

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.8 - phase: 0
(298 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
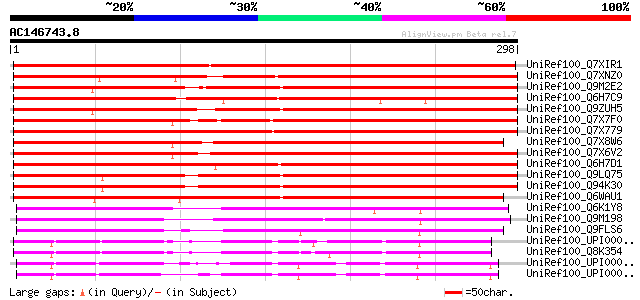
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XIR1 Carbonyl reductase-like protein [Oryza sativa] 361 1e-98
UniRef100_Q7XNZ0 OSJNBa0081C01.18 protein [Oryza sativa] 261 2e-68
UniRef100_Q9M2E2 Hypothetical protein T20K12.120 [Arabidopsis th... 258 1e-67
UniRef100_Q6H7C9 Short-chain dehydrogenase/reductase protein-lik... 254 1e-66
UniRef100_Q9ZUH5 Putative carbonyl reductase [Arabidopsis thaliana] 254 2e-66
UniRef100_Q7X7F0 OSJNBa0081C01.25 protein [Oryza sativa] 254 2e-66
UniRef100_Q7X779 OSJNBa0081C01.23 protein [Oryza sativa] 254 2e-66
UniRef100_Q7X8W6 OSJNBa0081C01.20 protein [Oryza sativa] 254 3e-66
UniRef100_Q7X6V2 OSJNBa0081C01.19 protein [Oryza sativa] 246 7e-64
UniRef100_Q6H7D1 Short-chain dehydrogenase/reductase protein-lik... 245 9e-64
UniRef100_Q9LQ75 T1N6.22 protein [Arabidopsis thaliana] 239 5e-62
UniRef100_Q94K30 Putative carbonyl reductase [Arabidopsis thaliana] 239 5e-62
UniRef100_Q6WAU1 (-)-isopiperitenone reductase [Mentha piperita] 238 1e-61
UniRef100_Q6K1Y8 Putative carbonyl reductase 3 [Oryza sativa] 180 4e-44
UniRef100_Q9M198 Hypothetical protein T16L24.260 [Arabidopsis th... 178 2e-43
UniRef100_Q9FLS6 Carbonyl reductase-like protein [Arabidopsis th... 172 1e-41
UniRef100_UPI00001D06CC UPI00001D06CC UniRef100 entry 151 2e-35
UniRef100_Q8K354 Carbonyl reductase 3 (Mus musculus adult male t... 150 5e-35
UniRef100_UPI00003A9CC2 UPI00003A9CC2 UniRef100 entry 149 9e-35
UniRef100_UPI00003A9CC1 UPI00003A9CC1 UniRef100 entry 148 2e-34
>UniRef100_Q7XIR1 Carbonyl reductase-like protein [Oryza sativa]
Length = 373
Score = 361 bits (927), Expect = 1e-98
Identities = 177/296 (59%), Positives = 229/296 (76%), Gaps = 2/296 (0%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKL-HQTGLSNVMFHQLDV 61
R AVVTG N+GIGLE+ +QLA GVTV+LTAR++ RG+DA+ L H++ LSN++FHQLD+
Sbjct: 77 RLAVVTGGNRGIGLEVCRQLALQGVTVILTARDEKRGKDAVESLCHESNLSNIIFHQLDI 136
Query: 62 LDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQ 121
LD S SLA++I +FG+LDIL+NNAG V VD++GL+ALN+DP WL+GK N L+Q
Sbjct: 137 LDGNSRASLARYINSRFGKLDILVNNAGVGGVAVDQDGLRALNIDPRVWLSGKAVN-LIQ 195
Query: 122 GVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLR 181
V+ QTY +A +CLNTNYYG+K +T ALLPLL+ SP+ ARIVN +SLR ELKRIPNE+LR
Sbjct: 196 SVIVQTYDEAVKCLNTNYYGLKWITEALLPLLKQSPSGARIVNTTSLRSELKRIPNEKLR 255
Query: 182 NELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAKKN 241
+EL ++D E +I+AM+ +FL D K E GW MLPAYS+SK +N YTR+LAK++
Sbjct: 256 DELRNIDIWDEARIEAMLNEFLLDLKNERLEEAGWPTMLPAYSMSKTVVNLYTRILAKRH 315
Query: 242 PHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYFDCTEIAE 297
P M INCVHPGFV+T+ NW+ G + +EGARG V +LLP DGPTGCYFD TE+ E
Sbjct: 316 PEMRINCVHPGFVNTEINWNTGIIPPEEGARGAVKAALLPQDGPTGCYFDQTELGE 371
>UniRef100_Q7XNZ0 OSJNBa0081C01.18 protein [Oryza sativa]
Length = 310
Score = 261 bits (667), Expect = 2e-68
Identities = 148/307 (48%), Positives = 199/307 (64%), Gaps = 21/307 (6%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGL--SNVMFHQLD 60
R A+VTG NKG+GLE +QLA G+ VVLTARN+ RG +A+ + ++G S+V+FHQLD
Sbjct: 13 RIALVTGGNKGVGLETCRQLASRGLRVVLTARNEARGLEAVDGIRRSGAADSDVVFHQLD 72
Query: 61 VLDALSIESLAKFIQHKFGRLDILINNAGASCVEVD-------KEGLKALNVDPAT-WLA 112
