

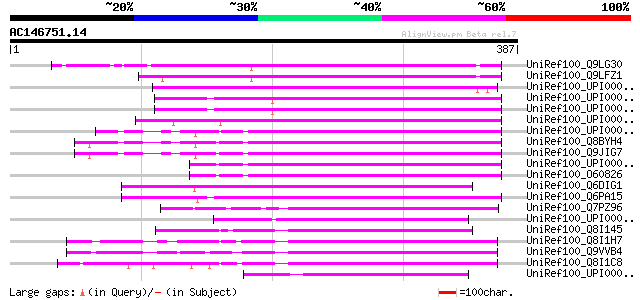
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146751.14 - phase: 1 /pseudo
(387 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LG30 F14J16.7 [Arabidopsis thaliana] 236 1e-60
UniRef100_Q9LFZ1 F20N2.20 [Arabidopsis thaliana] 230 4e-59
UniRef100_UPI00002BEBB2 UPI00002BEBB2 UniRef100 entry 126 1e-27
UniRef100_UPI0000363BF8 UPI0000363BF8 UniRef100 entry 123 1e-26
UniRef100_UPI0000363BF7 UPI0000363BF7 UniRef100 entry 123 1e-26
UniRef100_UPI0000439EB5 UPI0000439EB5 UniRef100 entry 122 2e-26
UniRef100_UPI00001CD73A UPI00001CD73A UniRef100 entry 121 3e-26
UniRef100_Q8BYH4 Mus musculus adult male spinal cord cDNA, RIKEN... 119 1e-25
UniRef100_Q9JIG7 DXImx40e protein (DNA segment, Chr X, Immunex 4... 119 1e-25
UniRef100_UPI000036F5CB UPI000036F5CB UniRef100 entry 119 2e-25
UniRef100_O60826 JM1 protein [Homo sapiens] 119 2e-25
UniRef100_Q6DIG1 MGC89563 protein [Xenopus tropicalis] 115 3e-24
UniRef100_Q6PA15 MGC68757 protein [Xenopus laevis] 113 1e-23
UniRef100_Q7PZ96 ENSANGP00000020101 [Anopheles gambiae str. PEST] 112 2e-23
UniRef100_UPI00004328CB UPI00004328CB UniRef100 entry 110 7e-23
UniRef100_Q8I145 CG9951-PA [Drosophila willistoni] 105 2e-21
UniRef100_Q8I1H7 CG9951-PA [Drosophila erecta] 99 2e-19
UniRef100_Q9VVB4 CG9951-PA [Drosophila melanogaster] 99 3e-19
UniRef100_Q8I1C8 CG9951-PA [Drosophila pseudoobscura] 96 1e-18
UniRef100_UPI0000363BF9 UPI0000363BF9 UniRef100 entry 95 4e-18
>UniRef100_Q9LG30 F14J16.7 [Arabidopsis thaliana]
Length = 585
Score = 236 bits (601), Expect = 1e-60
Identities = 156/386 (40%), Positives = 218/386 (56%), Gaps = 52/386 (13%)
Query: 33 MIILKLKAN*MISNMETQTPEIGSTSSDASISQANKDESNSVNDELNSVLNVNKLDTGEV 92
M I K++A ++ ++ + + S+ S A + AN D S+ D+ V +V D
Sbjct: 200 MHIQKVEA--VLKDLTMTSEKSHSSDSLAKNTSANVDFSSQKTDD--PVTDVRS-DLSLR 254
Query: 93 DAGRSSFGIEEDPSPIDDGHLTV-LQQKEKELIDEVTVRSSEVEHLEQELEMMKAAAEMA 151
D+ R EDP + + TV LQ + L++E+ SS++ LE ELE+++ AAE
Sbjct: 255 DSSRCEENSYEDP--FETNYETVELQNQHDVLLEELESGSSQLCSLESELELLQMAAERL 312
Query: 152 FNNQQSVDSYLEQLTEQILAKGNHLSTLELEW---------------------------- 183
++++ SYLEQL +Q++ K ++ L+ +W
Sbjct: 313 LDDKKPGGSYLEQLNQQLVVKRCNIMDLKKQWYEQYYIYFKLKASMHSVSWTTSGHDGRV 372
Query: 184 --------------DAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEKSISSEI 229
D +R LE +K L + L+ P+A KLR+ + + +S+SSEI
Sbjct: 373 MYRMITKYIMSSFRDDVRLTLETKKLLLLDQLHVEEPEAKEKFHKLRKTELDLQSLSSEI 432
Query: 230 RKREEEHSKLLADLEKQQKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQLESNS 289
+KRE+E L +LE+Q K A RKSY H I+EITKNSRK D DI+RI ETRE+QLE NS
Sbjct: 433 QKREDERCNLYNELERQPKAAPRKSYIHGIKEITKNSRKLDTDIQRISGETRELQLEKNS 492
Query: 290 IHERLHRTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRREVSEY 349
I ERLHR+YAV DEMV RE KKDP +QVYKLL +IH FEQISEKIL DR RRE +Y
Sbjct: 493 IQERLHRSYAVVDEMVTREVKKDPAVRQVYKLLTSIHSIFEQISEKILMTDRFRRETVDY 552
Query: 350 EMKLAAAPSTSRSLDLSKLQSDLDAV 375
E KL + T+R + L KLQ+DLDA+
Sbjct: 553 EKKLGSI--TARGMSLEKLQADLDAI 576
>UniRef100_Q9LFZ1 F20N2.20 [Arabidopsis thaliana]
Length = 520
Score = 230 bits (587), Expect = 4e-59
Identities = 137/321 (42%), Positives = 187/321 (57%), Gaps = 46/321 (14%)
Query: 99 FGIEEDPSPIDDGHLTV--LQQKEKELIDEVTVRSSEVEHLEQELEMMKAAAEMAFNNQQ 156
