALERG+PA++GV ++ RL
Sbjct: 217 SIRTAIDTGCGVASFGAYLMSRNIVTMSFAPRDTHEAQVQFALERGVPAIIGVLASIRLP 276
Query: 275 FPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQTEPT 334
FP+++FD+AHCSRCL+PW +G YL E+DR+LRPGG+W+LSGPPINW+ ++K W+
Sbjct: 277 FPARAFDIAHCSRCLIPWGQYNGTYLIEVDRVLRPGGYWILSGPPINWQRHWKGWERTRD 336
Query: 335 VLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSSDSDAG 394
L EQ+ +E +A +CW K+ + +A+WQKP NH+ C + L P FC+ + + G
Sbjct: 337 DLNSEQSQIERVARSLCWRKLVQREDLAVWQKPTNHVHCKRNRIALGRPPFCHRTLPNQG 396
Query: 395 WYTKMTACIFPLPEV--KDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTYSEDNM 452
WYTK+ C+ PLPEV +I E+AGG L +WP RLN PPR++ + + + + +
Sbjct: 397 WYTKLETCLTPLPEVTGSEIKEVAGGQLARWPERLNALPPRIKSGSLEGITEDEFVSNTE 456
Query: 453 IWKKRVSYYEVMLKSLS-SGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSNNLG 511
W++RVSYY+ + L+ +G+YRN +DMNA GGFA+ALV PVWVMNVVP +A N LG
Sbjct: 457 KWQRRVSYYKKYDQQLAETGRYRNFLDMNAHLGGFASALVDDPVWVMNVVPVEASVNTLG 516
Query: 512 IIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPEGTV 571
+IYERGLIGTY +WCE STYPRTYD IHA ++FS+Y D+CD+ DI++EM RILRP+G+V
Sbjct: 517 VIYERGLIGTYQNWCEAMSTYPRTYDFIHADSVFSLYKDRCDMEDILLEMDRILRPKGSV 576
Query: 572 IIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMVL 615
IIRD DV+ KVK+ITD M+WEG + D +N E I+ L
Sbjct: 577 IIRDDIDVLTKVKKITDAMQWEGR---IGDHENGPLEREKILFL 617
>UniRef100_Q8H636 Dehydration-responsive protein-like [Oryza sativa]
Length = 618
Score = 588 bits (1515), Expect = e-166
Identities = 273/524 (52%), Positives = 361/524 (68%), Gaps = 7/524 (1%)
Query: 95 CSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPKSKD 154
C Y+ + PCE + + P++ + +ERHCP ER CL+P P GY+ P WP+S+D
Sbjct: 90 CDAGYSEHTPCEGQRWSLRQPRRRFAYRERHCPPPAERRRCLVPAPRGYRAPLRWPRSRD 149
Query: 155 NAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNLDSG 214
AW++N P +LV K QNWI GD FPGGGT FP G Y+DD+ + L G
Sbjct: 150 AAWYANAPHEELVTEKGVQNWIRRDGDVLRFPGGGTMFPHGADRYIDDIAAAAGITLGGG 209
Query: 215 -RIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTHRL 273
+RT LD GCGVAS+GA L+ D+LTMS AP D H+AQV+FALERG+PAMLG+ +T RL
Sbjct: 210 GAVRTALDTGCGVASWGAYLLSRDVLTMSFAPKDTHEAQVLFALERGVPAMLGIMATKRL 269
Query: 274 TFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQTEP 333
+P+++FD+AHCSRCL+PW +GLY+ E+DR+LRPGG+WVLSGPP+NW ++K W+ P
Sbjct: 270 PYPARAFDMAHCSRCLIPWSKYNGLYMIEVDRVLRPGGYWVLSGPPVNWERHFKGWKRTP 329
Query: 334 TVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSS-DSD 392
L EQ+ +E +A +CW KV + G IA+WQK INH+ C N L FCNS+ D D
Sbjct: 330 EDLSSEQSAIEAIAKSLCWTKVQQMGDIAVWQKQINHVSCKASRNELGGLGFCNSNQDPD 389
