

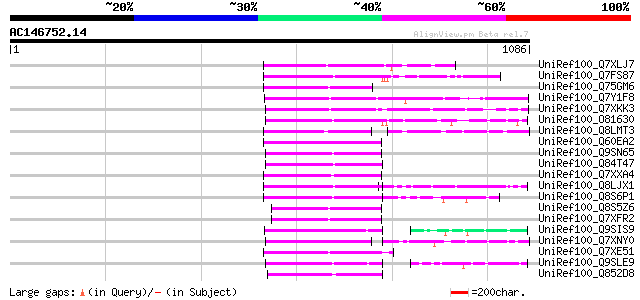
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.14 - phase: 2 /pseudo
(1086 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XLJ7 OSJNBa0009K15.13 protein [Oryza sativa] 205 7e-51
UniRef100_Q7FS87 Putative glycerol 3-phosphate permease [Zea mays] 197 2e-48
UniRef100_Q75GM6 Putative non-LTR retroelement reverse transcrip... 179 3e-43
UniRef100_Q7Y1F8 Putative reverse transcriptase [Oryza sativa] 172 7e-41
UniRef100_Q7XKK3 OSJNBa0038O10.4 protein [Oryza sativa] 168 9e-40
UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana] 167 1e-39
UniRef100_Q8LMT3 Putative retroelement [Oryza sativa] 167 2e-39
UniRef100_Q60EA2 Hypothetical protein OSJNBb0048K05.4 [Oryza sat... 160 2e-37
UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis tha... 152 5e-35
UniRef100_Q84T47 Putative reverse transcriptase [Oryza sativa] 152 5e-35
UniRef100_Q7XXA4 OSJNBa0019G23.12 protein [Oryza sativa] 152 7e-35
UniRef100_Q8LJX1 Putative reverse transcriptase [Sorghum bicolor] 152 7e-35
UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa] 151 9e-35
UniRef100_Q8S5Z6 Putative reverse transcriptase [Oryza sativa] 150 2e-34
UniRef100_Q7XFR2 Putative retroelement [Oryza sativa] 150 2e-34
UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcrip... 150 2e-34
UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa] 150 3e-34
UniRef100_Q7XE51 Putative non-LTR retroelement reverse transcrip... 148 1e-33
UniRef100_Q9SLE9 Putative non-LTR retroelement reverse transcrip... 148 1e-33
UniRef100_Q852D8 Putative retrotransposon protein [Oryza sativa] 147 2e-33
>UniRef100_Q7XLJ7 OSJNBa0009K15.13 protein [Oryza sativa]
Length = 779
Score = 205 bits (521), Expect = 7e-51
Identities = 132/427 (30%), Positives = 207/427 (47%), Gaps = 44/427 (10%)
Query: 532 VVGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLD 591
V V+ SQ+AF+ GR ILDG++I +E++ + K + + ++ K+DFEKAYD V W +L
Sbjct: 254 VANKVIGESQTAFLPGRFILDGVVILHEVLHELKNSGQSGIILKLDFEKAYDKVQWDFLF 313
Query: 592 EVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEG 651
+V+ + F W WI T + ++ +NG F +G+RQGDPLSP +F L A+
Sbjct: 314 DVLQRKGFCEKWIGWIKAATTKGSVAININGEVDQFFRTHKGVRQGDPLSPLMFYLVADA 373
Query: 652 LNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDV 711
L M++ G V VP ++HLQ+ADDT+L S N+ +K +L +E +
Sbjct: 374 LPEMLNKAKEAG-HLQGLVPHLVPGGLTHLQYADDTILFMTNSEENIVTVKFLLYCYEAM 432
Query: 712 SGLKVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHF--WHPL 769
SGLK+NF KS + + E A + +C+ G +P YLGLPI G +K H W L
Sbjct: 433 SGLKINFQKSEVIVLGVEEMEAQRVADLFNCKRGALPITYLGLPIKG-KKKYHLIKWEAL 491
Query: 770 VERIRRRLSGVLEDRESLWNVVLCAKY---------------------GEVGGRVQFSEG 808
G ++ + + N L K+ G+ GG Q S
Sbjct: 492 CRPKEDGGLGFIDTK--IMNEALLCKWIYKLESGNDSPCCNLLRKKYLGQGGGFFQSSPE 549
Query: 809 VGPIWWRQLNQIRCGVGLAENAWLVDNIVRKVGDGSTTLFWEDPWLLNVPLAVSFSRLFE 868
G +W+ L +++ W+ +G+G FW+D WL VPL + F ++
Sbjct: 550 GGSQFWKSLLEVK--------NWMKLGSAYNIGNGEAVRFWDDVWLGEVPLKIQFPYIYS 601
Query: 869 LAENKGVSVREMFLLGWGADGGAWRWRRRLFAWEEELVEECV--ARLSNFVLQADLSDRW 926
+ N +V +M + G W+ R R + EL E C ++LS L ++ D+
Sbjct: 602 ICANPDKTVSQMLI------EGEWKIRLRRSLGQAELNEWCELHSKLSTVQLTSE-EDKM 654
Query: 927 VWRLHSS 933
W+L S
Sbjct: 655 FWKLTKS 661
>UniRef100_Q7FS87 Putative glycerol 3-phosphate permease [Zea mays]
Length = 1279
Score = 197 bits (500), Expect = 2e-48
Identities = 152/546 (27%), Positives = 249/546 (44%), Gaps = 74/546 (13%)
Query: 531 MVVGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYL 590
+V+ +V +Q FIK R I D + A E + ++ KE+++ K+DFEK +D ++ L
Sbjct: 148 LVITKLVHQNQYGFIKDRSIQDCLPWAFEYISLCHKSRKEMVILKLDFEKTFDKLEHAAL 207
Query: 591 DEVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAE 650
E++ F W WI + + ++ VL+NG P FH RG+RQGDPLSP LF+LAA+
Sbjct: 208 LEILRHKGFGNKWIHWISMILGSGSSEVLLNGVPGKRFHCRRGVRQGDPLSPLLFVLAAD 267
Query: 651 GLNVMMSTVGSNGLF---APYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLL 707
L +++ G+ P G + P+ +Q+ DDT+LI + +K +L
Sbjct: 268 LLQTIVNKACKGGILRLPLPNNCGPDFPI----IQYVDDTILIMEACPRQLFFLKAMLNS 323
Query: 708 FEDVSGLKVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWH 767
F D +GL VN+HKS + +N+++ + A HC+ G + F YLGLP+G KL +
Sbjct: 324 FADSTGLHVNYHKSNIYPINVSDQKMEILANTFHCKIGSLSFTYLGLPLGIQRPKLGAFL 383
Query: 768 PLVERIRRRL---------SGVLE------------------------------DRESLW 788
PL+++I +RL +G LE R LW
