

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.7 + phase: 0
(376 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
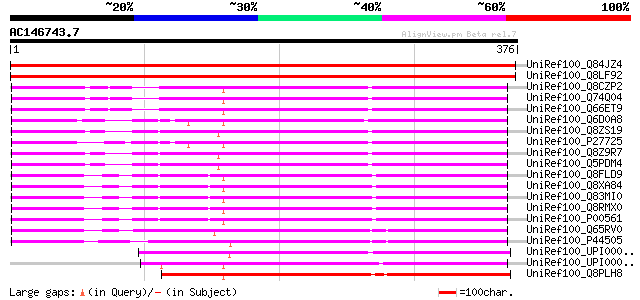
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84JZ4 Putative homoserine dehydrogenase [Arabidopsis ... 515 e-145
UniRef100_Q8LF92 Homoserine dehydrogenase-like protein [Arabidop... 512 e-144
UniRef100_Q8CZP2 Aspartokinase I, homoserine dehydrogenase I [Ye... 209 1e-52
UniRef100_Q74Q04 Bifunctional aspartokinase/homoserine dehydroge... 209 1e-52
UniRef100_Q66ET9 Bifunctional ThrA; aspartokinase I and homoseri... 209 1e-52
UniRef100_Q6D0A8 Bifunctional aspartokinase/homoserine dehydroge... 201 4e-50
UniRef100_Q8ZS19 Aspartokinase I [Salmonella typhimurium] 200 5e-50
UniRef100_P27725 Bifunctional aspartokinase/homoserine dehydroge... 200 5e-50
UniRef100_Q8Z9R7 Aspartokinase I/homoserine dehydrogenase I [Sal... 200 6e-50
UniRef100_Q5PDM4 Aspartokinase I/homoserine dehydrogenase I [Sal... 200 6e-50
UniRef100_Q8FLD9 Aspartokinase I [Escherichia coli O6] 199 8e-50
UniRef100_Q8XA84 Aspartokinase I, homoserine dehydrogenase I [Es... 199 1e-49
UniRef100_Q83MI0 Aspartokinase I, homoserine dehydrogenase I [Sh... 198 2e-49
UniRef100_Q8RMX0 Aspartokinase I-homoserine dehydrogenase I [Esc... 198 2e-49
UniRef100_P00561 Bifunctional aspartokinase/homoserine dehydroge... 198 2e-49
UniRef100_Q65RV0 LysC protein [Mannheimia succiniciproducens] 197 4e-49
UniRef100_P44505 Bifunctional aspartokinase/homoserine dehydroge... 196 1e-48
UniRef100_UPI0000254EC0 UPI0000254EC0 UniRef100 entry 193 6e-48
UniRef100_UPI00002A3B42 UPI00002A3B42 UniRef100 entry 192 2e-47
UniRef100_Q8PLH8 Aspartokinase [Xanthomonas axonopodis] 189 8e-47
>UniRef100_Q84JZ4 Putative homoserine dehydrogenase [Arabidopsis thaliana]
Length = 376
Score = 515 bits (1327), Expect = e-145
Identities = 259/375 (69%), Positives = 313/375 (83%)
Query: 1 MKTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDD 60
MK IP++LMGCGGVG HLLQHIVS RSLH+ G+ +RV+G+ DSKSLV D+L + +D
Sbjct: 1 MKKIPVLLMGCGGVGRHLLQHIVSCRSLHAKMGVHIRVIGVCDSKSLVAPMDVLKEELND 60
Query: 61 SFLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSD 120
L E+C +K G +LSKLG LG +V E + EIA LGK TGLA VDC+AS +
Sbjct: 61 ELLSEVCLIKSTGSALSKLGALGGYRVVHDSELSTETEEIAKLLGKSTGLAVVDCSASME 120
Query: 121 TIVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIRHESTVGAGLPVISSLNRII 180
TI +L + VDLGCC+V+ANKKP+TST+ ++KL +PR IRHESTVGAGLPVI+SLNRII
Sbjct: 121 TIEILMKAVDLGCCIVLANKKPVTSTLEHYDKLALHPRFIRHESTVGAGLPVIASLNRII 180
Query: 181 SSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVARKA 240
SSGDPV+RI+GSLSGTLGYVMSE+EDGKPLSQVV+AAK LGYTEPDPRDDLGGMDVARK
Sbjct: 181 SSGDPVHRIVGSLSGTLGYVMSELEDGKPLSQVVQAAKKLGYTEPDPRDDLGGMDVARKG 240
Query: 241 LILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAASNGN 300
LILAR+LG+RI MDSI+IESLYP+EMGP +M+ DDFL G++ LD++I+ERV+KA+S G
Sbjct: 241 LILARLLGKRIIMDSIKIESLYPEEMGPGLMSVDDFLHNGIVKLDQNIEERVKKASSKGC 300