Query: 393 AGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKEN-HDVFSLKTYSEDN 451
AGWY M CI PLPEV ++AGG +++WP RL PPR+ + ++ T+ +D+
Sbjct: 390 AGWYVNMEECITPLPEVSGPGDVAGGEVKRWPERLTSPPPRIAGGSLGSSVTVDTFIKDS 449
Query: 452 MIWKKRVSYYEVMLKSLS-SGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSNNL 510
+W++RV Y+ + L+ G+YRN++DMNAG GGFAAALV PVWVMNVVP A +N L
Sbjct: 450 EMWRRRVDRYKGVSGGLAEKGRYRNLLDMNAGLGGFAAALVDDPVWVMNVVPTAAVANTL 509
Query: 511 GIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPEGT 570
G+IYERGLIGTY DWCE STYPRTYDLIHAY+LF+MY D+C++ DI++EM R+LRPEGT
Sbjct: 510 GVIYERGLIGTYQDWCEAMSTYPRTYDLIHAYSLFTMYKDRCEMEDILLEMDRVLRPEGT 569
Query: 571 VIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
VI RD DV++K+K I D MRWE + D ++ E I+V
Sbjct: 570 VIFRDDVDVLVKIKNIADGMRWESR---IVDHEDGPMQREKILV 610
>UniRef100_Q8W396 Hypothetical protein OSJNBa0013O08.23 [Oryza sativa]
Length = 611
Score = 578 bits (1489), Expect = e-163
Identities = 270/528 (51%), Positives = 358/528 (67%), Gaps = 6/528 (1%)
Query: 86 TTKISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKN 145
+T++ F+ C YT+Y PCE+ KR FP+ N +ERHCP ++L CL+P P GY
Sbjct: 76 STEVKTFEPCDAQYTDYTPCEEQKRAMTFPRDNMIYRERHCPPEKDKLYCLVPAPKGYAA 135
Query: 146 PFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKK 205
PF WPKS+D ++N+P L K QNW+ G F FPGGGT FP G Y+D L
Sbjct: 136 PFHWPKSRDYVHYANIPHKSLTVEKAIQNWVHYEGKVFRFPGGGTQFPQGADKYIDHLAS 195
Query: 206 LLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAML 265
++P+ +G++RT LD GCGVAS GA L+ ++LTMS AP D H+AQV FALERG+PA +
Sbjct: 196 VIPIA--NGKVRTALDTGCGVASLGAYLLKKNVLTMSFAPRDNHEAQVQFALERGVPAYI 253
Query: 266 GVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVN 325
GV + +L+FPS+ FD+AHCSRCL+PW NDG+Y+ E+DR+LRPGG+WVLSGPPI W+++
Sbjct: 254 GVLGSMKLSFPSRVFDMAHCSRCLIPWSGNDGMYMMEVDRVLRPGGYWVLSGPPIGWKIH 313
Query: 326 YKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKF 385
YK WQ L+ EQ +E+ A +CW K++E IAIW+K IN C K K
Sbjct: 314 YKGWQRTKDDLQSEQRRIEQFAELLCWNKISEKDGIAIWRKRINDKSCPMKQENPKVDKC 373
Query: 386 CNSSDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLK 445
+ D+D WY KM C+ PLPEVK + E+AGG LE +P RLN PPR+ FS++
Sbjct: 374 ELAYDNDV-WYKKMEVCVTPLPEVKTMTEVAGGQLEPFPQRLNAVPPRITHGFVPGFSVQ 432
Query: 446 TYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDA 505
+Y +DN +W+K ++ Y+ + L +G+YRN+MDMNAG G FAAAL +WVMNVVP A
Sbjct: 433 SYQDDNKLWQKHINAYKKINNLLDTGRYRNIMDMNAGLGSFAAALESTKLWVMNVVPTIA 492
Query: 506 KSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRIL 565
++ LG+IYERGLIG Y DWCE FSTYPRTYDLIHA A+FS+Y +KC DI++EM RIL