V DA S+ LA F++ +FGRLDILINNAG S V+ D K+ ++ ++VD W+
Sbjct: 73 VTDAASVARLADFVRDQFGRLDILINNAGISGVDRDPVLVAKVKDQIEGMDVDQRVEWMR 132
Query: 113 GKVSNTLLQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGEL 172
+TY +A+ C+ TNYYG K VT ALLPLL LS + RIVN+SS G L
Sbjct: 133 ENSK---------ETYDEAKSCITTNYYGAKLVTEALLPLLLLS-SSGRIVNVSSGFGLL 182
Query: 173 KRIPNERLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMM-LPAYSISKASLN 231
+ +E LR E D+D L+E +++ ++ FL DFK N EA+GW AY ++KA+LN
Sbjct: 183 RNFNSEDLRKEFDDIDSLTEKRLEELLDLFLDDFKVNLIEAHGWPTGGSSAYKVAKAALN 242
Query: 232 AYTRVLAKKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYFD 291
AYTR+LAKK P + INC+ PG+V TD + H G +T +EGA V ++LLP DGPTG YFD
Sbjct: 243 AYTRILAKKYPTLRINCLTPGYVKTDISMHMGVLTPEEGASNSVKVALLPDDGPTGAYFD 302
Query: 292 CTEIAEF 298
A F
Sbjct: 303 RNGEASF 309
>UniRef100_Q9M2E2 Hypothetical protein T20K12.120 [Arabidopsis thaliana]
Length = 296
Score = 258 bits (659), Expect = 1e-67
Identities = 140/300 (46%), Positives = 196/300 (64%), Gaps = 14/300 (4%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH---QTGLSNVMFHQL 59
RYAVVTGAN+GIG EI +QLA G+ VVLT+R++ RG +A+ L + +++FHQL
Sbjct: 7 RYAVVTGANRGIGFEICRQLASEGIRVVLTSRDENRGLEAVETLKKELEISDQSLLFHQL 66
Query: 60 DVLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTL 119
DV D SI SLA+F++ +FG+LDIL+NNAG + D E L+A AGK
Sbjct: 67 DVADPASITSLAEFVKTQFGKLDILVNNAGIGGIITDAEALRAG--------AGK-EGFK 117
Query: 120 LQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNER 179
++T+TY+ EEC+ NYYG KR+ A +PLL+LS + RIVN+SS G+LK + NE
Sbjct: 118 WDEIITETYELTEECIKINYYGPKRMCEAFIPLLKLSDSP-RIVNVSSSMGQLKNVLNEW 176
Query: 180 LRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAK 239
+ L D + L+E +ID ++ + L+DFK + W + AY +SKASLN YTRVLAK
Sbjct: 177 AKGILSDAENLTEERIDQVINQLLNDFKEGTVKEKNWAKFMSAYVVSKASLNGYTRVLAK 236
Query: 240 KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLP-ADGPTGCYFDCTEIAEF 298
K+P +N V PGFV TD N+ G ++V+EGA PV L+LLP + P+GC+F +++EF
Sbjct: 237 KHPEFRVNAVCPGFVKTDMNFKTGVLSVEEGASSPVRLALLPHQETPSGCFFSRKQVSEF 296
>UniRef100_Q6H7C9 Short-chain dehydrogenase/reductase protein-like [Oryza sativa]
Length = 315
Score = 254 bits (650), Expect = 1e-66
Identities = 150/307 (48%), Positives = 190/307 (61%), Gaps = 17/307 (5%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
R AVVTG NKG+GLEI KQLA GVTVVLTAR++ RG A L Q GLS V+FHQ DV
Sbjct: 14 RVAVVTGGNKGLGLEICKQLAANGVTVVLTARSEERGAGAAAALRQLGLSEVLFHQFDVS 73
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQG 122
+ S LA FI+HKFG+LDIL+NNAG V D N+D + GK +N L+
Sbjct: 74 EPSSAAGLADFIKHKFGKLDILVNNAGILGVTFDFG-----NLDLNKAIEGKSANETLEW 128
Query: 123 VL---TQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNER 179
++ +T + AEECL NY+G K+ ALLPLLQ SP RIV +SS+ G+L E+
Sbjct: 129 LMQHTVETAENAEECLKINYHGNKKTIQALLPLLQSSP-DGRIVTVSSVFGQLSFFSGEK 187
Query: 180 LRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGW---GMMLPAYSISKASLNAYTRV 236
L+ EL D +LSE +ID + + F+ DFK + E+ GW AY SKA +AYTRV
Sbjct: 188 LKEELNDFSKLSEERIDELAELFVRDFKDGELESRGWPARADAFAAYKTSKALQHAYTRV 247
Query: 237 LAKKNPH-----MLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYFD 291
LA+K+ + +NCVHPG+V TD G +TV+EGA GPV L+L P G TG +F
Sbjct: 248 LARKHASSSSSPLRVNCVHPGYVKTDMTLGTGELTVEEGAAGPVALALSPPGGATGVFFI 307
Query: 292 CTEIAEF 298
TE A F
Sbjct: 308 QTEPASF 314
>UniRef100_Q9ZUH5 Putative carbonyl reductase [Arabidopsis thaliana]