Query: 301 VLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGNDTT 360
VLRYVCVIEG +VGI+E+ K+S LGRLRGSDN++E+Y+RCY QPLVIQGAGAGNDTT
Sbjct: 301 VLRYVCVIEGSSVQVGIREVSKDSPLGRLRGSDNIVEIYSRCYKEQPLVIQGAGAGNDTT 360
Query: 361 AAGVLADIVDIQDLF 375
AAGVLADI+D+QDLF
Sbjct: 361 AAGVLADIIDLQDLF 375
>UniRef100_Q8LF92 Homoserine dehydrogenase-like protein [Arabidopsis thaliana]
Length = 376
Score = 512 bits (1318), Expect = e-144
Identities = 258/375 (68%), Positives = 312/375 (82%)
Query: 1 MKTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDD 60
MK IP++LMGCGGVG HLLQHIVS RSLH+ G+ +RV+G+ DSKSLV D+L + +D
Sbjct: 1 MKKIPVLLMGCGGVGRHLLQHIVSCRSLHAKMGVHIRVIGVCDSKSLVAPMDVLKEELND 60
Query: 61 SFLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSD 120
L E+C +K G +LSKLG LG +V E + EIA LGK TGLA VDC+AS +
Sbjct: 61 ELLSEVCLIKSTGSALSKLGALGGYRVVHDSELSTETEEIAKLLGKSTGLAVVDCSASME 120
Query: 121 TIVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIRHESTVGAGLPVISSLNRII 180
TI +L + VDLGCC+V+ANKKP+TST+ ++KL +PR IRHESTVGAGLPVI+SLNRII
Sbjct: 121 TIEILMKAVDLGCCIVLANKKPVTSTLEHYDKLALHPRFIRHESTVGAGLPVIASLNRII 180
Query: 181 SSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVARKA 240
SSGDPV+RI+GSLS TLGYVMSE+EDGKPLSQVV+AAK LGYTEPDPRDDLGGMDVARK
Sbjct: 181 SSGDPVHRIVGSLSVTLGYVMSELEDGKPLSQVVQAAKKLGYTEPDPRDDLGGMDVARKG 240
Query: 241 LILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAASNGN 300
LILAR+LG+RI MDSI+IESLYP+EMGP +M+ DDFL G++ LD++I+ERV+KA+S G
Sbjct: 241 LILARLLGKRIIMDSIKIESLYPEEMGPGLMSVDDFLHNGIVKLDQNIEERVKKASSKGC 300
Query: 301 VLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGNDTT 360
VLRYVCVIEG +VGI+E+ K+S LGRLRGSDN++E+Y+RCY QPLVIQGAGAGNDTT
Sbjct: 301 VLRYVCVIEGSSVQVGIREVSKDSPLGRLRGSDNIVEIYSRCYKEQPLVIQGAGAGNDTT 360
Query: 361 AAGVLADIVDIQDLF 375
AAGVLADI+D+QDLF
Sbjct: 361 AAGVLADIIDLQDLF 375
>UniRef100_Q8CZP2 Aspartokinase I, homoserine dehydrogenase I [Yersinia pestis]
Length = 845
Score = 209 bits (532), Expect = 1e-52
Identities = 135/372 (36%), Positives = 202/372 (54%), Gaps = 29/372 (7%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG L++ I + + + LRV GI +SK+++ + D+
Sbjct: 490 QVIEVFVIGVGGVGGALIEQIYRQQPWLKQRHIDLRVCGIANSKAMLTNVHGIAL---DN 546
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL ++ +LS+L L + ++P VDCT+S
Sbjct: 547 WRQELAEVQEPF-NLSRLIRLVKEYHLLNP-------------------VIVDCTSSQAV 586
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP----RRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + ++ R+ +++ VGAGLPVI +L
Sbjct: 587 ADQYADFLTDGFHVVTPNKKANTSSMNYYRQMRAAATKSCRKFLYDTNVGAGLPVIENLQ 646
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + R G LSG+L ++ ++++G LS+ R AK+LGYTEPDPRDDL GMDVA
Sbjct: 647 NLLNAGDELMRFTGILSGSLSFIFGKLDEGMSLSEATRQAKALGYTEPDPRDDLSGMDVA 706
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR G ++ + I++ES+ P D D D L LD + V AA
Sbjct: 707 RKLLILAREAGYKLELADIEVESVLPASF--DASGDVDTFLARLPSLDAEFTRLVANAAE 764
Query: 298 NGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGN 357