Sbjct: 493 DTSTLGVIYERGLIGMYHDWCEGFSTYPRTYDLIHANAVFSLYENKCKFEDILLEMDRIL 552
Query: 566 RPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIM 613
RPEG VIIRD DV++KV++I + MRW+ + D + PE I+
Sbjct: 553 RPEGAVIIRDKVDVLVKVEKIANAMRWQ---TRLTDHEGGPHVPEKIL 597
>UniRef100_Q6EQ17 Dehydration-responsive protein-like [Oryza sativa]
Length = 616
Score = 578 bits (1489), Expect = e-163
Identities = 267/501 (53%), Positives = 350/501 (69%), Gaps = 3/501 (0%)
Query: 92 FQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPK 151
F+ C YT+Y PC+D R KFP++N +ERHCP E+L CLIP P GY PFPWPK
Sbjct: 83 FKPCPDRYTDYTPCQDQNRAMKFPRENMNYRERHCPPQKEKLHCLIPPPKGYVAPFPWPK 142
Query: 152 SKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNL 211
S+D F+N P+ L K QNW+ G+ F FPGGGT FP G Y+D L ++P+
Sbjct: 143 SRDYVPFANCPYKSLTVEKAIQNWVQFEGNVFRFPGGGTQFPQGADKYIDQLASVVPIA- 201
Query: 212 DSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTH 271
+G +RT LD GCGVAS+GA L+ ++L MS AP D H+AQV FALERG+PA++GV T
Sbjct: 202 -NGTVRTALDTGCGVASWGAYLLKRNVLAMSFAPRDSHEAQVQFALERGVPAVIGVLGTI 260
Query: 272 RLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQT 331
+L +PS++FD+AHCSRCL+PW AN G+Y+ E+DR+LRPGG+WVLSGPPINW+VNYK WQ
Sbjct: 261 KLPYPSRAFDMAHCSRCLIPWGANGGIYMMEVDRVLRPGGYWVLSGPPINWKVNYKGWQR 320
Query: 332 EPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSSDS 391
LE EQN +EE+A +CWEKV E G++AIW+K +N C + + SS + C+S+++
Sbjct: 321 TKKDLEAEQNKIEEIADLLCWEKVKEIGEMAIWRKRLNTESCPSRQDE-SSVQMCDSTNA 379
Query: 392 DAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTYSEDN 451
D WY KM C+ P+P+V D E+AGG ++ +P RLN PPR+ S + Y +D
Sbjct: 380 DDVWYKKMKPCVTPIPDVNDPSEVAGGAIKPFPSRLNAVPPRIANGLIPGVSSQAYQKDI 439
Query: 452 MIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSNNLG 511
+WKK V Y + K L +G+YRN+MDMNAGFGGFAAA+ WVMN VP +K + LG
Sbjct: 440 KMWKKHVKAYSSVNKYLLTGRYRNIMDMNAGFGGFAAAIESPKSWVMNAVPTISKMSTLG 499
Query: 512 IIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPEGTV 571
IYERGLIG Y DWCE FSTYPRTYDLIHA LF++Y +KC++ DI++EM R+LRPEG V
Sbjct: 500 AIYERGLIGIYHDWCEAFSTYPRTYDLIHASGLFTLYKNKCNMEDILLEMDRVLRPEGAV 559
Query: 572 IIRDSRDVILKVKEITDKMRW 592
I+RD D++ KV + M+W
Sbjct: 560 IMRDDVDILTKVNRLALGMKW 580
>UniRef100_Q68V16 Ankyrin protein kinase-like [Poa pratensis]
Length = 613
Score = 575 bits (1483), Expect = e-162
Identities = 266/508 (52%), Positives = 350/508 (68%), Gaps = 3/508 (0%)
Query: 85 ITTKISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYK 144
+ + + F+ C +T+Y PC+D R KFP++N +ERHCP E+L CL+P P GY
Sbjct: 75 LVSPVKKFKPCPDRFTDYTPCQDQNRAMKFPRENMNYRERHCPLQKEKLHCLVPPPKGYV 134