Length = 296
Score = 254 bits (649), Expect = 2e-66
Identities = 133/301 (44%), Positives = 198/301 (65%), Gaps = 16/301 (5%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH---QTGLSNVMFHQL 59
RYA+VTG N+GIG EI +QLA G+ V+LT+R++ +G +A+ L + +++FHQL
Sbjct: 7 RYAIVTGGNRGIGFEICRQLANKGIRVILTSRDEKQGLEAVETLKKELEISDQSIVFHQL 66
Query: 60 DVLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKA-LNVDPATWLAGKVSNT 118
DV D +S+ SLA+F++ FG+LDILINNAG V D + L+A + W
Sbjct: 67 DVSDPVSVTSLAEFVKTHFGKLDILINNAGVGGVITDVDALRAGTGKEGFKW-------- 118
Query: 119 LLQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNE 178
+ +T+TY+ AEEC+ NYYG KR+ A +PLLQLS + RI+N+SS G++K + NE
Sbjct: 119 --EETITETYELAEECIKINYYGPKRMCEAFIPLLQLSDSP-RIINVSSFMGQVKNLVNE 175
Query: 179 RLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLA 238
+ L D + L+E +ID ++ + L+D K + + W ++ AY +SKA LNAYTR+LA
Sbjct: 176 WAKGILSDAENLTEVRIDQVINQLLNDLKEDTAKTKYWAKVMSAYVVSKAGLNAYTRILA 235
Query: 239 KKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLP-ADGPTGCYFDCTEIAE 297
KK+P + +N V PGFV TD N+ G ++V+EGA PV L+LLP + P+GC+FD +++E
Sbjct: 236 KKHPEIRVNSVCPGFVKTDMNFKTGILSVEEGASSPVRLALLPHQESPSGCFFDRKQVSE 295
Query: 298 F 298
F
Sbjct: 296 F 296
>UniRef100_Q7X7F0 OSJNBa0081C01.25 protein [Oryza sativa]
Length = 307
Score = 254 bits (649), Expect = 2e-66
Identities = 140/300 (46%), Positives = 198/300 (65%), Gaps = 11/300 (3%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
R AVVTG NK IGLE+ +QLA G+TVVLTAR++TRG +A +L GLS+V+FHQL+V
Sbjct: 14 RVAVVTGGNKEIGLEVCRQLAADGITVVLTARDETRGVEAAERLRGMGLSSVVFHQLEVT 73
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVE----VDKEGLKALNVDPATWLAGKVSNT 118
D+ S+ LA F++ +FG+LDIL+NNA +E VD + + +D + ++
Sbjct: 74 DSSSVARLADFLKTRFGKLDILVNNAAVGGMEYAQGVDNNEEQFVGMD----VLQRLQWM 129
Query: 119 LLQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNE 178
QG +TY A+ + TNYYG K V LLPLL LS ++ +IVN+SS G L+ + NE
Sbjct: 130 RKQG--RETYDTAKNGVQTNYYGAKHVIQGLLPLL-LSSSEGKIVNVSSALGLLRFLGNE 186
Query: 179 RLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLA 238
LR EL D+D L+E ++D ++ FL DF+A + EA+GW M AY ++K ++NAYTR+ A
Sbjct: 187 DLRKELDDIDNLTEERLDEVLASFLKDFEAGELEAHGWPMGSAAYKVAKVAMNAYTRISA 246
Query: 239 KKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYFDCTEIAEF 298
+K+P + INC HPG+V TD + G +T +EGAR V ++LLP GPTG +FD + A F
Sbjct: 247 RKHPALRINCAHPGYVKTDLTINSGFLTPEEGARNVVTVALLPDGGPTGAFFDEGKEASF 306
>UniRef100_Q7X779 OSJNBa0081C01.23 protein [Oryza sativa]
Length = 309
Score = 254 bits (649), Expect = 2e-66
Identities = 143/298 (47%), Positives = 188/298 (62%), Gaps = 3/298 (1%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
R AVVTG NKGIGLE+ +QLA G TVVLTAR++ +G A+ KLH GLS+V+FHQLDV
Sbjct: 12 RIAVVTGGNKGIGLEVCRQLAGNGATVVLTARDEAKGAAAVEKLHGLGLSSVIFHQLDVT 71
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGAS-CVEVDKEGLKALNVDPA-TWLAGKVSNTLL 120
DA SI LA+F++ +FGRLDIL+NNA V VD L + + + G +
Sbjct: 72 DASSIARLAEFLESRFGRLDILVNNAAVGGIVPVDDPSFGLLPTEEKFSGMDGHQRIEWM 131
Query: 121 QGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERL 180
QTY A+ L TNYYG K VT ALLPLLQ S + RIVN++S G L+ NE L
Sbjct: 132 WKNCRQTYDAAKAGLKTNYYGTKNVTEALLPLLQ-SSSDGRIVNVASSFGLLRFFTNEEL 190
Query: 181 RNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAKK 240
+ EL D D LSE ++D ++ F+ DF+A GW AY ++KA+++AY R+LA+K