G VLRYV VIE RC+V ++ + N L +++ +N L YTR Y PLV++G GAGN
Sbjct: 765 QGKVLRYVGVIEDGRCKVRMEAVDGNDPLYKVKNGENALAFYTRYYQPIPLVLRGYGAGN 824
Query: 358 DTTAAGVLADIV 369
D TAAGV AD++
Sbjct: 825 DVTAAGVFADLL 836
>UniRef100_Q74Q04 Bifunctional aspartokinase/homoserine dehydrogenase I [Yersinia
pestis]
Length = 817
Score = 209 bits (532), Expect = 1e-52
Identities = 135/372 (36%), Positives = 202/372 (54%), Gaps = 29/372 (7%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG L++ I + + + LRV GI +SK+++ + D+
Sbjct: 462 QVIEVFVIGVGGVGGALIEQIYRQQPWLKQRHIDLRVCGIANSKAMLTNVHGIAL---DN 518
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL ++ +LS+L L + ++P VDCT+S
Sbjct: 519 WRQELAEVQEPF-NLSRLIRLVKEYHLLNP-------------------VIVDCTSSQAV 558
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP----RRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + ++ R+ +++ VGAGLPVI +L
Sbjct: 559 ADQYADFLTDGFHVVTPNKKANTSSMNYYRQMRAAATKSCRKFLYDTNVGAGLPVIENLQ 618
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + R G LSG+L ++ ++++G LS+ R AK+LGYTEPDPRDDL GMDVA
Sbjct: 619 NLLNAGDELMRFTGILSGSLSFIFGKLDEGMSLSEATRQAKALGYTEPDPRDDLSGMDVA 678
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR G ++ + I++ES+ P D D D L LD + V AA
Sbjct: 679 RKLLILAREAGYKLELADIEVESVLPASF--DASGDVDTFLARLPSLDAEFTRLVANAAE 736
Query: 298 NGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGN 357
G VLRYV VIE RC+V ++ + N L +++ +N L YTR Y PLV++G GAGN
Sbjct: 737 QGKVLRYVGVIEDGRCKVRMEAVDGNDPLYKVKNGENALAFYTRYYQPIPLVLRGYGAGN 796
Query: 358 DTTAAGVLADIV 369
D TAAGV AD++
Sbjct: 797 DVTAAGVFADLL 808
>UniRef100_Q66ET9 Bifunctional ThrA; aspartokinase I and homoserine dehydrogenase I
[Yersinia pseudotuberculosis]
Length = 819
Score = 209 bits (532), Expect = 1e-52
Identities = 135/372 (36%), Positives = 202/372 (54%), Gaps = 29/372 (7%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG L++ I + + + LRV GI +SK+++ + D+
Sbjct: 464 QVIEVFVIGVGGVGGALIEQIYRQQPWLKQRHIDLRVCGIANSKAMLTNVHGIAL---DN 520
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL ++ +LS+L L + ++P VDCT+S
Sbjct: 521 WRQELAEVQEPF-NLSRLIRLVKEYHLLNP-------------------VIVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP----RRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + ++ R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLTDGFHVVTPNKKANTSSMNYYRQMRAAATKSCRKFLYDTNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + R G LSG+L ++ ++++G LS+ R AK+LGYTEPDPRDDL GMDVA
Sbjct: 621 NLLNAGDELMRFTGILSGSLSFIFGKLDEGMSLSEATRQAKALGYTEPDPRDDLSGMDVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR G ++ + I++ES+ P D D D L LD + V AA
Sbjct: 681 RKLLILAREAGYKLELADIEVESVLPASF--DASGDVDTFLARLPSLDAEFTRLVANAAE 738
Query: 298 NGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGN 357
G VLRYV VIE RC+V ++ + N L +++ +N L YTR Y PLV++G GAGN
Sbjct: 739 QGKVLRYVGVIEDGRCKVRMEAVDGNDPLYKVKNGENALAFYTRYYQPIPLVLRGYGAGN 798
Query: 358 DTTAAGVLADIV 369
D TAAGV AD++
Sbjct: 799 DVTAAGVFADLL 810