Sbjct: 384 PLIQKIEKRLASTSIFLSQAGRLEMVNAVFSSLPTYYMCTLKLPKMVIRHIDRLRRHCLW 443
Query: 789 ------NVVLCAKYGEVGGRVQFSEGVGPIWWRQLNQIRCGVGLAENAWLVDNIVR-KVG 841
++L V + + VG WWR + + +I R ++
Sbjct: 444 RELPPPGLILFGPITTV--TLPSGKPVGSFWWRD---------VLKTLEPFKSIARVEIE 492
Query: 842 DGSTTLFWEDPWLLNVPLAVSFSRLFELAENKGVSVREMFLLGWGADGGAWRWRRRLFAW 901
DG TTL W D W +P + + L+ + K ++V + L+ + F
Sbjct: 493 DGKTTLLWHDNW-DGIPKSTQYLELWSFSACKDITVLQARLVD-SQELFHAPLSTEAFVQ 550
Query: 902 EEELVEECVARLSNFVLQADLSDRWVWRLHSSQLYTVQSAYSYLTAVDSNITAEFDRFLW 961
+EL +C+ L + + D+W S L++ Q Y++L + + T +LW
Sbjct: 551 LQEL-NDCLDNLPH----NNYKDKWCCSTPSG-LFSSQKIYAHL--MGNQWTHPIFSWLW 602
Query: 962 LKAVPLKVNIFVWRLFLNRLATKDNLRKRNVLEATNVSSGALCGKEEERDHLFFQCDYYG 1021
K +F W L +RL T+ LR+R++ + + EE HLFF+C++
Sbjct: 603 KSKCQPKHKVFFWLLLKDRLNTRAFLRQRSMFLDSYSCDNCILQVEETTIHLFFRCNFAK 662
Query: 1022 RLWLLI 1027
R W LI
Sbjct: 663 RCWSLI 668
>UniRef100_Q75GM6 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1614
Score = 179 bits (455), Expect = 3e-43
Identities = 93/228 (40%), Positives = 137/228 (59%), Gaps = 1/228 (0%)
Query: 532 VVGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLD 591
+ V+ +Q+AFI GR IL+G +I +E++ + R N E ++ K+DFEKAYD V W +L
Sbjct: 1158 IARDVISPTQTAFIPGRFILEGCVIIHEVLHEMNRKNLEGIILKIDFEKAYDKVSWDFLI 1217
Query: 592 EVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEG 651
EVM + FP W +WI CV + +NG TD F RGLRQGDPLSP LF L ++
Sbjct: 1218 EVMVRKGFPSKWVNWIKTCVMGGRVCININGERTDFFRTFRGLRQGDPLSPLLFNLISDA 1277
Query: 652 LNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDV 711
L M+ + G+ + V P ++HLQ+ADDT+L + A K +L FE++
Sbjct: 1278 LAAMLDSAKREGVLSG-LVPDIFPGGITHLQYADDTVLFVANDDKQIVATKFILYCFEEM 1336
Query: 712 SGLKVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIGGD 759
+ LKVN+HKS F + ++++ + A++ +C G+ P YLGLPIG D
Sbjct: 1337 AVLKVNYHKSEIFTLGLSDNDTNRVAMMFNCPVGQFPMKYLGLPIGPD 1384
Score = 44.7 bits (104), Expect = 0.016
Identities = 30/96 (31%), Positives = 42/96 (43%), Gaps = 8/96 (8%)
Query: 785 ESLWNVVLCAKYGEVGGRVQFSEGVGPIWWRQLNQIRCGVGLAENAWLVDNIVRKVGDGS 844
+SL +L KY + GG Q E +W+ L IR WL V +VGDGS
Sbjct: 1503 DSLCIELLRRKYLQDGGFFQCKERYASQFWKGLLNIR--------RWLSLGSVWQVGDGS 1554
Query: 845 TTLFWEDPWLLNVPLAVSFSRLFELAENKGVSVREM 880
FW D W PL F +F + + + V ++
Sbjct: 1555 HISFWRDVWWGQCPLRTLFPAIFAVCNQQDILVSQV 1590
>UniRef100_Q7Y1F8 Putative reverse transcriptase [Oryza sativa]
Length = 1014
Score = 172 bits (435), Expect = 7e-41
Identities = 152/568 (26%), Positives = 237/568 (40%), Gaps = 61/568 (10%)
Query: 533 VGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDE 592
+ V+ +QSAFI R I D L + L R+ + L FK+D KA+DSV W YL
Sbjct: 474 INKVISQAQSAFIITRCIQDNFLYVSNLAKKMHRSKQPTLFFKLDIAKAFDSVSWEYLLT 533
Query: 593 VMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGL 652
++ KM F WR W+ ++T+++ V +NG + RGLRQGDPLSP+LF+LA + L
Sbjct: 534 LLEKMGFSTRWRDWVALILSTSSSRVALNGIEGERIDHHRGLRQGDPLSPYLFILAMDPL 593
Query: 653 NVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDVS 712
+ ++ GL + A+ +ADDT + S +V ++ ++L F +
Sbjct: 594 HRLLKIATDEGLLSDL---ADRSARFRCSLYADDTAIFIKPSRQDVDSLISILNNFGSAT 650
Query: 713 GLKVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIG-GDPRKLHFWHPLVE 771
GL VN HKS ++ E L E G P YLG+P+ RK+H + L++
Sbjct: 651 GLLVNLHKSSVIPISCGEIDLDEMLHNFTGPRGTFPMRYLGMPLMISRLRKVHRQY-LID 709
Query: 772 RIRRRLSGVLEDRESLWNVVLCAKYGEVGGRVQFSEG----VGPIWWRQLNQIRCGVGL- 826
+I+ RL+ + S KY +G G + +W+ + + +G+
Sbjct: 710 KIKARLASWKKKLLSAGGRREPLKYDGLGVSNLTKMGRALRLRWLWYEWTSPDKPWIGMP 769
Query: 827 -----AENAWLVDNIVRKVGDGSTTLFWEDPWLLNVPLAVSFSRLFELAENKGVSVREMF 881
+N + +GDG LFWE PW + L LF A K +V E
Sbjct: 770 TPCDEQDNELFAASTSVTIGDGRKALFWESPWACSSTLKAMAPNLFHHARKKKRTVAEAL 829
Query: 882 LLGWGADGGAWRWRRRLFAWEEELVEECVARLSNFVLQADLSDRWVWRLHSSQLYTVQSA 941
D + L ++ E + S L D+ D W+ +S +YT +SA
Sbjct: 830 QDNKWIDDIRYNLTTVLVTDFFKVFE--IIWTSGITLTQDIEDNIRWKWTTSGVYTTKSA 887
Query: 942 YSYLTAVDSNITAEFDRFLWLKAVPLKVNIFVWRLFLNRLATKDNLRKRNVLEATNVSSG 1001
Y + +W T D L++R E N
Sbjct: 888 Y---------------QIIW---------------------TADRLQQR---EWPNNYFC 908
Query: 1002 ALCGKE-EERDHLFFQCDYYGRLWLLISNWLGFVTVFHDN----LYTHANQFCALGGFSK 1056
LC + E HLF++C ++W I+ LG + N + K
Sbjct: 909 QLCFRNLETAHHLFYECPITRKIWDAIATRLGMPQLRPQNWSKTTILEDRWTQTVSTMPK 968
Query: 1057 NYMKAFTIIWISVLFTIWKDRNRRIFQN 1084
K I I + + IWK+RN +F+N
Sbjct: 969 EKRKGAKSILILLSWEIWKERNNMVFKN 996
>UniRef100_Q7XKK3 OSJNBa0038O10.4 protein [Oryza sativa]