Sbjct: 191 KRELNDADSLSEERLDELLGMFVRDFEAGAVAERGWPTEFSAYKVAKAAMSAYARILARK 250
Query: 241 NPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYFDCTEIAEF 298
P + +NCV PG+V TD + G +T +EGA V ++LLPA GPTG FD + A F
Sbjct: 251 RPALRVNCVDPGYVKTDLTRNSGLLTPEEGASRVVAVALLPAGGPTGALFDGGKEASF 308
>UniRef100_Q7X8W6 OSJNBa0081C01.20 protein [Oryza sativa]
Length = 307
Score = 254 bits (648), Expect = 3e-66
Identities = 139/292 (47%), Positives = 189/292 (64%), Gaps = 10/292 (3%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
R AVVTG NKGIGLE+ +QLA G+TVVLTAR++TRG +A KL GLS+V+FHQL+V
Sbjct: 12 RVAVVTGGNKGIGLEVCRQLAADGITVVLTARDETRGVEAAEKLSGMGLSSVVFHQLEVT 71
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVE----VDKEGLKALNVDPATWLAGKVSNT 118
D+ S+ LA F++ +FG+LDIL+NNA +E VD + +++D LA
Sbjct: 72 DSSSVARLADFLKTRFGKLDILVNNAAVGGMEYVQGVDTNKEQFVSMDKKQRLAW----- 126
Query: 119 LLQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNE 178
L +TY A+ + TNYYG K V ALLPLL S + RIVN+SS G L+ + NE
Sbjct: 127 -LNKQGRETYDAAKNGVQTNYYGTKIVIQALLPLLLQSSGEGRIVNVSSDFGLLRVVNNE 185
Query: 179 RLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLA 238
LR EL DVD L+E ++D ++ FL DF+A EA+GW AY +K ++NAYTR+LA
Sbjct: 186 DLRKELDDVDNLTEERLDEVLDSFLKDFEAGALEAHGWPTAFAAYKTAKVAMNAYTRILA 245
Query: 239 KKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYF 290
+++P + +NC HPG+V TD G +T +EG R V ++LLP GPTG +F
Sbjct: 246 RRHPELRVNCAHPGYVKTDMTIDSGFLTPEEGGRNVVTVALLPDGGPTGAFF 297
>UniRef100_Q7X6V2 OSJNBa0081C01.19 protein [Oryza sativa]
Length = 309
Score = 246 bits (627), Expect = 7e-64
Identities = 141/302 (46%), Positives = 190/302 (62%), Gaps = 12/302 (3%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
R AVVTG NKGIGLE+ +QLA G+TVVLTAR++TRG +A KL GLS V+FH L+V
Sbjct: 12 RVAVVTGGNKGIGLEVCRQLAADGITVVLTARDETRGVEAAEKLRGMGLSCVIFHHLEVT 71
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVE----VDKEGLKALNVDPATWLAGKVSNT 118
D+ S+ LA F+ +FG+L+IL+NNA S +E VD + + +D L
Sbjct: 72 DSSSVSRLADFLTTRFGKLEILVNNAAVSGMEHAQRVDTNEEQFVGMDKQQRL------E 125
Query: 119 LLQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELK-RIPN 177
L +TY A+ + TNYYG K V LLPLL S + RIVN+SS G L+ + N
Sbjct: 126 WLNKQGRETYDAAKNGVQTNYYGTKLVIQTLLPLLLQSSGEGRIVNVSSDAGLLRWLVNN 185
Query: 178 ERLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMM-LPAYSISKASLNAYTRV 236
E LR EL DVD L+E ++D ++ FL DF+A EA+GW AY ++K ++NAYTR+
Sbjct: 186 EDLRKELDDVDNLTEERLDEVLDSFLKDFEAGALEAHGWPTAPFVAYKMAKVAMNAYTRI 245
Query: 237 LAKKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYFDCTEIA 296
LA+++P + +NCVHPG+V TD + G +T +EG R V ++LLP GPTG YFD A
Sbjct: 246 LARRHPELRVNCVHPGYVKTDMTINSGFLTPEEGGRNVVTVALLPDGGPTGAYFDEGREA 305
Query: 297 EF 298
F
Sbjct: 306 SF 307
>UniRef100_Q6H7D1 Short-chain dehydrogenase/reductase protein-like [Oryza sativa]
Length = 324
Score = 245 bits (626), Expect = 9e-64
Identities = 136/299 (45%), Positives = 192/299 (63%), Gaps = 4/299 (1%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
R AVVTG N+G+GLEI +QLA G+ VVLTAR++ +G A+ L Q+GLS V+FHQLDV
Sbjct: 27 RVAVVTGGNRGVGLEICRQLASNGILVVLTARDEKKGSQAVKALEQSGLSGVIFHQLDVT 86
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKAL-NVDPATWLAGKVSNTL-- 119
D SI L +FI+ KFG+ +IL+NNA +D E L+ L DP + L
Sbjct: 87 DRSSIMLLVEFIRTKFGKFNILVNNAAIGGTTIDPERLRELLEQDPKASFQEDLMGFLNS 146
Query: 120 LQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNER 179