>UniRef100_Q6D0A8 Bifunctional aspartokinase/homoserine dehydrogenase I [Erwinia
carotovora]
Length = 819
Score = 201 bits (510), Expect = 4e-50
Identities = 133/375 (35%), Positives = 201/375 (53%), Gaps = 35/375 (9%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ I + + + LRV GI +SK+++ +N D+
Sbjct: 464 QVIDVFVIGVGGVGGALLEQIHRQQPWLKDKHIDLRVCGIANSKAMLTN---INGVSMDT 520
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ +L + + P + G+++ + + + VDCT++
Sbjct: 521 WREDLAKARE-------------------PFNLGRLIRLVKEYHLLNPV-IVDCTSNQ-- 558
Query: 122 IVVLKQVVDL---GCCVVMANKKPLTSTMGDFEKLLTYP----RRIRHESTVGAGLPVIS 174
V Q VD G VV NKK T +M + +L + RR +++ VGAGLPVI
Sbjct: 559 -AVADQYVDFLADGFHVVTPNKKANTGSMNYYHQLRSAASKSRRRFLYDTNVGAGLPVIE 617
Query: 175 SLNRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGM 234
+L ++++GD + R G LSG+L ++ ++++G LS+ AK GYTEPDPRDDL GM
Sbjct: 618 NLQNLLNAGDELIRFTGILSGSLSFIFGKLDEGMSLSEATTQAKEKGYTEPDPRDDLSGM 677
Query: 235 DVARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEK 294
DVARK LILAR G ++ + I++ES+ P + D D L D + RV +
Sbjct: 678 DVARKLLILAREAGYQLELGDIEVESVLP--ISFDASGDVASFMQRLPAADDEFASRVAQ 735
Query: 295 AASNGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAG 354
A G VLRYV VIE RC+V I + N L +++ +N L Y+R Y PLV++G G
Sbjct: 736 ARDEGKVLRYVGVIEEGRCKVKISAVGGNDPLFKVKDGENALAFYSRYYQPLPLVLRGYG 795
Query: 355 AGNDTTAAGVLADIV 369
AGND TAAGV AD++
Sbjct: 796 AGNDVTAAGVFADLL 810
>UniRef100_Q8ZS19 Aspartokinase I [Salmonella typhimurium]
Length = 820
Score = 200 bits (509), Expect = 5e-50
Identities = 133/373 (35%), Positives = 195/373 (51%), Gaps = 30/373 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + ++ ++ + LRV G+ +SK+L+ LN D+
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQTWLKNKHIDLRVCGVANSKALLTNVHGLNL---DN 520
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL + P + G+++ + + + VDCT+S
Sbjct: 521 WQAELAQANA-------------------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKL----LTYPRRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + +L R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSMDYYHQLRFAAAQSRRKFLYDTNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + + G LSG+L ++ ++E+G LSQ A+ +GYTEPDPRDDL GMDVA
Sbjct: 621 NLLNAGDELQKFSGILSGSLSFIFGKLEEGMSLSQATALAREMGYTEPDPRDDLSGMDVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR GR + + I IE + P E D D L LD RV KA
Sbjct: 681 RKLLILARETGRELELSDIVIEPVLPNEF--DASGDVTAFMAHLPQLDDAFAARVAKARD 738
Query: 298 NGNVLRYVCVIEGPR-CEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAG 356
G VLRYV IE C V I E+ N L +++ +N L Y+ Y PLV++G GAG
Sbjct: 739 EGKVLRYVGNIEEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGAG 798
Query: 357 NDTTAAGVLADIV 369
ND TAAGV AD++
Sbjct: 799 NDVTAAGVFADLL 811
>UniRef100_P27725 Bifunctional aspartokinase/homoserine dehydrogenase I (AKI-HDI)
[Includes: Aspartokinase I (EC 2.7.2.4); Homoserine
dehydrogenase I (EC 1.1.1.3)] [Serratia marcescens]
Length = 819
Score = 200 bits (509), Expect = 5e-50