Length = 1045
Score = 168 bits (425), Expect = 9e-40
Identities = 147/571 (25%), Positives = 241/571 (41%), Gaps = 72/571 (12%)
Query: 535 SVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDEVM 594
S+V +QSAFIKGR I D L + R N+ +L+FK+D KA+DS+ W YL ++
Sbjct: 493 SLVSINQSAFIKGRSIHDNFLFVRNMARRFHRTNRPMLLFKLDITKAFDSIRWDYLMALL 552
Query: 595 AKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGLNV 654
++ FP WR W+ + T+T+ VL+NG P RGLRQGDPLSP LF+LA L
Sbjct: 553 QRLGFPVKWRYWLGALLYTSTSQVLLNGIPGQRITHGRGLRQGDPLSPLLFVLAINPLQR 612
Query: 655 MMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDVSGL 714
+++ G + + + +ADD + + +V+A+ +L F +GL
Sbjct: 613 LLTKATDLGSLSKL---RGIMPRLRTSMYADDAAIFVNPTRGDVQALTEILQRFGTATGL 669
Query: 715 KVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLP-IGGDPRKLHFWHPLVERI 773
NF KS + N L + + P YLGLP I G RK+ H + ++I
Sbjct: 670 VTNFQKSQVAAIKCNNIDLDDILEGVPAVRACFPIKYLGLPLILGRMRKVDVQH-IFDKI 728
Query: 774 RRRLSGVLEDRESLW---NVVLCAKYGEVG--GRVQFSEGVGPIW----WRQLNQIRCGV 824
R++G W N+ L + G +G +F + W W+ N+ G+
Sbjct: 729 TGRITG--------WRGKNMGLAGRQGGLGVLNLEKFMRALRLRWLWHEWKDPNKPWVGL 780
Query: 825 GL----AENAWLVDNIVRKVGDGSTTLFWEDPWLLNVPLAVSFSRLFELAENKGVSVREM 880
+ + + + +GDG+ FW+ W+ ++ +++ + ++R+
Sbjct: 781 EVPCDEVDKSLFAASTKITIGDGNIARFWDSAWIDGRRPKDLMPLVYAISKKRKKTLRQ- 839
Query: 881 FLLGWGADGGAWRWRRRLF---AWEEELVEECVA---RLSNFVLQADLSDRWVWRLHSSQ 934
G + AW L LVE+ VA + N L+ + SD+ W+
Sbjct: 840 -----GKENDAWVGDLALETNPVITVPLVEQLVALWTAIRNVQLEEEESDQIAWKFTPHG 894
Query: 935 LYTVQSAYSYLTAVDSNITAEFDRFLWLKAVPLKVNIFVWRLFLNRLATKDNLRKRNVLE 994
Y+ SAY + T FD +W P K + W + NR+ T D L R
Sbjct: 895 HYSASSAYKAQCVGAPDTT--FDSLIWKVWAPGKCKLHAWLIIQNRVWTSDRLATRG--- 949
Query: 995 ATNVSSGALCGKEEERDHLFFQCDYYGRLWLLISNWLGFVTVFHDNLYTHANQFCALGGF 1054
GR+ + LG D ++ G
Sbjct: 950 ----------------------WQNNGRVPPARTPTLGVAQSVKD----WWEMLASVSGV 983
Query: 1055 SKNYMKAFTIIWISVLFTIWKDRNRRIFQNQ 1085
K ++ ++ ++ +WK+RNRRIF ++
Sbjct: 984 PKKGLRTLILL---IVLEVWKERNRRIFDHK 1011
>UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana]
Length = 1662
Score = 167 bits (424), Expect = 1e-39
Identities = 165/613 (26%), Positives = 262/613 (41%), Gaps = 109/613 (17%)
Query: 535 SVVFASQSAFIKGRQILDGILIANELVDDAK---RNNKELLMFKVDFEKAYDSVDWRYLD 591
++V SQ+AFI GR + D ++IA+E++ K R ++ + K D KAYD V+W +L+
Sbjct: 913 AIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLE 972
Query: 592 EVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEG 651
M F W WIM V + SVLVNG P +RG+RQGDPLSP+LF+L A+
Sbjct: 973 TTMRLFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADI 1032
Query: 652 LNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDV 711
LN ++ + G +G VP V+HLQFADD+L + N +A+K V ++E
Sbjct: 1033 LNHLIKNRVAEGDIRGIRIGNGVP-GVTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYY 1091
Query: 712 SGLKVNFHKSMF-FGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWHPLV 770
SG K+N KSM FG ++ + + ++ + YLGLP +K ++ ++
Sbjct: 1092 SGQKINMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYII 1151
Query: 771 ERIRRRLS-----------------------------------GVLEDRESL-----WNV 790
ER+++R S ++ + E+L W
Sbjct: 1152 ERVKKRTSSWSAKYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNIVSEIEALLMNFWWE- 1210
Query: 791 VLCAKYGEV----GGRVQFSEGVGPIWWRQLNQIRCGVGLAENAW-LVDNIVRKVGDGST 845
AK E+ R+Q+S+ G + +R L + + LA+ W +++N
Sbjct: 1211 -KNAKKREIPWIAWKRLQYSKKEGGLGFRDLAKFNDAL-LAKQVWRMINNPNSLFARIMK 1268
Query: 846 TLFWEDPWLLNVPLA--VSFSRLFELAENKGVSVREMFLLGWGADGGAWRWRRRLFAWEE 903
++ + +L+ S+ LA + F++G G G++R+ W
Sbjct: 1269 ARYFREDSILDAKRQRYQSYGWTSMLAGLDVIKKGSRFIVGDGKT-GSYRY------WNA 1321
Query: 904 ELVEECVARLSN-FVLQADLS-----DRWVWRLHSSQLYTVQSAYSYLTAVDSNITAEFD 957
L+ + V+ + FV+ LS D+ VW SS YT
Sbjct: 1322 HLISQLVSPDDHRFVMNHHLSRIVHQDKLVWNYSSSGDYT-------------------- 1361
Query: 958 RFLWLKAVPLKVNIFVWRLFLNRLATKDNLRKRNVLEATNVSSGALCGKEEER-DHLFFQ 1016
LW + K+ +WR L T+ L R + + C EEE +H+ F
Sbjct: 1362 --LWKLPIIPKIKYMLWRTISKALPTRSRLLTRGMDIDPHCPR---CPTEEETINHVLFT 1416
Query: 1017 CDYYGRLWLLIS-NWLGFVTVFHDNLYTHANQFCALGGFSKNYMKA------FTIIWISV 1069
C Y +W L + WL T D T N + FS N + F +IW
Sbjct: 1417 CPYAASIWGLSNFPWLPGHTFSQD---TEENISFLINSFSNNTLNTEQRLAPFWLIW--- 1470
Query: 1070 LFTIWKDRNRRIF 1082
+WK RN +F
Sbjct: 1471 --RLWKARNNLVF 1481
>UniRef100_Q8LMT3 Putative retroelement [Oryza sativa]
Length = 764
Score = 167 bits (423), Expect = 2e-39