F ++ P+ D + LQ + L++E+ SS++ LE ELE+++ AAE ++++
Sbjct: 193 FSSQKTDDPVTDTNYETVELQNQHDVLLEELESGSSQLCSLESELELLQMAAERLLDDKK 252
Query: 157 SVDSYLEQLTEQILAKGNHLSTLELEW--------------------------------- 183
SYLEQL +Q++ K ++ L+ +W
Sbjct: 253 PGGSYLEQLNQQLVVKRCNIMDLKKQWYEQYYIYFKLKASMHSVSWTTSGHDGRVMYRMI 312
Query: 184 ---------DAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEKSISSEIRKREE 234
D +R LE +K L + L+ P+A KLR+ + + +S+SSEI+KRE+
Sbjct: 313 TKYIMSSFRDDVRLTLETKKLLLLDQLHVEEPEAKEKFHKLRKTELDLQSLSSEIQKRED 372
Query: 235 EHSKLLADLEKQQKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQLESNSIHERL 294
E L +LE+Q K A RKSY H I+EITKNSRK D DI+RI ETRE+QLE NSI ERL
Sbjct: 373 ERCNLYNELERQPKAAPRKSYIHGIKEITKNSRKLDTDIQRISGETRELQLEKNSIQERL 432
Query: 295 HRTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRREVSEYEMKLA 354
HR+YAV DEMV RE KKDP +QVYKLL +IH FEQISEKIL DR RRE +YE KL
Sbjct: 433 HRSYAVVDEMVTREVKKDPAVRQVYKLLTSIHSIFEQISEKILMTDRFRRETVDYEKKLG 492
Query: 355 AAPSTSRSLDLSKLQSDLDAV 375
+ T+R + L KLQ+DLDA+
Sbjct: 493 SI--TARGMSLEKLQADLDAI 511
>UniRef100_UPI00002BEBB2 UPI00002BEBB2 UniRef100 entry
Length = 362
Score = 126 bits (316), Expect = 1e-27
Identities = 76/268 (28%), Positives = 133/268 (49%), Gaps = 5/268 (1%)
Query: 110 DGHLTVLQQKEKELIDEVTVRSSEVEHLEQELEMMKAAAEMAFNNQQSVDSYLEQLTEQI 169
D + K KE ++ + + LE + K A + ++++ V+ +L + +
Sbjct: 84 DDEIASCADKVKEAMEATEEKHALTADLESNYLLHKQALGIVISSERPVEESEAELNKVL 143
Query: 170 LAKGNHLSTLELEWDAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEKSISSEI 229
+ L+ EW++ + PL + ++ +A + L+++ + + E K S+ +
Sbjct: 144 AVAKERMEQLQKEWESAKAPLLQAIDAHAKAADEKRANAKIQLEEIEKWRSEGKETSALL 203
Query: 230 RKREEEHSKLLADLEKQQKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQLESNS 289
R +E E +LL + E K R S+ R+ EI KN +KQ+ +I +I+ +TR VQ E S
Sbjct: 204 RAKEHEQRQLLEEYESAPKNVHRPSFVRRVNEIIKNIKKQEGEINKIVGDTRTVQREIQS 263
Query: 290 IHERLHRTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRREVSEY 349
E L RTY V +E +FREA+ D +Q YK L +H GF + K+ A RR ++
Sbjct: 264 AQELLERTYTVVEETLFREARSDDLCRQAYKHLHGMHSGFADLLLKVEATGAARRAQTDI 323
Query: 350 EMKLAA---APSTSRSL--DLSKLQSDL 372
+ KL A P+ S + DLS +Q +
Sbjct: 324 QRKLTALSKQPNNSERVARDLSLIQEQI 351
>UniRef100_UPI0000363BF8 UPI0000363BF8 UniRef100 entry
Length = 631
Score = 123 bits (308), Expect = 1e-26
Identities = 74/268 (27%), Positives = 142/268 (52%), Gaps = 7/268 (2%)
Query: 111 GHLTVLQQKEKELIDEVTVRSSEVEHLEQELEMMKAAAEMAFNNQQSVDSYLEQLTEQIL 170
G + ++ ++++E+ R+ E+++EM K A ++ D+ L QL +
Sbjct: 354 GDMKLMSVSCTQVLNELKQRNLGNSEKEEKMEMKKKAIDLL----PDADNNLVQLQLVVE 409
Query: 171 AKGNHLSTLELEWDAIRKPLEERKKTLEE---SLYSSSPDAHVMLQKLREMQQEEKSISS 227
+ L +W+ R PL + + L+E L ++ L +++ + ++ + +
Sbjct: 410 TSVKRVVNLAAQWEKHRAPLIQEHRRLKEILVPLVKFQLESSRKLSEIKSLHEKIRVSTE 469
Query: 228 EIRKREEEHSKLLADLEKQQKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQLES 287
E +K+E+ + +L+A +++ + ASR +YT RI EI N +KQ +I +IL +T+ +Q E
Sbjct: 470 EAKKKEDMYKQLVAQVDRLPQEASRSAYTQRILEIVSNIKKQKEEITKILMDTKHLQKEI 529
Query: 288 NSIHERLHRTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRREVS 347
NS+ +L RT+AV DEMVF++AKKD + ++ YK L +H+ Q+ + I I RE+
Sbjct: 530 NSLTGKLDRTFAVTDEMVFKDAKKDESVRKSYKYLAALHENCNQLIQTIEDTGTILREIR 589
Query: 348 EYEMKLAAAPSTSRSLDLSKLQSDLDAV 375