G L Q Y+ A+ECL N+YG K VT L+PLL LS + +++NL+S +L+ I NE
Sbjct: 147 YMGSLQQNYEMAKECLEINFYGTKDVTDCLMPLLLLSNS-GKVINLTSKISQLQFISNEG 205
Query: 180 LRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAK 239
+ L D+D LS+ K+ + FL DFK + EA+GW ++ AY++SK +NAY+R+LAK
Sbjct: 206 VIKVLSDIDNLSDEKLKDVASIFLKDFKDGNLEAHGWQPVVSAYAVSKTLVNAYSRLLAK 265
Query: 240 KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYFDCTEIAEF 298
++P + + CV+PGFV TD N+ G ++V+EGA PV L+L A + YF+ EI+EF
Sbjct: 266 RHPSLEVCCVNPGFVKTDMNYGIGLISVEEGANAPVRLALQEACSDSCLYFEQCEISEF 324
>UniRef100_Q9LQ75 T1N6.22 protein [Arabidopsis thaliana]
Length = 325
Score = 239 bits (611), Expect = 5e-62
Identities = 130/299 (43%), Positives = 197/299 (65%), Gaps = 11/299 (3%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH-QTGLSN--VMFHQL 59
R AVVTG+NKGIG EI +QLA G+TVVLTAR++ +G A+ KL + G S+ + FH L
Sbjct: 35 RVAVVTGSNKGIGFEICRQLANNGITVVLTARDENKGLAAVQKLKTENGFSDQAISFHPL 94
Query: 60 DVLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTL 119
DV + +I SLA F++ +FG+LDIL+NNAG V+ + LKA +A + T
Sbjct: 95 DVSNPDTIASLAAFVKTRFGKLDILVNNAGVGGANVNVDVLKAQ-------IAEAGAPTD 147
Query: 120 LQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNER 179
+ +++ TY+ EEC+ TNYYGVKR+ A++PLLQ S + RIV+++S G+L+ + NE
Sbjct: 148 ISKIMSDTYEIVEECVKTNYYGVKRMCEAMIPLLQSSDSP-RIVSIASTMGKLENVSNEW 206
Query: 180 LRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAK 239
+ L D + L+E KID ++ ++L D+K + GW ++ Y +SKA++ A TRVLAK
Sbjct: 207 AKGVLSDAENLTEEKIDEVINEYLKDYKEGALQVKGWPTVMSGYILSKAAVIALTRVLAK 266
Query: 240 KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYFDCTEIAEF 298
++ +IN V PGFV+T+ N++ G ++V+EGA PV L+L+P P+G +FD ++ F
Sbjct: 267 RHKSFIINSVCPGFVNTEINFNTGILSVEEGAASPVKLALVPNGDPSGLFFDRANVSNF 325
>UniRef100_Q94K30 Putative carbonyl reductase [Arabidopsis thaliana]
Length = 295
Score = 239 bits (611), Expect = 5e-62
Identities = 130/299 (43%), Positives = 197/299 (65%), Gaps = 11/299 (3%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH-QTGLSN--VMFHQL 59
R AVVTG+NKGIG EI +QLA G+TVVLTAR++ +G A+ KL + G S+ + FH L
Sbjct: 5 RVAVVTGSNKGIGFEICRQLANNGITVVLTARDENKGLAAVQKLKTENGFSDQAISFHPL 64
Query: 60 DVLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTL 119
DV + +I SLA F++ +FG+LDIL+NNAG V+ + LKA +A + T
Sbjct: 65 DVSNPDTIASLAAFVKTRFGKLDILVNNAGVGGANVNVDVLKAQ-------IAEAGAPTD 117
Query: 120 LQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNER 179
+ +++ TY+ EEC+ TNYYGVKR+ A++PLLQ S + RIV+++S G+L+ + NE
Sbjct: 118 ISKIMSDTYEIVEECVKTNYYGVKRMCEAMIPLLQSSDSP-RIVSIASTMGKLENVSNEW 176
Query: 180 LRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAK 239
+ L D + L+E KID ++ ++L D+K + GW ++ Y +SKA++ A TRVLAK
Sbjct: 177 AKGVLSDAENLTEEKIDEVINEYLKDYKEGALQVKGWPTVMSGYILSKAAVIALTRVLAK 236
Query: 240 KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYFDCTEIAEF 298
++ +IN V PGFV+T+ N++ G ++V+EGA PV L+L+P P+G +FD ++ F
Sbjct: 237 RHKSFIINSVCPGFVNTEINFNTGILSVEEGAASPVKLALVPNGDPSGLFFDRANVSNF 295
>UniRef100_Q6WAU1 (-)-isopiperitenone reductase [Mentha piperita]
Length = 314
Score = 238 bits (608), Expect = 1e-61
Identities = 136/301 (45%), Positives = 187/301 (61%), Gaps = 14/301 (4%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQ---TGLSNVMFHQL 59
RYA+VTGANKGIG EI +QLA G+ V+LT+RN+ RG +A KL + + ++FHQL
Sbjct: 6 RYALVTGANKGIGFEICRQLAEKGIIVILTSRNEKRGLEARQKLLKELNVSENRLVFHQL 65