Identities = 88/225 (39%), Positives = 132/225 (58%), Gaps = 7/225 (3%)
Query: 532 VVGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLD 591
+ V+ SQ+AFI GRQ LDG++I +E++ + K+ K ++FK+DFEKAYD V W +L
Sbjct: 75 IAQKVIGESQTAFIPGRQNLDGVVILHEVLHELKKEKKSGIIFKLDFEKAYDKVQWSFLF 134
Query: 592 EVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEG 651
+V+ K F W W+ +V +NG D F RG+RQGDPLSP LF L A+
Sbjct: 135 DVLHKKGFSDRWIQWVKMATIGGKMAVNINGEVKDFFKTYRGVRQGDPLSPLLFNLVADA 194
Query: 652 LNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDV 711
L+ M++ A V VP ++HLQ+ADDT+L + N+ +K +L +E +
Sbjct: 195 LSEMLNN-------AKQAVPHLVPGGLTHLQYADDTILFMTNTEENIVTVKFLLYCYEAM 247
Query: 712 SGLKVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPI 756
SGLK+N+ KS + +E A + +C+ G++PF YLG+PI
Sbjct: 248 SGLKINYQKSEIMVIGGDEMETQRVADLFNCQAGKMPFTYLGIPI 292
Score = 82.8 bits (203), Expect = 5e-14
Identities = 71/303 (23%), Positives = 124/303 (40%), Gaps = 33/303 (10%)
Query: 790 VVLCAKY-GEVGGRVQFSEGVGPIWWRQLNQIRCGVGLAENAWLVDNIVRKVGDGSTTLF 848
V+L KY + GG Q +W+ L++++ W+ KVG+G T F
Sbjct: 420 VLLRNKYMKDGGGFFQSKAEESSQFWKGLHEVK--------KWMDLGSSYKVGNGKATNF 471
Query: 849 WEDPWLLNVPLAVSFSRLFELAENKGVSVREMFLLG-WGADGGAWRWRRRLFAWEEELVE 907
W D W+ PL + ++ + +K +V +M L G W + R L W +
Sbjct: 472 WSDVWIGETPLKTQYPNIYRMCADKEKTVSQMCLEGDWYIELRRSLGERDLNEWND---- 527
Query: 908 ECVARLSNFVLQADLS---DRWVWRLHSSQLYTVQSAYSYLTAVDSNITAEFDRFLWLKA 964
L N + L D +W+L + Y ++ Y L+ + + + LW +
Sbjct: 528 -----LHNTPREIHLKEERDCIIWKLTKNGFYKAKTLYQALSF--GGVKDKVMQDLWRSS 580
Query: 965 VPLKVNIFVWRLFLNRLATKDNLRKRNVLEATNVSSGALCGKEEERDHLFFQCDYYGRLW 1024
+PLKV I W + R+ L+K + N LCG+ E+ DHL F+C +W
Sbjct: 581 IPLKVKILFWLMLKGRIQAAGQLKKMKWSGSPNCK---LCGQTEDVDHLMFRCHISQFMW 637
Query: 1025 LLISNWLGFVTVFHDNLYTHANQFCALGGFSKNYMKAFTIIWISV-LFTIWKDRNRRIFQ 1083
+ G+ DN+ + K+ K+ + ++ + IW RN +F
Sbjct: 638 CCYRDAFGW-----DNIPSSREDLLEKLNIQKDGQKSVLLACLAAGTWAIWLMRNDWVFN 692
Query: 1084 NQI 1086
++
Sbjct: 693 GKL 695
>UniRef100_Q60EA2 Hypothetical protein OSJNBb0048K05.4 [Oryza sativa]
Length = 1275
Score = 160 bits (405), Expect = 2e-37
Identities = 86/246 (34%), Positives = 138/246 (55%), Gaps = 1/246 (0%)
Query: 532 VVGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLD 591
V SV+ +Q+AFI GR IL+G +I +E++ + + ++ ++ K+DFEK YD V W +L
Sbjct: 884 VADSVISPTQTAFIPGRFILEGCVILHEVLHEMRTKAQKGVILKIDFEKTYDRVSWDFLF 943
Query: 592 EVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEG 651
E +++ FP+ W +W+ CV V +NG +D F RGLRQGDPLSP LF L ++
Sbjct: 944 EFLSRKGFPKKWINWVKACVMRGRVCVNINGERSDFFRTYRGLRQGDPLSPLLFNLVSDA 1003
Query: 652 LNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDV 711
L + G+ + P ++HLQ+ADDT++ + A K +L FE++
Sbjct: 1004 LAAIFDNAKRAGILTGLVLEI-FPNGITHLQYADDTVIFIPFDTNQIVASKFLLYCFEEM 1062
Query: 712 SGLKVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWHPLVE 771
+G+K+N+HKS F V + A +++ G P YLGLPI + + L+E
Sbjct: 1063 AGMKINYHKSEVFTVGLEIDETERVAHMLNFPIGTFPMKYLGLPISPEKILTQDLNFLLE 1122
Query: 772 RIRRRL 777
++ +RL
Sbjct: 1123 KMEKRL 1128
>UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis thaliana]
Length = 1294
Score = 152 bits (384), Expect = 5e-35
Identities = 90/248 (36%), Positives = 138/248 (55%), Gaps = 5/248 (2%)
Query: 535 SVVFASQSAFIKGRQILDGILIANELVDDAK---RNNKELLMFKVDFEKAYDSVDWRYLD 591
++V SQ+AFI GR + D ++IA+E++ K R ++ + K D KAYD V+W +L+
Sbjct: 893 AIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLE 952
Query: 592 EVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEG 651
M F W WIM V + SVLVNG P +RG+RQGDPLSP+LF+L A+
Sbjct: 953 TTMRLFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADI 1012
Query: 652 LNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDV 711
LN ++ + G +G VP V+HLQFADD+L + N +A+K V ++E
Sbjct: 1013 LNHLIKNRVAEGDIRGIRIGNGVP-GVTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYY 1071
Query: 712 SGLKVNFHKSMF-FGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWHPLV 770
SG K+N KSM FG ++ + + ++ + YLGLP +K ++ ++
Sbjct: 1072 SGQKINMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYII 1131
Query: 771 ERIRRRLS 778
ER+++R S
Sbjct: 1132 ERVKKRTS 1139
>UniRef100_Q84T47 Putative reverse transcriptase [Oryza sativa]
Length = 746
Score = 152 bits (384), Expect = 5e-35
Identities = 86/247 (34%), Positives = 142/247 (56%), Gaps = 6/247 (2%)
Query: 536 VVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDEVMA 595