+ E ++ +L ++ D AV
Sbjct: 590 DLEEQIETENGNKTVANLERILEDYRAV 617
>UniRef100_UPI0000363BF7 UPI0000363BF7 UniRef100 entry
Length = 658
Score = 123 bits (308), Expect = 1e-26
Identities = 74/268 (27%), Positives = 142/268 (52%), Gaps = 7/268 (2%)
Query: 111 GHLTVLQQKEKELIDEVTVRSSEVEHLEQELEMMKAAAEMAFNNQQSVDSYLEQLTEQIL 170
G + ++ ++++E+ R+ E+++EM K A ++ D+ L QL +
Sbjct: 381 GDMKLMSVSCTQVLNELKQRNLGNSEKEEKMEMKKKAIDLL----PDADNNLVQLQLVVE 436
Query: 171 AKGNHLSTLELEWDAIRKPLEERKKTLEE---SLYSSSPDAHVMLQKLREMQQEEKSISS 227
+ L +W+ R PL + + L+E L ++ L +++ + ++ + +
Sbjct: 437 TSVKRVVNLAAQWEKHRAPLIQEHRRLKEILVPLVKFQLESSRKLSEIKSLHEKIRVSTE 496
Query: 228 EIRKREEEHSKLLADLEKQQKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQLES 287
E +K+E+ + +L+A +++ + ASR +YT RI EI N +KQ +I +IL +T+ +Q E
Sbjct: 497 EAKKKEDMYKQLVAQVDRLPQEASRSAYTQRILEIVSNIKKQKEEITKILMDTKHLQKEI 556
Query: 288 NSIHERLHRTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRREVS 347
NS+ +L RT+AV DEMVF++AKKD + ++ YK L +H+ Q+ + I I RE+
Sbjct: 557 NSLTGKLDRTFAVTDEMVFKDAKKDESVRKSYKYLAALHENCNQLIQTIEDTGTILREIR 616
Query: 348 EYEMKLAAAPSTSRSLDLSKLQSDLDAV 375
+ E ++ +L ++ D AV
Sbjct: 617 DLEEQIETENGNKTVANLERILEDYRAV 644
>UniRef100_UPI0000439EB5 UPI0000439EB5 UniRef100 entry
Length = 629
Score = 122 bits (305), Expect = 2e-26
Identities = 82/296 (27%), Positives = 149/296 (49%), Gaps = 17/296 (5%)
Query: 97 SSFGIEEDPSPIDDGHLTVLQQKEKEL----------IDEVTVRSSEVEHLEQELEMMKA 146
SS EED + L+ LQQ+ ++L I ++TV +V Q E+ A
Sbjct: 321 SSQQSEEDLKAQQEAELSALQQQLQQLSVQMEEVGGDIKQLTVSIQQVTDELQTREVTNA 380
Query: 147 AAEMAFN-NQQSVD------SYLEQLTEQILAKGNHLSTLELEWDAIRKPLEERKKTLEE 199
E + +Q++D + L +L + + + L +W+ R PL + + L+E
Sbjct: 381 ERENSVKIKRQTIDLLPDAENNLLKLQSLVESSSKRVVQLASQWEKHRVPLIDEHRRLKE 440
Query: 200 SLYSSSPDAHVMLQKLREMQQEEKSISSEIRKREEEHSKLLADLEKQQKVASRKSYTHRI 259
+ ++ L +++++ + + + E +K+E + +LL + E K SR +YT RI
Sbjct: 441 LCSNRESESSRKLSEIKDLHDKIRQSAEEAKKKESLYKQLLTEFETLSKDVSRSAYTIRI 500
Query: 260 EEITKNSRKQDADIERILKETREVQLESNSIHERLHRTYAVADEMVFREAKKDPTGQQVY 319
EI N +KQ +I +IL +T+++Q E N + +L RT+AV DE+VF++AKKD + ++ Y
Sbjct: 501 LEIVGNIKKQKEEITKILSDTKDLQKEINGLTGKLDRTFAVTDELVFKDAKKDESVRKSY 560
Query: 320 KLLVNIHKGFEQISEKILAADRIRREVSEYEMKLAAAPSTSRSLDLSKLQSDLDAV 375
K L +H+ Q+ + I I RE+ + E ++ +L K+ D A+
Sbjct: 561 KYLAALHENCTQLIQTIEDTGTIMREIRDLEEQIETENGKRTVSNLEKILEDYKAI 616
>UniRef100_UPI00001CD73A UPI00001CD73A UniRef100 entry
Length = 677
Score = 121 bits (304), Expect = 3e-26
Identities = 92/312 (29%), Positives = 156/312 (49%), Gaps = 29/312 (9%)
Query: 66 ANKDESNSVNDELNSVLNVNKLDTGEVDAGRSSFGIEEDPSPIDDGHLTVLQQKEKELID 125
A ++E S+ ++L SV + + EV+A S G+ L Q E E
Sbjct: 379 AQEEELESLREQLASVNH----NIEEVEANMKSLGMN-------------LVQVETEC-- 419
Query: 126 EVTVRSSEVEHLEQE--LEMMKAAAEMAFNNQQSVDSYLEQLTEQILAKGNHLSTLELEW 183
R SE+ EQE L + E+ + ++ L+ + E + HL++ +W
Sbjct: 420 ----RQSELSVAEQEQALRLKSRTVELLPDGAANLTK-LQLVVESSAQRLIHLAS---QW 471
Query: 184 DAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEKSISSEIRKREEEHSKLLADL 243
+ R PL + L + ++ L +++E+ Q ++ + E R++EE + +L+++L
Sbjct: 472 EKHRVPLLAEYRHLRKLQDCRELESSRRLVEIQELHQSVRAAAEEARRKEEVYKQLVSEL 531
Query: 244 EKQQKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQLESNSIHERLHRTYAVADE 303
E K SR +YT RI EI N RKQ +I +IL +T+E+Q E NS+ +L RT+AV DE
Sbjct: 532 ETLPKDVSRLAYTQRILEIVGNIRKQKEEITKILSDTKELQKEINSLSGKLDRTFAVTDE 591
Query: 304 MVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRREVSEYEMKLAAAPSTSRSL 363
+VF++AKKD ++ YK L +H+ Q+ + I I REV + E ++
Sbjct: 592 LVFKDAKKDDAVRKAYKYLAALHENCSQLIQTIEDTGTIMREVRDLEEQIETEMGKKTLS 651
Query: 364 DLSKLQSDLDAV 375
+L K+ D A+
Sbjct: 652 NLEKICEDYRAL 663
>UniRef100_Q8BYH4 Mus musculus adult male spinal cord cDNA, RIKEN full-length
enriched library, clone:A330065E14 product:JM1 PROTEIN
(SIMILAR TO JM1 PROTEIN) homolog [Mus musculus]
Length = 627
Score = 119 bits (299), Expect = 1e-25
Identities = 94/330 (28%), Positives = 161/330 (48%), Gaps = 31/330 (9%)
Query: 50 QTPEIGSTSS--DASISQANKDESNSVNDELNSVLNVNKLDTGEVDAGRSSFGIEEDPSP 107
Q ++ +TS + A + E S+ ++L SV + + EV+A + GI
Sbjct: 311 QVADVPATSQRLEQDTRAAQEQELESLREQLASVNH----NIEEVEADMKTLGIN----- 361
Query: 108 IDDGHLTVLQQKEKELIDEVTVRSSEVEHLEQE--LEMMKAAAEMAFNNQQSVDSYLEQL 165
L Q E E R SE+ EQE L + E+ + ++ + L+ +
Sbjct: 362 --------LVQVETEC------RQSELSVAEQEQALRLKSRTVELLPDGAANL-AKLQLV 406
Query: 166 TEQILAKGNHLSTLELEWDAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEKSI 225
E + HL++ +W+ R PL + L ++ L +++E+ ++
Sbjct: 407 VESSAQRLIHLAS---QWEKHRVPLLAEYRHLRRLQDCRELESSRRLAEIQELHHSVRAA 463
Query: 226 SSEIRKREEEHSKLLADLEKQQKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQL 285
+ E R++EE + +L+++LE K SR +YT RI EI N RKQ +I +IL +T+E+Q
Sbjct: 464 AEEARRKEEVYKQLVSELETLPKDVSRLAYTQRILEIVGNIRKQKEEITKILSDTKELQK 523
Query: 286 ESNSIHERLHRTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRRE 345
E NS+ +L RT+AV DE+VF++AKKD ++ YK L +H+ Q+ + I I RE
Sbjct: 524 EINSLFGKLDRTFAVTDELVFKDAKKDDAVRKAYKYLAALHENCSQLIQTIEDTGTIMRE 583
Query: 346 VSEYEMKLAAAPSTSRSLDLSKLQSDLDAV 375
V + E ++ +L K+ D A+
Sbjct: 584 VRDLEEQIETXMGKKTLSNLEKICEDYRAL 613
>UniRef100_Q9JIG7 DXImx40e protein (DNA segment, Chr X, Immunex 40, expressed) (Mus
musculus ES cells cDNA, RIKEN full-length enriched
library, clone:2410018C10 product:JM1 PROTEIN (SIMILAR
TO JM1 PROTEIN) homolog) [Mus musculus]
Length = 627
Score = 119 bits (298), Expect = 1e-25
Identities = 94/330 (28%), Positives = 161/330 (48%), Gaps = 31/330 (9%)
Query: 50 QTPEIGSTSS--DASISQANKDESNSVNDELNSVLNVNKLDTGEVDAGRSSFGIEEDPSP 107
Q ++ +TS + A + E S+ ++L SV + + EV+A + GI
Sbjct: 311 QVADVPATSQRLEQDTRAAQEQELESLREQLASVNH----NIEEVEADMKTLGIN----- 361
Query: 108 IDDGHLTVLQQKEKELIDEVTVRSSEVEHLEQE--LEMMKAAAEMAFNNQQSVDSYLEQL 165
L Q E E R SE+ EQE L + E+ + ++ + L+ +
Sbjct: 362 --------LVQVETEC------RQSELSVAEQEQALRLKSRTVELLPDGAANL-AKLQLV 406
Query: 166 TEQILAKGNHLSTLELEWDAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEKSI 225
E + HL++ +W+ R PL + L ++ L +++E+ ++
Sbjct: 407 VESSAQRLIHLAS---QWEKHRVPLLAEYRHLRRLQDCRELESSRRLAEIQELHHSVRAA 463
Query: 226 SSEIRKREEEHSKLLADLEKQQKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQL 285
+ E R++EE + +L+++LE K SR +YT RI EI N RKQ +I +IL +T+E+Q
Sbjct: 464 AEEARRKEEVYKQLVSELETLPKDVSRLAYTQRILEIVGNIRKQKEEITKILSDTKELQK 523
Query: 286 ESNSIHERLHRTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRRE 345
E NS+ +L RT+AV DE+VF++AKKD ++ YK L +H+ Q+ + I I RE
Sbjct: 524 EINSLSGKLDRTFAVTDELVFKDAKKDDAVRKAYKYLAALHENCSQLIQTIEDTGTIMRE 583