Query: 145 NPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLK 204
PFPWPKS+D F+N P+ L K QNW+ G+ F FPGGGT FP G Y+D L
Sbjct: 135 APFPWPKSRDYVPFANCPYKSLTVEKAIQNWVQYEGNVFRFPGGGTQFPQGADKYIDQLA 194
Query: 205 KLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAM 264
++P+ +G +RT LD GCGVAS+GA L+ ++L M AP D H+AQV FALERG+PA+
Sbjct: 195 AVIPIA--NGTVRTALDTGCGVASWGAYLLKRNVLAMPFAPRDSHEAQVQFALERGVPAV 252
Query: 265 LGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRV 324
+GV T +L +PS++FD+AHCSRCL+PW NDGLY+ E+DR+LRPGG+WVLSGPPINW+V
Sbjct: 253 IGVLGTIKLPYPSRAFDMAHCSRCLIPWGLNDGLYMMEVDRVLRPGGYWVLSGPPINWKV 312
Query: 325 NYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPK 384
NYK WQ LE EQN +EE+A +CWEKV+E G+ AIW+K +N C + + S+ +
Sbjct: 313 NYKGWQRTKKDLEAEQNKIEEIAELLCWEKVSEKGETAIWRKRVNTESCPSR-HEESTVQ 371
Query: 385 FCNSSDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSL 444
C S+++D WY M AC+ PLP+V++ E+AGG ++ +P RLN PPR+ S
Sbjct: 372 MCKSTNADDVWYKTMKACVTPLPDVENPSEVAGGAIKPFPSRLNAIPPRIANGLIPGVSS 431
Query: 445 KTYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFD 504
+ Y +DN +WKK V Y + K L +G+YRN+MDMNAGFGGFAAA+ WVMNVVP
Sbjct: 432 QAYEKDNKMWKKHVKAYSNVNKYLLTGRYRNIMDMNAGFGGFAAAIESPKSWVMNVVPTI 491
Query: 505 AKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRI 564
K LG +Y RGLIG Y DWCE FSTYPRTYDLIHA LF++Y +KC + DI++EM RI
Sbjct: 492 GKIATLGSVYGRGLIGIYHDWCEAFSTYPRTYDLIHASGLFTLYKNKCSLEDILLEMDRI 551
Query: 565 LRPEGTVIIRDSRDVILKVKEITDKMRW 592
LRPEG VI+RD D++ KV + MRW
Sbjct: 552 LRPEGAVIMRDDVDILTKVDKFARGMRW 579
>UniRef100_Q9M609 Hypothetical protein [Malus domestica]
Length = 608
Score = 570 bits (1468), Expect = e-161
Identities = 269/523 (51%), Positives = 357/523 (67%), Gaps = 12/523 (2%)
Query: 92 FQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPK 151
F+ C YT+Y PC+D KR FP+++ +ERHCP E+L CLIP P GY PFPWPK
Sbjct: 84 FEPCHHRYTDYTPCQDQKRAMTFPREDMNYRERHCPPEEEKLHCLIPAPKGYVTPFPWPK 143
Query: 152 SKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNL 211
S+D ++N P+ L K QNWI G+ F FPGGGT FP G Y+D L ++P+
Sbjct: 144 SRDYVPYANAPYKSLTVEKAVQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLAAVIPIK- 202
Query: 212 DSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTH 271
+G +RT LD GCGVAS+GA L+ ++L MS AP D H+AQV FALERG+PA++GV T
Sbjct: 203 -NGTVRTALDTGCGVASWGAYLLSRNVLAMSFAPRDSHEAQVQFALERGVPAVIGVLGTI 261
Query: 272 RLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQT 331
+L +PS++FD+AHCSRCL+PW NDG YL+E+DR+LRPGG+WVLSGPPINW+ NY+AWQ
Sbjct: 262 KLPYPSRAFDMAHCSRCLIPWGINDGKYLKEVDRVLRPGGYWVLSGPPINWKNNYQAWQR 321