Query: 60 DVLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKE----------GLKALNVDPAT 109
DV D S+ ++A FI+ KFG+LDIL+NNAG S VE+ + KAL A
Sbjct: 66 DVTDLASVAAVAVFIKSKFGKLDILVNNAGVSGVEMVGDVSVFNEYIEADFKALQALEAG 125
Query: 110 WLAGKVSNTLLQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLR 169
G + + ++ A++C+ TNYYG KR+T AL+PLLQLSP+ RIVN+SS
Sbjct: 126 AKEEPPFKPKANGEMIEKFEGAKDCVVTNYYGPKRLTQALIPLLQLSPSP-RIVNVSSSF 184
Query: 170 GELKRIPNERLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKAS 229
G L + NE + LGD D L+E ++D +V+ FL D K E + W A +SKA+
Sbjct: 185 GSLLLLWNEWAKGVLGDEDRLTEERVDEVVEVFLKDIKEGKLEESQWPPHFAAERVSKAA 244
Query: 230 LNAYTRVLAKKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCY 289
LNAYT++ AKK P IN + PG+ TD +H G ++V E A+ PV L+LLP GP+GC+
Sbjct: 245 LNAYTKIAAKKYPSFRINAICPGYAKTDITFHAGPLSVAEAAQVPVKLALLPDGGPSGCF 304
Query: 290 F 290
F
Sbjct: 305 F 305
>UniRef100_Q6K1Y8 Putative carbonyl reductase 3 [Oryza sativa]
Length = 298
Score = 180 bits (457), Expect = 4e-44
Identities = 117/297 (39%), Positives = 159/297 (53%), Gaps = 36/297 (12%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
AVVTGAN+GIG + +LA G+ VVLTAR+ RG A L GL +V F +LDV D
Sbjct: 23 AVVTGANRGIGHALAARLAEQGLAVVLTARDGARGEAAAAALRARGLRSVRFRRLDVSDP 82
Query: 65 LSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGVL 124
S+ + A +++ + G LDIL+NNA S E+D
Sbjct: 83 ASVAAFASWLRDELGGLDILVNNAAVSFNEID---------------------------- 114
Query: 125 TQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLRNEL 184
T + + AE L TN+YG K + ALLPL + S A +RI+N+SS G L ++ + LR+ L
Sbjct: 115 TNSVEHAETVLRTNFYGAKMLIEALLPLFRRSAANSRILNISSQLGLLNKVRDPSLRSML 174
Query: 185 GDVDELSEGKIDAMVKKFLHDFKANDHEA--NGWGMMLPAYSISKASLNAYTRVLAKK-- 240
D L+EGKI+ M +FL + K A GW + Y++SK +LNAY+RVLA +
Sbjct: 175 LDEASLTEGKIERMASRFLAEVKDGTWSAPGRGWPAVWTDYAVSKLALNAYSRVLAARLA 234
Query: 241 --NPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLL-PADGPTGCYFD-CT 293
+ +NC PGF TD GT T +E R L+LL P D PTG +F CT
Sbjct: 235 RGGDRVAVNCFCPGFTRTDMTRGWGTRTAEEAGRVAAGLALLPPGDLPTGKFFKWCT 291
>UniRef100_Q9M198 Hypothetical protein T16L24.260 [Arabidopsis thaliana]
Length = 302
Score = 178 bits (451), Expect = 2e-43
Identities = 114/293 (38%), Positives = 155/293 (51%), Gaps = 32/293 (10%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
AVVTGANKGIG +VK+L LG+TVVLTARN G A L + G NV F LD+ D
Sbjct: 31 AVVTGANKGIGFAVVKRLLELGLTVVLTARNAENGSQAAESLRRIGFGNVHFCCLDISDP 90
Query: 65 LSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGVL 124
SI + A + G LDIL+NNA S V
Sbjct: 91 SSIAAFASWFGRNLGILDILVNNAAVS----------------------------FNAVG 122
Query: 125 TQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLRNEL 184
K+ E + TN+YG K +T ALLPL + S + +RI+N+SS G L ++ + +R L
Sbjct: 123 ENLIKEPETIIKTNFYGAKLLTEALLPLFRRSVSVSRILNMSSRLGTLNKLRSPSIRRIL 182
Query: 185 GDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAKK--NP 242
+ ++L+ +IDA + +FL D K+ E GW P Y+ISK +LNAY+RVLA++
Sbjct: 183 -ESEDLTNEQIDATLTQFLQDVKSGTWEKQGWPENWPDYAISKLALNAYSRVLARRYDGK 241
Query: 243 HMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADG-PTGCYFDCTE 294
+ +NC+ PGF T +GT T DE A L LLP + TG ++ C E
Sbjct: 242 KLSVNCLCPGFTRTSMTGGQGTHTADEAAAIVAKLVLLPPEKLATGKFYICVE 294
>UniRef100_Q9FLS6 Carbonyl reductase-like protein [Arabidopsis thaliana]
Length = 304
Score = 172 bits (435), Expect = 1e-41