Query: 346 VSEYEMKLAAAPSTSRSLDLSKLQSDLDAV 375
V + E ++ +L K+ D A+
Sbjct: 584 VRDLEEQIETEMGKKTLSNLEKICEDYRAL 613
>UniRef100_UPI000036F5CB UPI000036F5CB UniRef100 entry
Length = 324
Score = 119 bits (297), Expect = 2e-25
Identities = 73/238 (30%), Positives = 129/238 (53%), Gaps = 4/238 (1%)
Query: 138 EQELEMMKAAAEMAFNNQQSVDSYLEQLTEQILAKGNHLSTLELEWDAIRKPLEERKKTL 197
EQ L + A E+ + ++ + L+ + E + HL+ +W+ R PL + L
Sbjct: 77 EQALRLKSRAVELLPDGTANL-AKLQLVVENSAQRVIHLAG---QWEKHRVPLLAEYRHL 132
Query: 198 EESLYSSSPDAHVMLQKLREMQQEEKSISSEIRKREEEHSKLLADLEKQQKVASRKSYTH 257
+ ++ L +++E+ Q ++ + E R++EE + +L+++LE + SR +YT
Sbjct: 133 RKLQDCRELESSRRLAEIQELHQSVRAAAEEARRKEEVYKQLMSELETLPRDVSRLAYTQ 192
Query: 258 RIEEITKNSRKQDADIERILKETREVQLESNSIHERLHRTYAVADEMVFREAKKDPTGQQ 317
RI EI N RKQ +I +IL +T+E+Q E NS+ +L RT+AV DE+VF++AKKD ++
Sbjct: 193 RILEIVGNIRKQKEEITKILSDTKELQKEINSLSGKLDRTFAVTDELVFKDAKKDDAVRK 252
Query: 318 VYKLLVNIHKGFEQISEKILAADRIRREVSEYEMKLAAAPSTSRSLDLSKLQSDLDAV 375
YK L +H+ Q+ + I I REV + E ++ +L K++ D A+
Sbjct: 253 AYKYLAALHENCSQLIQTIEDTGTIMREVRDLEEQIETELGKKTLSNLEKIREDYRAL 310
>UniRef100_O60826 JM1 protein [Homo sapiens]
Length = 627
Score = 119 bits (297), Expect = 2e-25
Identities = 73/238 (30%), Positives = 129/238 (53%), Gaps = 4/238 (1%)
Query: 138 EQELEMMKAAAEMAFNNQQSVDSYLEQLTEQILAKGNHLSTLELEWDAIRKPLEERKKTL 197
EQ L + A E+ + ++ + L+ + E + HL+ +W+ R PL + L
Sbjct: 380 EQALRLKSRAVELLPDGTANL-AKLQLVVENSAQRVIHLAG---QWEKHRVPLLAEYRHL 435
Query: 198 EESLYSSSPDAHVMLQKLREMQQEEKSISSEIRKREEEHSKLLADLEKQQKVASRKSYTH 257
+ ++ L +++E+ Q ++ + E R++EE + +L+++LE + SR +YT
Sbjct: 436 RKLQDCRELESSRRLAEIQELHQSVRAAAEEARRKEEVYKQLMSELETLPRDVSRLAYTQ 495
Query: 258 RIEEITKNSRKQDADIERILKETREVQLESNSIHERLHRTYAVADEMVFREAKKDPTGQQ 317
RI EI N RKQ +I +IL +T+E+Q E NS+ +L RT+AV DE+VF++AKKD ++
Sbjct: 496 RILEIVGNIRKQKEEITKILSDTKELQKEINSLSGKLDRTFAVTDELVFKDAKKDDAVRK 555
Query: 318 VYKLLVNIHKGFEQISEKILAADRIRREVSEYEMKLAAAPSTSRSLDLSKLQSDLDAV 375
YK L +H+ Q+ + I I REV + E ++ +L K++ D A+
Sbjct: 556 AYKYLAALHENCSQLIQTIEDTGTIMREVRDLEEQIETELGKKTLSNLEKIREDYRAL 613
>UniRef100_Q6DIG1 MGC89563 protein [Xenopus tropicalis]
Length = 604
Score = 115 bits (287), Expect = 3e-24
Identities = 70/271 (25%), Positives = 136/271 (49%), Gaps = 3/271 (1%)
Query: 86 KLDTGEVDAGRSSFGIEEDPSPIDDGHLTVLQQKEKELIDEVTVRSSEVEHLEQ---ELE 142
K D + DA + ++ + + L + K L V+ EV + Q E E
Sbjct: 326 KKDAQDTDAEQQELSSLQEQIDSIEQEIRGLSESNKRLQLTVSQVEGEVNEMRQSCEEKE 385
Query: 143 MMKAAAEMAFNNQQSVDSYLEQLTEQILAKGNHLSTLELEWDAIRKPLEERKKTLEESLY 202
+ + A D+ L +L + A + ++ L +W++ + L E + L+
Sbjct: 386 KIVRVKKRAVELLPDADNNLVKLQALVDASSHRMANLVGQWESHQVRLSEEYRELKRVQQ 445
Query: 203 SSSPDAHVMLQKLREMQQEEKSISSEIRKREEEHSKLLADLEKQQKVASRKSYTHRIEEI 262
++ ++ +++ ++ + + E +++EE + +LL++ E K SR +YT RI EI
Sbjct: 446 EQEDESSRWMKDAKDLYEKIRGAADEAKRKEELYKQLLSEYESLPKEVSRAAYTQRILEI 505
Query: 263 TKNSRKQDADIERILKETREVQLESNSIHERLHRTYAVADEMVFREAKKDPTGQQVYKLL 322
N +KQ +I +IL +T+E+Q E N++ ++ RT+ V DE+VF++AKKD ++ YK L
Sbjct: 506 VSNIKKQKEEITKILSDTKELQKEINNLTGKVDRTFVVTDELVFKDAKKDEPVRKAYKYL 565
Query: 323 VNIHKGFEQISEKILAADRIRREVSEYEMKL 353
+H+ Q+ + I I RE+ + E ++
Sbjct: 566 AALHENCSQLIQTIEDTGTILREIRDLEEQI 596
>UniRef100_Q6PA15 MGC68757 protein [Xenopus laevis]
Length = 632
Score = 113 bits (282), Expect = 1e-23