Query: 332 EPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSSDS 391
L++EQ +EE A +CWEK +E G+ AIWQK ++ C + + S FC + ++
Sbjct: 322 PKEDLQEEQRQIEEAAKLLCWEKKSEKGETAIWQKRVDSDSCGDRQDD-SRANFCKADEA 380
Query: 392 DAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTYSEDN 451
D+ WY KM CI P P+V + G L+ +P RL PPR+ + S++ Y EDN
Sbjct: 381 DSVWYKKMEGCITPYPKV------SSGELKPFPKRLYAVPPRISSGSVPGVSVEDYEEDN 434
Query: 452 MIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSNNLG 511
WKK V+ Y+ + K + +G+YRN+MDMNAG GGFAAA+ +WVMNV+P A+ N LG
Sbjct: 435 NKWKKHVNAYKRINKLIDTGRYRNIMDMNAGLGGFAAAIESPKLWVMNVMPTIAEKNTLG 494
Query: 512 IIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPEGTV 571
++YERGLIG Y DWCE FSTYPRTYDLIHA+ +FSMY KC+ DI++EM RILRPEG V
Sbjct: 495 VVYERGLIGIYHDWCEGFSTYPRTYDLIHAHGVFSMYNGKCNWEDILLEMDRILRPEGAV 554
Query: 572 IIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
I RD DV++KVK+I MRW+ + D ++ PE ++V
Sbjct: 555 IFRDEVDVLIKVKKIVGGMRWD---TKLVDHEDGPLVPEKVLV 594
>UniRef100_Q6L446 Putative methyltransferase [Solanum demissum]
Length = 612
Score = 560 bits (1443), Expect = e-158
Identities = 270/529 (51%), Positives = 353/529 (66%), Gaps = 15/529 (2%)
Query: 88 KISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPF 147
++ + C YT+Y PC+D KR FP++N +ERHCP E+L CLIP P GY PF
Sbjct: 80 EVEELKPCDPQYTDYTPCQDQKRAMTFPRENMNYRERHCPPQEEKLHCLIPAPKGYVTPF 139
Query: 148 PWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLL 207
PWPKS+D ++N P+ L K QNW+ G+ F FPGGGT FP G Y+D L ++
Sbjct: 140 PWPKSRDYVPYANAPYKSLTVEKAIQNWVQYEGNMFRFPGGGTQFPQGADKYIDQLASVV 199
Query: 208 PVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGV 267
P+ ++G +RT LD GCGVAS+GA L +++ MS AP D H+AQV FALERG+PA++GV
Sbjct: 200 PI--ENGTVRTALDTGCGVASWGAYLWKRNVIAMSFAPRDSHEAQVQFALERGVPAVIGV 257
Query: 268 FSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYK 327
T ++ +PSK+FD+AHCSRCL+PW A DG+ + E+DR+LRPGG+WVLSGPPINW+VN+K
Sbjct: 258 LGTIKMPYPSKAFDMAHCSRCLIPWGAADGILMMEVDRVLRPGGYWVLSGPPINWKVNFK 317
Query: 328 AWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCN 387
AWQ LE+EQ +EE A +CWEK++E G+ AIWQK + C + S+ + C
Sbjct: 318 AWQRPKEDLEEEQRKIEEAAKLLCWEKISEKGETAIWQKRKDSASC-RSAQENSAARVCK 376
Query: 388 SSDSDAGWYTKMTACIFPLPEVKDIDEIAGG--VLEKWPIRLNDSPPRLRKENHDVFSLK 445
SD D+ WY KM CI P + GG L+ +P RL PPR+ S+
Sbjct: 377 PSDPDSVWYNKMEMCITP-------NNGNGGDESLKPFPERLYAVPPRIANGLVSGVSVA 429
Query: 446 TYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDA 505
Y ED+ WKK +S Y+ + K L +G+YRN+MDMNAG GGFAAAL WVMNV+P A