Identities = 116/300 (38%), Positives = 160/300 (52%), Gaps = 42/300 (14%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKL-HQTGLSNVMFHQLDVLD 63
AVVTG+N+GIG EI +QLA G+TVVLTARN G +A+ L HQ V FHQLDV D
Sbjct: 25 AVVTGSNRGIGFEIARQLAVHGLTVVLTARNVNAGLEAVKSLRHQEEGLKVYFHQLDVTD 84
Query: 64 ALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGV 123
+ SI +++ FG LDIL+NNAG + L D
Sbjct: 85 SSSIREFGCWLKQTFGGLDILVNNAGVN---------YNLGSD----------------- 118
Query: 124 LTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLR------GELKRIPN 177
T + AE ++TNY G K +T A++PL++ SP AR+VN+SS EL+R+ N
Sbjct: 119 --NTVEFAETVISTNYQGTKNMTKAMIPLMRPSPHGARVVNVSSRLENLVEIHELQRLAN 176
Query: 178 ERLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVL 237
LR++L D L+E ID V KF++ K E+ GW YS+SK ++NAYTR++
Sbjct: 177 VELRDQLSSPDLLTEELIDRTVSKFINQVKDGTWESGGWPQTFTDYSMSKLAVNAYTRLM 236
Query: 238 AK------KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSL-LPADGPTGCYF 290
AK + + +N PG+V T + G M ++ A V LSL L + TG +F
Sbjct: 237 AKELERRGEEEKIYVNSFCPGWVKTAMTGYAGNMPPEDAADTGVWLSLVLSEEAVTGKFF 296
>UniRef100_UPI00001D06CC UPI00001D06CC UniRef100 entry
Length = 354
Score = 151 bits (381), Expect = 2e-35
Identities = 113/295 (38%), Positives = 158/295 (53%), Gaps = 54/295 (18%)
Query: 3 RYAVVTGANKGIGLEIVKQLA--FLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLD 60
R A+VTGANKGIG I + L F G VVLTAR++ RGR A+ +L GLS FHQLD
Sbjct: 83 RVALVTGANKGIGFAITRDLCRKFSG-DVVLTARDEARGRAAVKQLQAEGLSP-RFHQLD 140
Query: 61 VLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLL 120
+ + SI +L F++ ++G L++L+NNAG + +D DP
Sbjct: 141 IDNPQSIRALRDFLRKEYGGLNVLVNNAGIA-FRMD---------DP------------- 177
Query: 121 QGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPN--- 177
T +AE L TN++ + V LLP+++ R+VN+SSL+G LK + N
Sbjct: 178 ----TPFDVQAEVTLKTNFFATRNVCTELLPIMK---PHGRVVNVSSLQG-LKALENCSE 229
Query: 178 ---ERLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYT 234
ER R D L+EG + ++KKF+ D K HE GW AY +SK + T
Sbjct: 230 DLQERFR-----CDTLTEGDLVDLMKKFVEDTKNEVHEREGWPDS--AYGVSKLGVTVLT 282
Query: 235 RVLAK------KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPAD 283
R+LA+ K +L+N PG+V TD +G+ TV+EGA PV L+LLP D
Sbjct: 283 RILARQLDEKRKADRILLNACCPGWVKTDMARDQGSRTVEEGAETPVYLALLPPD 337
>UniRef100_Q8K354 Carbonyl reductase 3 (Mus musculus adult male tongue cDNA, RIKEN
full- length enriched library, clone:2310037K07
product:CARBONYL REDUCTASE (EC 1.1.1.184) (CARBONYL
REDUCTASE 3) homolog) [Mus musculus]
Length = 277
Score = 150 bits (378), Expect = 5e-35
Identities = 112/292 (38%), Positives = 158/292 (53%), Gaps = 48/292 (16%)
Query: 3 RYAVVTGANKGIGLEIVKQLA--FLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLD 60
R A+VTGANKGIG I + L F G VVLTAR++ RGR A+ +L GLS FHQLD
Sbjct: 6 RVALVTGANKGIGFAITRDLCRKFSG-DVVLTARDEARGRAAVQQLQAEGLSP-RFHQLD 63
Query: 61 VLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLL 120
+ D SI +L F++ ++G L++L+NNAG + +D DP
Sbjct: 64 IDDPQSIRALRDFLRKEYGGLNVLVNNAGIA-FRMD---------DP------------- 100
Query: 121 QGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERL 180
T +AE L TN++ + V LLP+++ R+VN+SSL+G LK + N
Sbjct: 101 ----TPFDIQAEVTLKTNFFATRNVCTELLPIMK---PHGRVVNISSLQG-LKALEN--C 150
Query: 181 RNELGD---VDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVL 237
R +L + D L+E + ++KKF+ D K HE GW AY +SK + TR+L