Identities = 133/375 (35%), Positives = 198/375 (52%), Gaps = 35/375 (9%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG L++ I + + + LRV GI +S+ ++
Sbjct: 464 QVIEVFVIGVGGVGGALIEQIYRQQPWLKQKHIDLRVCGIANSRVMLTN----------- 512
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
HG S L Q P + G+++ + + + VDCT+S
Sbjct: 513 --------VHGIALDSWRDALAGAQ---EPFNLGRLIRLVKEYHLLNPV-IVDCTSSQ-- 558
Query: 122 IVVLKQVVDL---GCCVVMANKKPLTSTMGDFEKLLTYP----RRIRHESTVGAGLPVIS 174
V Q VD G VV NKK TS+M +++L R+ +++ VGAGLPVI
Sbjct: 559 -AVADQYVDFLADGFHVVTPNKKANTSSMNYYQQLRAAAAGSHRKFLYDTNVGAGLPVIE 617
Query: 175 SLNRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGM 234
+L ++++GD + R G LSG+L ++ ++++G LS A++ GYTEPDPRDDL GM
Sbjct: 618 NLQNLLNAGDELVRFSGILSGSLSFIFGKLDEGLSLSAATLQARANGYTEPDPRDDLSGM 677
Query: 235 DVARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEK 294
DVARK LILAR G ++ + I++E + P D D D L LDK+ V
Sbjct: 678 DVARKLLILAREAGYKLELSDIEVEPVLPPSF--DASGDVDTFLARLPELDKEFARNVAN 735
Query: 295 AASNGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAG 354
AA G VLRYV +I+ RC+V I+ + N L +++ +N L Y+R Y PLV++G G
Sbjct: 736 AAEQGKVLRYVGLIDEGRCKVRIEAVDGNDPLYKVKNGENALAFYSRYYQPLPLVLRGYG 795
Query: 355 AGNDTTAAGVLADIV 369
AGND TAAGV AD++
Sbjct: 796 AGNDVTAAGVFADLL 810
>UniRef100_Q8Z9R7 Aspartokinase I/homoserine dehydrogenase I [Salmonella typhi]
Length = 820
Score = 200 bits (508), Expect = 6e-50
Identities = 133/373 (35%), Positives = 195/373 (51%), Gaps = 30/373 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + ++ ++ + LRV G+ +SK+L+ LN D+
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQTWLKNKHIDLRVCGVANSKALLTNVHGLNL---DN 520
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL + P + G+++ + + + VDCT+S
Sbjct: 521 WQAELAQANA-------------------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKL----LTYPRRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + +L R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSMDYYHQLRFAAAQSRRKFLYDTNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + + G LSG+L ++ ++E+G LSQ A+ +GYTEPDPRDDL GMDVA
Sbjct: 621 NLLNAGDELQKFSGILSGSLSFIFGKLEEGMSLSQATALAREMGYTEPDPRDDLSGMDVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR GR + + I IE + P E D D L LD RV KA
Sbjct: 681 RKLLILARETGRELELSDIVIEPVLPDEF--DASGDVTAFMAHLPQLDDAFAARVAKARD 738
Query: 298 NGNVLRYVCVIEGPR-CEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAG 356
G VLRYV IE C V I E+ N L +++ +N L Y+ Y PLV++G GAG
Sbjct: 739 EGKVLRYVGNIEEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGAG 798
Query: 357 NDTTAAGVLADIV 369
ND TAAGV AD++
Sbjct: 799 NDVTAAGVFADLL 811
>UniRef100_Q5PDM4 Aspartokinase I/homoserine dehydrogenase I [Salmonella enterica
subsp. enterica serovar Paratypi A str. ATCC 9150]
Length = 820
Score = 200 bits (508), Expect = 6e-50
Identities = 133/373 (35%), Positives = 195/373 (51%), Gaps = 30/373 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + ++ ++ + LRV G+ +SK+L+ LN D+