Identities = 73/297 (24%), Positives = 142/297 (47%), Gaps = 11/297 (3%)
Query: 86 KLDTGEVDAGRSSFGIEEDPSPIDDGHLTVLQQKEKELIDEVTVRSSEVEHLEQELE--- 142
K D + DA + ++ + + L + K L + EV + Q E
Sbjct: 326 KKDAQDADAEQQEISSLQEQIDSIEQEIRGLSESNKLLQLTIGQVEGEVNDMHQSCEEKA 385
Query: 143 ----MMKAAAEMAFNNQQSVDSYLEQLTEQILAKGNHLSTLELEWDAIRKPLEERKKTLE 198
+ K A E+ D+ L++L + A ++ L +W++ + L + L
Sbjct: 386 NVVRVKKRAVELL----PDADNNLDKLQTLVDASAQRMANLIGQWESHQARLSDEYMELN 441
Query: 199 ESLYSSSPDAHVMLQKLREMQQEEKSISSEIRKREEEHSKLLADLEKQQKVASRKSYTHR 258
++ ++ +++ ++ + + E +++EE + +LL++ E K SR +YT R
Sbjct: 442 RVQQEQEDESSRWMKDAKDLYEKIQGSADEAKRKEELYKQLLSEYESLPKEVSRAAYTQR 501
Query: 259 IEEITKNSRKQDADIERILKETREVQLESNSIHERLHRTYAVADEMVFREAKKDPTGQQV 318
I EI N +KQ +I +IL +T+E+Q E N++ ++ RT+ V DE+VF++AKKD ++
Sbjct: 502 ILEIVSNIKKQKEEITKILSDTKELQKEINNLTGKVDRTFVVTDELVFKDAKKDEPVRKA 561
Query: 319 YKLLVNIHKGFEQISEKILAADRIRREVSEYEMKLAAAPSTSRSLDLSKLQSDLDAV 375
YK L +H+ Q+ + I I RE+ + E ++ + +L K+ D A+
Sbjct: 562 YKYLAALHENCSQLIQTIEDTGTILREIRDLEEQIETETTKKTLSNLQKILEDYRAI 618
>UniRef100_Q7PZ96 ENSANGP00000020101 [Anopheles gambiae str. PEST]
Length = 556
Score = 112 bits (280), Expect = 2e-23
Identities = 73/258 (28%), Positives = 132/258 (50%), Gaps = 13/258 (5%)
Query: 116 LQQKEKELIDEVTVRSSEVEHLEQELEMMKAAAEMAFNNQQSVDSYLEQLTEQILAKGNH 175
L K + + + S E L++E + +K + N + + LE + I A G
Sbjct: 290 LHAKRRTVAESAQAVESVAEKLKEE-KKIKERTHILLENPEVNVAKLESI---IAAAGEK 345
Query: 176 LSTLELEWDAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEKSISSEIRKREEE 235
+ L+ +W+A R PL TLEE +S Q + + + + + +++ +
Sbjct: 346 MKKLQSQWEAHRAPLVA---TLEEHQAKNSD------QVIESARHKSEEVIVDLQTKSAL 396
Query: 236 HSKLLADLEKQQKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQLESNSIHERLH 295
H++L+ + E+ + SR +YT RI EI N RKQ DI++IL +TR +Q E NSI +L
Sbjct: 397 HARLVQEYERMGRTVSRTAYTSRILEIIGNIRKQKTDIDKILHDTRSLQKEINSITGQLD 456
Query: 296 RTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRREVSEYEMKLAA 355
R + V D+++FR AK+D ++ Y LLV +H +++ + ++REV E E ++
Sbjct: 457 RQFTVTDDLIFRNAKRDEYCKRAYILLVALHTECSELTALVQETGTVKREVRELEDQIEN 516
Query: 356 APSTSRSLDLSKLQSDLD 373
+ +L+++ DL+
Sbjct: 517 EKDRNVVTNLAQIGQDLE 534
>UniRef100_UPI00004328CB UPI00004328CB UniRef100 entry
Length = 208
Score = 110 bits (275), Expect = 7e-23
Identities = 57/195 (29%), Positives = 109/195 (55%), Gaps = 3/195 (1%)
Query: 156 QSVDSYLEQLTEQILAKGNHLSTLELEWDAIRKPLEERKKTLEESLYSSSPDAHVMLQKL 215
Q + +++L I N L L ++W+ R PL ++ + E S L +
Sbjct: 15 QDGEENIKKLEAAIEITTNKLINLGIQWEKHRIPLIQKYRQEREK---HSTKLQKKLDDI 71
Query: 216 REMQQEEKSISSEIRKREEEHSKLLADLEKQQKVASRKSYTHRIEEITKNSRKQDADIER 275
+ ++++E+ + E R +++++S+L+ +++K K +R +YT RI EI N RKQ +I +
Sbjct: 72 KLLKEKERELQEECRNKDQQYSQLVTEVQKLSKEVNRSAYTQRILEIINNIRKQRDEISK 131
Query: 276 ILKETREVQLESNSIHERLHRTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEK 335
+L +TRE+Q E N++ RL R++ VADE++FR+A+ + ++ YKLL +H ++
Sbjct: 132 VLVDTREIQKEINTLTGRLERSFTVADELIFRDARTNEASRKAYKLLATLHSDCSELVNL 191
Query: 336 ILAADRIRREVSEYE 350
+ RE+ + E
Sbjct: 192 VEETGATIREIRDLE 206
>UniRef100_Q8I145 CG9951-PA [Drosophila willistoni]
Length = 570
Score = 105 bits (263), Expect = 2e-21
Identities = 67/243 (27%), Positives = 125/243 (50%), Gaps = 14/243 (5%)