+V +Q FIK R I D + A E + +++ + +++ K+DFEKA+DSV+ + ++
Sbjct: 291 LVHKNQYGFIKSRTIQDCLAWAFEFIHQCQQSRRPIVIIKIDFEKAFDSVEHNAILAMLK 350
Query: 596 KMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGLNVM 655
F W E ++T+++SVL+NG P F +RG++QGDP+SP LF+LAA+ L M
Sbjct: 351 AKGFDDTWIHCTKEILSTSSSSVLLNGVPGKFFDCKRGVKQGDPISPLLFVLAADLLQSM 410
Query: 656 MSTVGSNGLFA-PY-TVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDVSG 713
+ SNG+ + P T + P+ +Q+ADDTL+I + +K +L + + +G
Sbjct: 411 TNRALSNGILSLPIPTHDLKFPI----IQYADDTLIIMKACQKEIFNLKGILQSYSESTG 466
Query: 714 LKVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWHPLVERI 773
LK+N+ KS +N+ E + C+ G +PF YLGLP+G K+ + PL++R+
Sbjct: 467 LKINYQKSCMIHINVLEEDKDILSKTFGCQSGNLPFTYLGLPVGTTKPKIIDFAPLIDRV 526
Query: 774 RRRLSGV 780
RRL V
Sbjct: 527 ERRLPAV 533
>UniRef100_Q7XXA4 OSJNBa0019G23.12 protein [Oryza sativa]
Length = 1140
Score = 152 bits (383), Expect = 7e-35
Identities = 88/250 (35%), Positives = 133/250 (53%), Gaps = 7/250 (2%)
Query: 532 VVGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLD 591
V ++V +Q+AF++GR ILDG+ I +E V + R ++FK+DFEKAYD V W +L
Sbjct: 764 VADNLVNPTQTAFMRGRNILDGVAIIHETVHELHRKKLNGVIFKIDFEKAYDKVKWPFLL 823
Query: 592 EVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEG 651
+ + F W SWI + + +V VN F ++GL+QGDPLSP LF +
Sbjct: 824 QTLRMKGFSPKWISWIKSFIVGGSVAVKVNDDVGPFFQTKKGLQQGDPLSPILFNFIVDM 883
Query: 652 LNVMMSTVGSNGL---FAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLF 708
L +++ + G P+ + +S LQ+ DD +L + MK+VLL F
Sbjct: 884 LATLINRAKTQGQVDGLIPHLIDG----GLSILQYVDDIVLFMNHDLEKAQNMKSVLLAF 939
Query: 709 EDVSGLKVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWHP 768
E +SGLK+NFHKS + + + A + C+ G PF YLG+PI + W
Sbjct: 940 EQLSGLKINFHKSELYCFGEALEYRDQYAQLFGCQVGNFPFRYLGIPIHCRKLRNAEWKE 999
Query: 769 LVERIRRRLS 778
+VER ++LS
Sbjct: 1000 VVERFEKKLS 1009
>UniRef100_Q8LJX1 Putative reverse transcriptase [Sorghum bicolor]
Length = 1998
Score = 152 bits (383), Expect = 7e-35
Identities = 84/253 (33%), Positives = 135/253 (53%), Gaps = 7/253 (2%)
Query: 531 MVVGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYL 590
M + +V +Q FI+ R I D + + E + + K +++ K+DFEKA+D ++ + +
Sbjct: 1341 MKITDLVHKNQYGFIRQRTIQDCLAWSFEYLHLCHHSRKAIVIIKLDFEKAFDKMNHKAM 1400
Query: 591 DEVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAE 650
+M F + W +W+ T+ T++VL+NG P H RG+RQGDPLSP LF+LAA+
Sbjct: 1401 LTIMKAKGFGQKWLNWMKSIFTSGTSNVLLNGTPGKTIHCLRGVRQGDPLSPLLFVLAAD 1460
Query: 651 GLNVMMSTVGSNGLF---APYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLL 707
L +++ L P + P+ +Q+ADDTL+I + +K+++
Sbjct: 1461 FLQSLINKAKCMDLLKLPIPMQTNQDFPI----IQYADDTLIIAEGDTRQLLILKSIINT 1516
Query: 708 FEDVSGLKVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWH 767
F + +GLKVN KSM +N++E L A C G PF YLGLP+G + +
Sbjct: 1517 FSEATGLKVNLQKSMMLPINMDEERLDTLARTFGCSKGSFPFTYLGLPLGITKPTIQDYQ 1576
Query: 768 PLVERIRRRLSGV 780
PL+ + RL V
Sbjct: 1577 PLINKCEARLGSV 1589
Score = 61.2 bits (147), Expect = 2e-07
Identities = 63/314 (20%), Positives = 133/314 (42%), Gaps = 34/314 (10%)
Query: 773 IRRRLSGVLEDRESLWNVVLCAKY---GEVGGRVQFSEGVGPIWWRQLNQIRCGVGLAEN 829
+ + L +++ W ++ K+ G++ G+++ G WWR + ++ +
Sbjct: 1676 LMKNLDKFFNKKDTPWVQMIWDKHYAKGKLPGQIR----KGSFWWRDILKLL-------D 1724
Query: 830 AWLVDNIVRKVGDGSTTLFWEDPWLLNVPLAVSFSRLFELAENKGVSVREMFLLGWGADG 889
+ ++++ G G + +FW+D W + L + LF ++K +S E+ G +
Sbjct: 1725 PYKEMSLIQMKG-GESIVFWQDKWGMQ-KLRLEMPELFSFVQHKFLSAAEVL----GQED 1778
Query: 890 GAWRWRRRLFAWEEELVEECVARLSNFVLQADLSDRWVWRLHSSQLYTVQSAYSYLTAVD 949
+ + +++ R+ N + D+W ++ ++ ++ AYS L
Sbjct: 1779 FTRLLQLPISETAFHQLQQLTTRIDNLERTQE-QDQWKYKWGNN--FSATKAYSELMGHQ 1835
Query: 950 SNITAEFDRFLWLKAVPLKVNIFVWRLFLNRLATKDNLRKRNVLEATNVSSGALCGKEEE 1009
E +++W K +F W L +RL+T + L+++N+ + L EE
Sbjct: 1836 Q--IHEVHKWIWKSFCQPKHKVFFWLLIKDRLSTCNILKRKNMQLQSYNCVLCLQNSEET 1893
Query: 1010 RDHLFFQCDYYGRLWLLISNWLGFVTVFHDNLYTHANQFCALGGFSKNYMKAFTIIWISV 1069
HLF C Y + W L+ + + F + + H GF+ + TI+ +
Sbjct: 1894 TSHLFLHCAYARQCWQLLDMDIPPNSDFPE-ITDHFKH-----GFNNQFFMVTTIL---L 1944
Query: 1070 LFTIWKDRNRRIFQ 1083
+ IW RN IF+
Sbjct: 1945 CWAIWSTRNNLIFR 1958
>UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa]
Length = 1509
Score = 151 bits (382), Expect = 9e-35
Identities = 94/253 (37%), Positives = 142/253 (55%), Gaps = 7/253 (2%)
Query: 532 VVGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELL---MFKVDFEKAYDSVDWR 588