Sbjct: 430 KYQEDSKKWKKHISAYKKINKLLDTGRYRNIMDMNAGLGGFAAALHSPKFWVMNVMPTIA 489
Query: 506 KSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRIL 565
+ N LG+I+ERGLIG Y DWCE FSTYPRTYDLIHA LFS+Y DKC+ DI++EM RIL
Sbjct: 490 EKNTLGVIFERGLIGIYHDWCEAFSTYPRTYDLIHASGLFSLYKDKCEFEDILLEMDRIL 549
Query: 566 RPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
RPEG VI+RD+ DV++KVK+I MRW + D ++ PE I+V
Sbjct: 550 RPEGAVILRDNVDVLIKVKKIIGGMRW---NFKLMDHEDGPLVPEKILV 595
>UniRef100_Q60D37 Putative methyltransferase [Solanum tuberosum]
Length = 612
Score = 560 bits (1442), Expect = e-158
Identities = 269/527 (51%), Positives = 351/527 (66%), Gaps = 11/527 (2%)
Query: 88 KISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPF 147
++ + C YT+Y PC+D KR FP++N +ERHCP E+L CLIP P GY PF
Sbjct: 80 EVEELKPCDPQYTDYTPCQDQKRAMTFPRENMNYRERHCPPQEEKLHCLIPAPKGYVTPF 139
Query: 148 PWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLL 207
PWPKS+D ++N P+ L K QNW+ G+ F FPGGGT FP G Y+D L ++
Sbjct: 140 PWPKSRDYVPYANAPYKSLTVEKAIQNWVQYEGNVFRFPGGGTQFPQGADKYIDQLASVV 199
Query: 208 PVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGV 267
P+ ++G +RT LD GCGVAS+GA L +++ MS AP D H+AQV FALERG+PA++GV
Sbjct: 200 PI--ENGTVRTALDTGCGVASWGAYLWKRNVIAMSFAPRDSHEAQVQFALERGVPAVIGV 257
Query: 268 FSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYK 327
T ++ +PSK+FD+AHCSRCL+PW A DG+ + E+DR+LRPGG+WVLSGPPINW+VN+K
Sbjct: 258 LGTIKMPYPSKAFDMAHCSRCLIPWGAADGILMMEVDRVLRPGGYWVLSGPPINWKVNFK 317
Query: 328 AWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCN 387
AWQ LE+EQ +EE A +CWEK++E G+ AIWQK + C + S+ + C
Sbjct: 318 AWQRPKEDLEEEQRKIEEAAKLLCWEKISEKGETAIWQKRKDSASC-RSAQENSAARVCK 376
Query: 388 SSDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTY 447
SD D+ WY KM CI P + L+ +P RL PPR+ S+ Y
Sbjct: 377 PSDPDSVWYNKMEMCITP-----NTGNGGDESLKPFPERLYAVPPRIANGLVSGVSVAKY 431
Query: 448 SEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKS 507
ED+ WKK VS Y+ + K L +G+YRN+MDMNAG GGFAAAL WVMNV+P A+
Sbjct: 432 QEDSKKWKKHVSAYKKINKLLDTGRYRNIMDMNAGLGGFAAALHSPKFWVMNVMPTIAEK 491
Query: 508 NNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRP 567
N LG+I+ERGLIG Y DWCE FSTYPRTYDLIHA LFS+Y DKC+ DI++EM RILRP
Sbjct: 492 NTLGVIFERGLIGIYHDWCEAFSTYPRTYDLIHASGLFSLYKDKCEFEDILLEMDRILRP 551
Query: 568 EGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
EG VI+RD+ DV++KVK+I MRW + D ++ PE I+V
Sbjct: 552 EGAVILRDNVDVLIKVKKIIGGMRW---NFKLMDHEDGPLVPEKILV 595
>UniRef100_Q5XWR1 Hypothetical protein [Solanum tuberosum]
Length = 592
Score = 560 bits (1442), Expect = e-158