Query: 112 HLTVLQQKEKELIDEVTVRSSEVEHLEQELEMMKAAAEMAFNNQQSVDSYLEQLTEQILA 171
H ++ K EL T S E E +E +L+ + + + +++V LE L E A
Sbjct: 298 HRKMVTSKLNELKIRETTASQEKETIESKLKSHEKLSLVLSEPEENVGK-LEALVE---A 353
Query: 172 KGNHLSTLELEWDAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEKSISSEIRK 231
N TL +W R+PL + TL + + Q+++ ++ + + EI
Sbjct: 354 TENKRQTLNQQWQDYRRPLLDTLSTLRNAQEA---------QQVQIIRNSIEKLEQEIIA 404
Query: 232 REEEHSKLLADLEKQ-QKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQLESNSI 290
+ ++H++L + L+ Q VA R+ YT RI E KN RKQ DI ++L +TR++Q + N +
Sbjct: 405 KSQQHNELNSSLKNATQSVAPRQEYTRRIHEFIKNIRKQREDIFKVLDDTRQLQKQLNVV 464
Query: 291 HERLHRTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRREVSEYE 350
+ +L R + D+++F+ AK D ++ YKLL +H ++ E + + +E+ + E
Sbjct: 465 NAQLQRQFNYTDDLLFQSAKHDLHAKKAYKLLAQLHSNCSELVECVSLTGNVTKEIRDLE 524
Query: 351 MKL 353
+++
Sbjct: 525 VQI 527
>UniRef100_Q8I1H7 CG9951-PA [Drosophila erecta]
Length = 553
Score = 99.4 bits (246), Expect = 2e-19
Identities = 76/330 (23%), Positives = 155/330 (46%), Gaps = 36/330 (10%)
Query: 44 ISNMETQTPEIGSTSSDASISQANKDESNSVNDELNSVLNVNKLDTGEVDAGRSSFGIEE 103
+ + E P I + DAS +E S EL+ + ++ + A R + +
Sbjct: 235 VDSEEPAPPPISTVKPDASA-----EEEASPIQELSDQVEELRVQCETLLAERKAHAVA- 288
Query: 104 DPSPIDDGHLTVLQQKEKELIDEVTVRSSEVEHLEQELEMMKAAAEMAFNNQQSVDSYLE 163
+ L+Q+E T S E+ ++ L++ + + + ++++++ LE
Sbjct: 289 ---------IAALKQRE-------TKASEEISRIQPTLKLHERTSLVLADSEENLTK-LE 331
Query: 164 QLTEQILAKGNHLSTLELEWDAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEK 223
L + +K TL +W RKPL E + L+ + + Q+++ ++ +
Sbjct: 332 ALLKSTQSKR---ITLTQQWQDYRKPLLESLEKLKTAKEA---------QEVQGIRNNIE 379
Query: 224 SISSEIRKREEEHSKLLADLEK-QQKVASRKSYTHRIEEITKNSRKQDADIERILKETRE 282
+ E+ + ++H++L A L Q +A RK YT RI E N RKQ ADI ++L +TR+
Sbjct: 380 QLEQELLAKTQQHNELNATLRNASQSLAPRKEYTRRIHEFIGNIRKQRADIYKVLDDTRQ 439
Query: 283 VQLESNSIHERLHRTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRI 342
+Q + N + +L R + D+++F+ AK D ++ YKLL +H ++ E + +
Sbjct: 440 LQKQLNVVGAQLQRQFNYTDDLLFQSAKHDLHAKRAYKLLAQLHANCNELVECVSLTGNV 499
Query: 343 RREVSEYEMKLAAAPSTSRSLDLSKLQSDL 372
+++ E E+++ + L ++ D+
Sbjct: 500 TKQIRELEVQIDGEKLKNVLTSLQQITGDI 529
>UniRef100_Q9VVB4 CG9951-PA [Drosophila melanogaster]
Length = 555
Score = 98.6 bits (244), Expect = 3e-19
Identities = 77/330 (23%), Positives = 154/330 (46%), Gaps = 34/330 (10%)
Query: 44 ISNMETQTPEIGSTSSDASISQANKDESNSVNDELNSVLNVNKLDTGEVDAGRSSFGIEE 103
+S+ + P I + AS A K+E S EL+ + ++ + A R + +
Sbjct: 235 VSSEDPAPPLIPTVKPPAS---AEKEEEPSPIQELSDQVQELRVQCENLLAERKAHAVA- 290
Query: 104 DPSPIDDGHLTVLQQKEKELIDEVTVRSSEVEHLEQELEMMKAAAEMAFNNQQSVDSYLE 163
+ L+Q+E + +E++ R L + ++ A +E N +++ L+
Sbjct: 291 ---------IAALKQRETKAREEIS-RIQPTLKLHERTSLVLADSE---ENLAKLEALLK 337
Query: 164 QLTEQILAKGNHLSTLELEWDAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEK 223
+ L TL +W RKPL E + L+ + + Q+++ ++ +
Sbjct: 338 STQSKRL-------TLTQQWQDYRKPLLESLEKLKTAKEA---------QEVQGIRNSIE 381
Query: 224 SISSEIRKREEEHSKLLADLEK-QQKVASRKSYTHRIEEITKNSRKQDADIERILKETRE 282
+ E+ + ++H++L A L Q +A RK YT RI E N RKQ ADI ++L +TR+
Sbjct: 382 QLEQELLAKTQQHNELNATLRSASQSLAPRKEYTRRIHEFIGNIRKQRADIYKVLDDTRQ 441
Query: 283 VQLESNSIHERLHRTYAVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRI 342