++ V+ +QSAF+ GR I D ILIA E+ + + FK+D KAYD V+W
Sbjct: 729 ILPDVISPAQSAFVPGRLISDNILIAYEMTHYMRNKRSGQVGYAAFKLDMSKAYDRVEWS 788
Query: 589 YLDEVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLA 648
+L ++M K+ F W + IM+CV+T T + VNG ++ F ERGLRQGDPLSP+LFLL
Sbjct: 789 FLHDMMLKLGFHTDWVNLIMKCVSTVTYRIRVNGELSESFSPERGLRQGDPLSPYLFLLC 848
Query: 649 AEGLNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLF 708
AEG + ++S G + P SVSHL FADD+L++ + + ++T+L ++
Sbjct: 849 AEGFSALLSKTEEEGRLHGIRICQGAP-SVSHLLFADDSLILCRANGGEAQQLQTILQIY 907
Query: 709 EDVSGLKVNFHKSMFFGVNINESWLHEAAVV--MHCRHGRIPFIYLGLPIGGDPRKLHFW 766
E+ SG +N KS + N S L + AV+ ++ + YLGLP+ + +
Sbjct: 908 EECSGQVINKDKSAVM-FSPNTSSLEKGAVMAALNMQRETTNEKYLGLPVFVGRSRTKIF 966
Query: 767 HPLVERIRRRLSG 779
L ERI +R+ G
Sbjct: 967 SYLKERIWQRIQG 979
Score = 78.6 bits (192), Expect = 1e-12
Identities = 69/267 (25%), Positives = 114/267 (41%), Gaps = 50/267 (18%)
Query: 780 VLEDRESLWNVVLCAKYGEVGG--RVQFSEGVGPIWWRQLNQIRCGVGLAENAWLVDNIV 837
+++D +SL + VL AKY +G R + + V W I+ G+ + +N ++
Sbjct: 1074 LIQDPDSLCSRVLRAKYFPLGDCFRPKQTSNVSYTW----RSIQKGLRVLQNG-----MI 1124
Query: 838 RKVGDGSTTLFWEDPWLLNVPLAVSFSRLFELAENKGVSVREMFLLGWGADGGAWRWRRR 897
+VGDGS W DPW +P S + N V E+ G
Sbjct: 1125 WRVGDGSKINIWADPW---IPRGWSRKPMTPRGANLVTKVEELIDPYTGT---------- 1171
Query: 898 LFAWEEELV-----EECVARLSNFVLQADLSDRWVWRLHSSQLYTVQSAYSYLTAVDSNI 952
W+E+L+ EE VA + + + ++ D W + +TV+SAY ++
Sbjct: 1172 ---WDEDLLSQTFWEEDVAAIKSIPVHVEMEDVLAWHFDARGCFTVKSAYKVQREMERRA 1228
Query: 953 TA--------------EFDRFLWLKAVPLKVNIFVWRLFLNRLATKDNLRKRNVLEATNV 998
+ +F + LW VP K+ F+WR+ N LA + NL R + T
Sbjct: 1229 SRNGCPGVSNWESGDDDFWKKLWKLGVPGKIKHFLWRMCHNTLALRANLHHRGMDVDTRC 1288
Query: 999 SSGALCGK-EEERDHLFFQCDYYGRLW 1024
+CG+ E+ HLFF+C ++W
Sbjct: 1289 ---VMCGRYNEDAGHLFFKCKPVKKVW 1312
>UniRef100_Q8S5Z6 Putative reverse transcriptase [Oryza sativa]
Length = 1259
Score = 150 bits (380), Expect = 2e-34
Identities = 83/229 (36%), Positives = 134/229 (58%), Gaps = 1/229 (0%)
Query: 549 QILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDEVMAKMNFPRLWRSWIM 608
++ G +I +E + + KR ++ ++FK+DFEKAYD V+W +L EVM K NF SW+
Sbjct: 974 RVARGCVILHETLHELKRKGEKGVIFKIDFEKAYDRVNWGFLYEVMKKNNFSPELISWVK 1033
Query: 609 ECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGLNVMMSTVGSNGLFAPY 668
+ A S+ + G F RGLRQGDPLSP LF L + L V++ G+
Sbjct: 1034 KFNEGAKVSINIYGDQGPFFRTFRGLRQGDPLSPLLFNLVGDALAVILEKAKERGILKG- 1092
Query: 669 TVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDVSGLKVNFHKSMFFGVNI 728
V VP +SHLQ+ADDT+L NVR +K +L FE++SG+K+N+ KS + + +
Sbjct: 1093 LVPDLVPRGLSHLQYADDTILFIKVGDDNVRVLKFLLFCFEEMSGMKINYQKSEVYVLGV 1152
Query: 729 NESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWHPLVERIRRRL 777
++ A +++C+ G +PF YLGLP+ + + + +++++ +RL
Sbjct: 1153 DKEEEIRIANMLNCKVGTMPFTYLGLPMHCEKVGMKDFRLIIQKVEKRL 1201
>UniRef100_Q7XFR2 Putative retroelement [Oryza sativa]
Length = 1265
Score = 150 bits (380), Expect = 2e-34
Identities = 83/229 (36%), Positives = 134/229 (58%), Gaps = 1/229 (0%)
Query: 549 QILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDEVMAKMNFPRLWRSWIM 608
++ G +I +E + + KR ++ ++FK+DFEKAYD V+W +L EVM K NF SW+
Sbjct: 980 RVARGCVILHETLHELKRKGEKGVIFKIDFEKAYDRVNWGFLYEVMKKNNFSPELISWVK 1039
Query: 609 ECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGLNVMMSTVGSNGLFAPY 668
+ A S+ + G F RGLRQGDPLSP LF L + L V++ G+
Sbjct: 1040 KFNEGAKVSINIYGDQGPFFRTFRGLRQGDPLSPLLFNLVGDALAVILEKAKERGILKG- 1098
Query: 669 TVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDVSGLKVNFHKSMFFGVNI 728
V VP +SHLQ+ADDT+L NVR +K +L FE++SG+K+N+ KS + + +
Sbjct: 1099 LVPDLVPRGLSHLQYADDTILFIKVGDDNVRVLKFLLFCFEEMSGMKINYQKSEVYVLGV 1158
Query: 729 NESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWHPLVERIRRRL 777
++ A +++C+ G +PF YLGLP+ + + + +++++ +RL
Sbjct: 1159 DKEEEIRIANMLNCKVGTMPFTYLGLPMHCEKVGMKDFRLIIQKVEKRL 1207
>UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1715
Score = 150 bits (380), Expect = 2e-34
Identities = 90/254 (35%), Positives = 137/254 (53%), Gaps = 11/254 (4%)
Query: 533 VGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNK---ELLMFKVDFEKAYDSVDWRY 589
+G V+ SQ+AF+ G+ I D +L+A+EL+ K + + K D KAYD V+W +
Sbjct: 891 LGDVISDSQAAFVPGQNISDNVLVAHELLHSLKSRRECQSGYVAVKTDISKAYDRVEWNF 950
Query: 590 LDEVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAA 649