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQTWLKNKHIDLRVCGVANSKALLTNVHGLNL---DN 520
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL + P + G+++ + + + VDCT+S
Sbjct: 521 WQAELAQANA-------------------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKL----LTYPRRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + +L R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSMDYYHQLRFAAAQSRRKFLYDTNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + + G LSG+L ++ ++E+G LSQ A+ +GYTEPDPRDDL GMDVA
Sbjct: 621 NLLNAGDELQKFSGILSGSLSFIFGKLEEGMSLSQATALAREMGYTEPDPRDDLSGMDVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR GR + + I IE + P E D D L LD RV KA
Sbjct: 681 RKLLILARETGRELELSDIVIEPVLPDEF--DASGDVTAFMAHLPQLDDAFAARVAKARD 738
Query: 298 NGNVLRYVCVIEGPR-CEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAG 356
G VLRYV IE C V I E+ N L +++ +N L Y+ Y PLV++G GAG
Sbjct: 739 EGKVLRYVGNIEEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGAG 798
Query: 357 NDTTAAGVLADIV 369
ND TAAGV AD++
Sbjct: 799 NDVTAAGVFADLL 811
>UniRef100_Q8FLD9 Aspartokinase I [Escherichia coli O6]
Length = 841
Score = 199 bits (507), Expect = 8e-50
Identities = 133/374 (35%), Positives = 198/374 (52%), Gaps = 32/374 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + +S ++ + LRV G+ +SK+L+ LN
Sbjct: 485 QVIEVFVIGVGGVGGALLEQLKRQQSWLKNKHIDLRVCGVANSKALLTNVHGLN------ 538
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L++ E L++ + P + G+++ + + + VDCT+S
Sbjct: 539 -------LENWQEELAQAKE---------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 581
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP-----RRIRHESTVGAGLPVISSL 176
+ G VV NKK TS+M D+ L Y R+ +++ VGAGLPVI +L
Sbjct: 582 ADQYADFLREGFHVVTPNKKANTSSM-DYYHQLRYAAEKSRRKFLYDTNVGAGLPVIENL 640
Query: 177 NRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDV 236
++++GD + + G LSG+L Y+ ++++G S+ A+ +GYTEPDPRDDL GMDV
Sbjct: 641 QNLLNAGDELMKFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDV 700
Query: 237 ARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAA 296
ARK LILAR GR + + I+IE + P E + D L LD RV KA
Sbjct: 701 ARKLLILARETGRELELADIEIEPVLPAEFNAE--GDVAAFMANLSQLDNLFAARVAKAR 758
Query: 297 SNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV I E C V I E+ N L +++ +N L Y+ Y PLV++G GA
Sbjct: 759 DEGKVLRYVGNIDEDGVCRVKIAEVDSNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGA 818
Query: 356 GNDTTAAGVLADIV 369
GND TAAGV AD++
Sbjct: 819 GNDVTAAGVFADLL 832
>UniRef100_Q8XA84 Aspartokinase I, homoserine dehydrogenase I [Escherichia coli
O157:H7]
Length = 820
Score = 199 bits (505), Expect = 1e-49
Identities = 133/374 (35%), Positives = 199/374 (52%), Gaps = 32/374 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + +S ++ + LRV G+ +SK+L+ LN
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQSWLKNKHIDLRVCGVANSKALLTNVHGLN------ 517
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L++ E L++ + P + G+++ + + + VDCT+S