+Q + N + +L R + D+++F+ AK D ++ YKLL +H ++ E + +
Sbjct: 442 LQKQLNVVGAQLQRQFNYTDDLLFQSAKHDLHAKRAYKLLAQLHANCNELVECVSLTGNV 501
Query: 343 RREVSEYEMKLAAAPSTSRSLDLSKLQSDL 372
+++ E E+++ + L ++ D+
Sbjct: 502 TKQIRELEVQIDGEKLKNVLTSLQQITGDI 531
>UniRef100_Q8I1C8 CG9951-PA [Drosophila pseudoobscura]
Length = 553
Score = 96.3 bits (238), Expect = 1e-18
Identities = 91/373 (24%), Positives = 166/373 (44%), Gaps = 51/373 (13%)
Query: 37 KLKAN*MISNMETQTPEIGSTSSDASISQANKDESNSVNDELNSVLNVNKLDT---GEVD 93
KL N +N+E +T E+ D + ++S D + SVL+ N L+ G D
Sbjct: 171 KLNLNVPSANLEERTKEV-QQYFDQQAPNLFQQTASSSYDLIASVLHKNDLERWGQGLTD 229
Query: 94 AGRSSFGIEEDPSPI----DDGHLTVLQQKE-KELIDEVTVRSSEVEHL----------- 137
+G E+ P+ + T + +EL DEV ++ E L
Sbjct: 230 SGMFLLASEDPVIPMVLATEKPSATAAEMTPLEELTDEVQKLRTQCETLLGERKAYTARI 289
Query: 138 ----EQELEMMKAAAEMA-------------FNNQQSVDSYLEQLTEQILAKGNHLSTLE 180
++E E K A++ + +Q+V LE L + AK TL
Sbjct: 290 GALKQRESEATKELADIQPLLKLHERTCLVLADPEQNVTK-LEALLQATEAKRR---TLT 345
Query: 181 LEWDAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEKSISSEIRKREEEHSKLL 240
+W R PL E +TL+ + + Q ++ ++ + + E++++ ++H+ L
Sbjct: 346 QQWQDYRLPLLETLETLKTAKEA---------QHVQSIRSGIEQLEQELKEKTQQHNDLN 396
Query: 241 ADLEKQ-QKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQLESNSIHERLHRTYA 299
L Q +A RK YT RI E N RKQ ADI ++L +TR++Q + N + +L R +
Sbjct: 397 TALRTATQSLAPRKEYTRRIHEFIGNIRKQRADIFKVLDDTRQLQKQLNVVGAQLQRQFN 456
Query: 300 VADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRREVSEYEMKLAAAPST 359
D+++F+ AK D ++ YKLL +H ++ E + + +++ E E+++
Sbjct: 457 YTDDLLFQSAKHDLHAKKAYKLLAQLHANCSELVECVALTGNVTKQIRELEVQIDGEKLK 516
Query: 360 SRSLDLSKLQSDL 372
+ L ++ D+
Sbjct: 517 NVLTSLQQITGDI 529
>UniRef100_UPI0000363BF9 UPI0000363BF9 UniRef100 entry
Length = 508
Score = 94.7 bits (234), Expect = 4e-18
Identities = 55/172 (31%), Positives = 91/172 (51%), Gaps = 9/172 (5%)
Query: 179 LELEWDAIRKPLEERKKTLEESLYSSSPDAHVMLQKLREMQQEEKSISSEIRKREEEHSK 238
L +W+ R PL + + L+E + H + + ++ E R
Sbjct: 342 LAAQWEKHRAPLIQEHRRLKEMCSNHHVRLHTHMSE---------QVAMETRSALTFSQS 392
Query: 239 LLADLEKQQKVASRKSYTHRIEEITKNSRKQDADIERILKETREVQLESNSIHERLHRTY 298
+A +++ + ASR +YT RI EI N +KQ +I +IL +T+ +Q E NS+ +L RT+
Sbjct: 393 QVAQVDRLPQEASRSAYTQRILEIVSNIKKQKEEITKILMDTKHLQKEINSLTGKLDRTF 452
Query: 299 AVADEMVFREAKKDPTGQQVYKLLVNIHKGFEQISEKILAADRIRREVSEYE 350
AV DEMVF++AKKD + ++ YK L +H+ Q+ + I I RE+ + E
Sbjct: 453 AVTDEMVFKDAKKDESVRKSYKYLAALHENCNQLIQTIEDTGTILREIRDLE 504
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.312 0.128 0.331
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 545,139,831
Number of Sequences: 2790947
Number of extensions: 21934728
Number of successful extensions: 160930
Number of sequences better than 10.0: 9551
Number of HSP's better than 10.0 without gapping: 561
Number of HSP's successfully gapped in prelim test: 9349
Number of HSP's that attempted gapping in prelim test: 127229
Number of HSP's gapped (non-prelim): 26293
length of query: 387
length of database: 848,049,833
effective HSP length: 129
effective length of query: 258
effective length of database: 488,017,670
effective search space: 125908558860
effective search space used: 125908558860
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146751.14