L++VM ++ F W WIM CVT+ + VL+NG P + RG+RQGDPLSP+LFL A
Sbjct: 951 LEKVMIQLGFAPRWVKWIMTCVTSVSYEVLINGSPYGKIFPSRGIRQGDPLSPYLFLFCA 1010
Query: 650 EGLNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFE 709
E L+ M+ N + + +++SHL FADD+L S N+ + + +E
Sbjct: 1011 EVLSNMLRKAEVNKQIHGMKITKDC-LAISHLLFADDSLFFCRASNQNIEQLALIFKKYE 1069
Query: 710 DVSGLKVNFHK-SMFFGVNI---NESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHF 765
+ SG K+N+ K S+ FG I LH + + R G YLGLP RK+
Sbjct: 1070 EASGQKINYAKSSIIFGQKIPTMRRQRLHRLLGIDNVRGGG---KYLGLPEQLGRRKVEL 1126
Query: 766 WHPLVERIRRRLSG 779
+ +V +++ R G
Sbjct: 1127 FEYIVTKVKERTEG 1140
Score = 71.2 bits (173), Expect = 2e-10
Identities = 63/270 (23%), Positives = 107/270 (39%), Gaps = 55/270 (20%)
Query: 839 KVGDGSTTLFWEDPWLLNVPLAVSFSRLFELAENKGVSVREMFLLGWGADGGAWRWRRRL 898
++GDG TT WEDPWL +P + + + + + V ++ WR
Sbjct: 1269 RLGDGQTTKIWEDPWLPTLPPRPARGPILD----EDMKVADL-------------WRENK 1311
Query: 899 FAWEEELVEECV----------ARLSNFVLQADLSDRWVWRLHSSQLYTVQSAYSYLTAV 948
W+ + E + LSN+ + D + W + YTV+S Y T V
Sbjct: 1312 REWDPVIFEGVLNPEDQQLAKSLYLSNYAAR----DSYKWAYTRNTQYTVRSGYWVATHV 1367
Query: 949 DSNITAE-----------FDRFLWLKAVPLKVNIFVWRLFLNRLATKDNLRKRNVLEATN 997
N+T E + +W + K+ F+WR L+T LR RN+ +
Sbjct: 1368 --NLTEEEIINPLEGDVPLKQEIWRLKITPKIKHFIWRCLSGALSTTTQLRNRNI--PAD 1423
Query: 998 VSSGALCGKEEERDHLFFQCDYYGRLWLLI----SNWLGFVTVFHDNLYTHANQFCALGG 1053
+ C +E +H+ F C Y +W SN L F +N+ + G
Sbjct: 1424 PTCQRCCNADETINHIIFTCSYAQVVWRSANFSGSNRLCFTDNLEENI-----RLILQGK 1478
Query: 1054 FSKNYMKAFTIIWISVLFTIWKDRNRRIFQ 1083
++N ++ +++ +WK RN +FQ
Sbjct: 1479 KNQNLPILNGLMPFWIMWRLWKSRNEYLFQ 1508
>UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa]
Length = 1026
Score = 150 bits (378), Expect = 3e-34
Identities = 79/221 (35%), Positives = 126/221 (56%), Gaps = 2/221 (0%)
Query: 536 VVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDEVMA 595
++ Q+AF+K R I++G++I +E+++ + + ++FKVDFEKAYD V+W ++ ++
Sbjct: 343 IISKQQTAFLKNRFIMEGVVILHEVLNSIHQKKQSGILFKVDFEKAYDKVNWVFIYRMLK 402
Query: 596 KMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGLNVM 655
FP W WIM+ V +V VN F +GLRQGDPLSP LF LAAE L ++
Sbjct: 403 AKGFPDQWCDWIMKVVMGGKVAVKVNDQIGSFFKTHKGLRQGDPLSPLLFNLAAEALTLL 462
Query: 656 MSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDVSGLK 715
+ N L + +++ LQ+ADDT+ + + + +K +L LFE +SGLK
Sbjct: 463 VQRAEENSLIEGLGTNGDNKIAI--LQYADDTIFLINDKLDHAKNLKYILCLFEQLSGLK 520
Query: 716 VNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPI 756
+NF+KS F + + + C+ G +P YLG+PI
Sbjct: 521 INFNKSEVFCFGEAKEKQDLYSNIFTCKVGSLPLKYLGIPI 561
Score = 104 bits (260), Expect = 1e-20
Identities = 90/319 (28%), Positives = 142/319 (44%), Gaps = 46/319 (14%)
Query: 781 LEDRESLWNVVLCAKY--GEVGGRVQFSEGVGPIWWRQLNQIRCGVGLAENAWLVDNIVR 838
LE+ E W ++ AKY + ++ G W GV ++ + +
Sbjct: 679 LENEEGWWQEIIYAKYCSDKPLSGLRLKAGSSHFWQ--------GVMEVKDDFF-SFCTK 729
Query: 839 KVGDGSTTLFWEDPWLLNVPLAVSFSRLFELAENKGVSVREMFLLGWGA-------DGGA 891
VG+G TLFWED WL PLA+ F L+ + K +++ ++ G G
Sbjct: 730 IVGNGEKTLFWEDSWLGGKPLAIQFPSLYGIVITKRITIADLNRKGIDCMKFRRDLHGDK 789
Query: 892 WR-WRRRLFAWEE-ELVEECVARLSNFVLQADLSDRWVWRLHSSQLYTVQSAYSYLTAVD 949
R WR+ + +WE LVE C D+ W L +TV+S Y L
Sbjct: 790 LRDWRKIVNSWEGLNLVENC-------------KDKLWWTLSKDGKFTVRSFYRALKLQQ 836
Query: 950 SNITAEFDRFLWLKAVPLKVNIFVWRLFLNRLATKDNLRKRNVLEATNVSSGALCGKEEE 1009
++ ++ +W VPLK+ IF+W N++ TKDNL KR + N C K E
Sbjct: 837 TSFP---NKKIWKFRVPLKIRIFIWFFTKNKILTKDNLLKRGWRKGDNKCQ--FCDKVET 891
Query: 1010 RDHLFFQCDYYGRLWLLISNWLGFVTVF-HDNLYTHANQFCALGGFSKNYMKAFTIIWI- 1067
HLFF C +W +I+ L V +L+ + ++ F+KN I+ I
Sbjct: 892 VQHLFFDCPLARLIWNIIACALNVKPVLSRQDLF--GSWIQSMDKFTKN----LVIVGIA 945
Query: 1068 SVLFTIWKDRNRRIFQNQI 1086
+VL++IWK RN+ F+ ++
Sbjct: 946 AVLWSIWKCRNKACFERKL 964
>UniRef100_Q7XE51 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1652
Score = 148 bits (373), Expect = 1e-33
Identities = 93/273 (34%), Positives = 136/273 (49%), Gaps = 13/273 (4%)
Query: 531 MVVGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYL 590
+V V+ SQ+ F+ GR I++G++I +E + + + K ++ K+DFEKAYD VDW++L
Sbjct: 1246 LVAQKVIKPSQTTFLSGRNIMEGVVILHETLHELHKKKKNGVILKLDFEKAYDKVDWKFL 1305
Query: 591 DEVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAE 650
+ + F W WI V + +V VN F +GLRQGDPLSP LF L +
Sbjct: 1306 QQSLRMKGFSSKWCDWIDSIVRGGSVAVKVNDEIGSYFQTRKGLRQGDPLSPILFNLVVD 1365