Sbjct: 151 REDLQEKFRCDTLTEVDLVDLMKKFVEDTKNEVHEREGWPDS--AYGVSKLGVTVLTRIL 208
Query: 238 AK------KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPAD 283
A+ K +L+N PG+V TD +G+ TV+EGA PV L+LLP D
Sbjct: 209 ARQLDEKRKADRILLNACCPGWVKTDMARDQGSRTVEEGAETPVYLALLPPD 260
>UniRef100_UPI00003A9CC2 UPI00003A9CC2 UniRef100 entry
Length = 265
Score = 149 bits (376), Expect = 9e-35
Identities = 111/298 (37%), Positives = 161/298 (53%), Gaps = 54/298 (18%)
Query: 5 AVVTGANKGIGLEIVKQLA--FLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
AVVTG+NKGIGL IV+ L F G V LTAR+ RG++A+ KL + GL + +FHQLD+
Sbjct: 2 AVVTGSNKGIGLAIVRDLCKQFKG-DVYLTARDPARGQEAVAKLQEEGL-HPLFHQLDID 59
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQG 122
D SI+ L F++ K+G L++L+NNAG + K +VD WL G
Sbjct: 60 DLQSIKVLRDFLKEKYGGLNVLVNNAGIA--------FKGKSVD---WL----------G 98
Query: 123 VLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSL-----RGELKRIPN 177
+ +AE L TN++G + + LLPL++ R+VN+SS+ G +
Sbjct: 99 I------QAEVTLKTNFFGTRNICTELLPLIK---PYGRVVNVSSMVSISALGGCSQELQ 149
Query: 178 ERLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVL 237
++ R++ DEL E ++ KF+ D K + HE GW AY +SK + +R+
Sbjct: 150 KKFRSDTITEDELVE-----LMTKFVEDTKKSVHEKEGWPN--TAYGVSKIGVTVLSRIQ 202
Query: 238 A------KKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLP--ADGPTG 287
A +K H+L+N PG+V TD K + +EGA PV L+LLP ADGP G
Sbjct: 203 ARMLNEKRKGDHILLNACCPGWVRTDMAGPKAPKSPEEGAETPVYLALLPSDADGPHG 260
>UniRef100_UPI00003A9CC1 UPI00003A9CC1 UniRef100 entry
Length = 276
Score = 148 bits (374), Expect = 2e-34
Identities = 110/298 (36%), Positives = 159/298 (52%), Gaps = 54/298 (18%)
Query: 5 AVVTGANKGIGLEIVKQLA--FLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
AVVTG+NKGIGL IV+ L F G V LTAR+ RG++A+ KL + GL + +FHQLD+
Sbjct: 7 AVVTGSNKGIGLAIVRDLCKQFKG-DVYLTARDPARGQEAVAKLQEEGL-HPLFHQLDID 64
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQG 122
D SI+ L F++ K+G L++L+NNAG +A KVS+
Sbjct: 65 DLQSIKVLRDFLKEKYGGLNVLVNNAG---------------------IAFKVSDR---- 99
Query: 123 VLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSL-----RGELKRIPN 177
T +AE L TN++G + + LLPL++ R+VN+SS+ G +
Sbjct: 100 --TPFAVQAEVTLKTNFFGTRNICTELLPLIK---PYGRVVNVSSMVSISALGGCSQELQ 154
Query: 178 ERLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVL 237
++ R++ DEL E ++ KF+ D K + HE GW AY +SK + +R+
Sbjct: 155 KKFRSDTITEDELVE-----LMTKFVEDTKKSVHEKEGWPN--TAYGVSKIGVTVLSRIQ 207
Query: 238 A------KKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLP--ADGPTG 287
A +K H+L+N PG+V TD K + +EGA PV L+LLP ADGP G
Sbjct: 208 ARMLNEKRKGDHILLNACCPGWVRTDMAGPKAPKSPEEGAETPVYLALLPSDADGPHG 265
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 472,710,462
Number of Sequences: 2790947
Number of extensions: 18709203
Number of successful extensions: 65058
Number of sequences better than 10.0: 7236
Number of HSP's better than 10.0 without gapping: 1800
Number of HSP's successfully gapped in prelim test: 5440
Number of HSP's that attempted gapping in prelim test: 56263
Number of HSP's gapped (non-prelim): 8926
length of query: 298
length of database: 848,049,833
effective HSP length: 126
effective length of query: 172
effective length of database: 496,390,511
effective search space: 85379167892
effective search space used: 85379167892
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146743.8