Identities = 271/529 (51%), Positives = 353/529 (66%), Gaps = 15/529 (2%)
Query: 88 KISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPF 147
++ + C YT+Y PC+D KR FP++N +ERHCP E+L CLIP P GY PF
Sbjct: 60 EVEELKPCDPQYTDYTPCQDQKRAMTFPRENMNYRERHCPPQEEKLHCLIPAPKGYVTPF 119
Query: 148 PWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLL 207
PWPKS+D ++N P+ L K QNW+ G+ F FPGGGT FP G Y+D L ++
Sbjct: 120 PWPKSRDYVPYANAPYKSLTVEKAIQNWVQYEGNVFRFPGGGTQFPQGADKYIDQLASVV 179
Query: 208 PVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGV 267
P+ ++G +RT LD GCGVAS+GA L +++ MS AP D H+AQV FALERG+PA++GV
Sbjct: 180 PI--ENGTVRTALDTGCGVASWGAYLWKRNVIAMSFAPRDSHEAQVQFALERGVPAVIGV 237
Query: 268 FSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYK 327
T ++ +PSK+FD+AHCSRCL+PW A DG+ + E+DR+LRPGG+WVLSGPPINW+VN+K
Sbjct: 238 LGTIKMPYPSKAFDMAHCSRCLIPWGAADGILMMEVDRVLRPGGYWVLSGPPINWKVNFK 297
Query: 328 AWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCN 387
AWQ LE+EQ +EE A +CWEK++E G+ AIWQK + C + S+ + C
Sbjct: 298 AWQRPKEDLEEEQRKIEEAAKLLCWEKISEKGETAIWQKRKDSASC-RSAQENSAARVCK 356
Query: 388 SSDSDAGWYTKMTACIFPLPEVKDIDEIAGG--VLEKWPIRLNDSPPRLRKENHDVFSLK 445
SD D+ WY KM CI P + GG L+ +P RL PPR+ S+
Sbjct: 357 PSDPDSVWYNKMEMCITP-------NNGNGGDESLKPFPERLYAVPPRIANGLVSGVSVA 409
Query: 446 TYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDA 505
Y ED+ WKK VS Y+ + K L +G+YRN+MDMNAG GGFAAAL WVMNV+P A
Sbjct: 410 KYQEDSKKWKKHVSAYKKINKLLDTGRYRNIMDMNAGLGGFAAALHNPKFWVMNVMPTIA 469
Query: 506 KSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRIL 565
+ N LG+I+ERGLIG Y DWCE FSTYPRTYDLIHA LFS+Y DKC+ DI++EM RIL
Sbjct: 470 EKNTLGVIFERGLIGIYHDWCEAFSTYPRTYDLIHASGLFSLYKDKCEFEDILLEMDRIL 529
Query: 566 RPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
RPEG VI+RD+ DV++KVK+I MRW + D ++ PE I+V
Sbjct: 530 RPEGAVILRDNVDVLIKVKKIIGGMRW---NFKLMDHEDGPLVPEKILV 575
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.140 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,137,932,260
Number of Sequences: 2790947
Number of extensions: 52276559
Number of successful extensions: 165850
Number of sequences better than 10.0: 290
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 199
Number of HSP's that attempted gapping in prelim test: 165013
Number of HSP's gapped (non-prelim): 405
length of query: 620
length of database: 848,049,833
effective HSP length: 134
effective length of query: 486
effective length of database: 474,062,935
effective search space: 230394586410
effective search space used: 230394586410
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC146752.3