Query: 651 GLNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFED 710
L +++ G F V V +S LQ+ADDT+L R +K VL FE
Sbjct: 1366 MLAILIQRAKDQGRFKG-VVPHLVDNGLSILQYADDTILFMDHDLDEARDLKLVLSTFEK 1424
Query: 711 VSGLKVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWHPLV 770
+S LK+NF+KS F + HE + C PF YLG+ + W +
Sbjct: 1425 LSSLKINFYKSELFCYGKAKDVEHEYVKLFGCDTEDYPFKYLGIRMHHKRINNKDWQGVE 1484
Query: 771 ERIRRRLSGVLEDRESLWNVVLCAKYGEVGGRV 803
ERI+++LS W K+ VGGR+
Sbjct: 1485 ERIQKKLSS--------WK----GKFLSVGGRL 1505
>UniRef100_Q9SLE9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1319
Score = 148 bits (373), Expect = 1e-33
Identities = 95/252 (37%), Positives = 136/252 (53%), Gaps = 11/252 (4%)
Query: 535 SVVFASQSAFIKGRQILDGILIANELVDDAKRNNK---ELLMFKVDFEKAYDSVDWRYLD 591
S+V SQSAF R I D ILIA+E+V + N+K E ++FK D KAYD V+W +L
Sbjct: 483 SIVSPSQSAFFAERLISDNILIAHEIVHSLRTNDKISKEFMVFKTDMSKAYDRVEWSFLQ 542
Query: 592 EVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEG 651
E++ + F W SWIM CVT+ T SVL+NG ERG+RQGDP+SPFLF+L E
Sbjct: 543 EILVALGFNDKWISWIMGCVTSVTYSVLINGQHFGHITPERGIRQGDPISPFLFVLCTEA 602
Query: 652 LNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDV 711
L ++ ++ + P SV+HL F DDT L+ + ++ M L + +
Sbjct: 603 LIHILQQAENSKKVSGIQFNGSGP-SVNHLLFVDDTQLVCRATKSDCEQMMLCLSQYGHI 661
Query: 712 SGLKVNFHK-SMFFGVNINES---WLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWH 767
SG +N K S+ FGV ++E W+ + + H G YLGLP K +
Sbjct: 662 SGQLINVEKSSITFGVKVDEDTKRWIKNRSGI-HLEGGTGK--YLGLPENLSGSKQDLFG 718
Query: 768 PLVERIRRRLSG 779
+ E+++ LSG
Sbjct: 719 YIKEKLQSHLSG 730
Score = 63.2 bits (152), Expect = 4e-08
Identities = 68/262 (25%), Positives = 108/262 (40%), Gaps = 36/262 (13%)
Query: 838 RKVGDGSTTLFWEDPWLLNVPLAVSFSRLFELAENKGVSVREMFLLGWGADGGAWRWRRR 897
R +G+G T W D WL + R L + ++ L+ D W +
Sbjct: 876 RLIGNGEQTNVWIDKWLFDG----HSRRPMNLHSLMNIHMKVSHLI----DPLTRNWNLK 927
Query: 898 LFAWEEELVEECVARLSNFVLQADLSDRWVWRLHSSQLYTVQSAYSYLTA---------- 947
E E+ V + + D + W ++ LYTV+S Y +
Sbjct: 928 KLT--ELFHEKDVQLIMHQRPLISSEDSYCWAGTNNGLYTVKSGYERSSRETFKNLFKEA 985
Query: 948 -VDSNITAEFDRFLWLKAVPLKVNIFVWRLFLNRLATKDNLRKRNVLEATNVSSGALCGK 1006
V ++ FD+ L+ VP K+ +F+W+ LA +D LR R + A G L K
Sbjct: 986 DVYPSVNPLFDKVWSLETVP-KIKVFMWKALKGALAVEDRLRSRGIRTA----DGCLFCK 1040
Query: 1007 EEER--DHLFFQCDYYGRLW---LLISNWLGFVTVFHDNLYTHANQFCALGGFSKNYMKA 1061
EE +HL FQC + ++W L+ + GF T N+ H Q G ++
Sbjct: 1041 EEIETINHLLFQCPFARQVWALSLIQAPATGFGTSIFSNI-NHVIQNSQNFGIPRHMRTV 1099
Query: 1062 FTIIWISVLFTIWKDRNRRIFQ 1083
W+ L+ IWK+RN+ +FQ
Sbjct: 1100 SP--WL--LWEIWKNRNKTLFQ 1117
>UniRef100_Q852D8 Putative retrotransposon protein [Oryza sativa]
Length = 543
Score = 147 bits (371), Expect = 2e-33
Identities = 80/240 (33%), Positives = 132/240 (54%), Gaps = 3/240 (1%)
Query: 540 SQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDEVMAKMNF 599
+Q+AFI+GR I + + LV R +KE+++ K+D KA+D+V W +L +++ F
Sbjct: 16 TQNAFIRGRSIHENFIFVKGLVQQYHRQHKEMILLKLDISKAFDTVSWCFLLDMLKWRGF 75
Query: 600 PRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGLNVMMSTV 659
WR W++ TA TS+L+NG ++ F RGLRQGDPLSP LF+LA + L +++
Sbjct: 76 GAKWRLWLVSLFLTAETSILINGNESNPFKPARGLRQGDPLSPLLFVLAMDALQAVVAQA 135
Query: 660 GSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDVSGLKVNFH 719
++GL + VP S+S +ADD +L S + +K +L +F SGL N++
Sbjct: 136 KASGLISEPAPRRPVP-SIS--IYADDAVLFFKPSQQEAKVVKAILQIFGAASGLMTNYN 192
Query: 720 KSMFFGVNINESWLHEAAVVMHCRHGRIPFIYLGLPIGGDPRKLHFWHPLVERIRRRLSG 779
KS + ++ L A + C P IYLGLP+ P+++++ +++G
Sbjct: 193 KSAITPIQCSQEQLQVVANELQCNIQLFPIIYLGLPLSTRKPTKAEVQPILDKLANKVAG 252
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.347 0.155 0.542
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,672,261,757
Number of Sequences: 2790947
Number of extensions: 65997106
Number of successful extensions: 281250
Number of sequences better than 10.0: 840
Number of HSP's better than 10.0 without gapping: 531
Number of HSP's successfully gapped in prelim test: 309
Number of HSP's that attempted gapping in prelim test: 279306
Number of HSP's gapped (non-prelim): 1358
length of query: 1086
length of database: 848,049,833
effective HSP length: 138
effective length of query: 948
effective length of database: 462,899,147
effective search space: 438828391356
effective search space used: 438828391356
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 80 (35.4 bits)
Medicago: description of AC146752.14