Sbjct: 518 -------LENWQEELAQAKE---------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP-----RRIRHESTVGAGLPVISSL 176
+ G VV NKK TS+M D+ LL + R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSM-DYYHLLRHAAEKSRRKFLYDTNVGAGLPVIENL 619
Query: 177 NRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDV 236
++++GD + + G LSG+L Y+ ++++G S+ A+ +GYTEPDPRDDL GMDV
Sbjct: 620 QNLLNAGDELMKFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDV 679
Query: 237 ARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAA 296
ARK LILAR GR + + I+IE + P E + D L LD RV KA
Sbjct: 680 ARKLLILARETGRELELADIEIEPVLPAEFNAE--GDVAAFMANLSQLDDLFAARVAKAR 737
Query: 297 SNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV I E C V I E+ N L +++ +N L Y+ Y PLV++G GA
Sbjct: 738 DEGKVLRYVGNIDEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGA 797
Query: 356 GNDTTAAGVLADIV 369
GND TAAGV AD++
Sbjct: 798 GNDVTAAGVFADLL 811
>UniRef100_Q83MI0 Aspartokinase I, homoserine dehydrogenase I [Shigella flexneri]
Length = 834
Score = 198 bits (504), Expect = 2e-49
Identities = 133/374 (35%), Positives = 198/374 (52%), Gaps = 32/374 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + +S ++ + LRV G+ +SK+L+ LN
Sbjct: 478 QVIEVFVIGVGGVGGALLEQLKRQQSWLKNKHIDLRVCGVANSKALLTSVHGLN------ 531
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L++ E L++ + P + G+++ + + + VDCT+S
Sbjct: 532 -------LENWQEELAQAKE---------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 574
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP-----RRIRHESTVGAGLPVISSL 176
+ G VV NKK TS+M D+ L Y R+ +++ VGAGLPVI +L
Sbjct: 575 ADQYADFLREGFHVVTPNKKANTSSM-DYYHQLRYAAEKSRRKFLYDTNVGAGLPVIENL 633
Query: 177 NRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDV 236
++++GD + + G LSG+L Y+ ++++G S+ A+ +GYTEPDPRDDL GMDV
Sbjct: 634 QNLLNAGDELVKFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDV 693
Query: 237 ARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAA 296
ARK LILAR GR + + I+IE + P E + D L LD RV KA
Sbjct: 694 ARKLLILARETGRELELADIEIEPVLPAEFNAE--GDVAAFMANLSQLDDLFAARVAKAR 751
Query: 297 SNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV I E C V I E+ N L +++ +N L Y+ Y PLV++G GA
Sbjct: 752 DEGKVLRYVGNIDEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGA 811
Query: 356 GNDTTAAGVLADIV 369
GND TAAGV AD++
Sbjct: 812 GNDVTAAGVFADLL 825
>UniRef100_Q8RMX0 Aspartokinase I-homoserine dehydrogenase I [Escherichia coli]
Length = 820
Score = 198 bits (504), Expect = 2e-49
Identities = 133/374 (35%), Positives = 198/374 (52%), Gaps = 32/374 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + +S ++ + LRV G+ +SK+L+ LN
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQSWLKNKHIDLRVCGVANSKALLTNVHGLN------ 517
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L++ E L++ + P + G+++ + + + VDCT+S
Sbjct: 518 -------LENWQEELAQAKE---------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP-----RRIRHESTVGAGLPVISSL 176
+ G VV NKK TS+M D+ L Y R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSM-DYYHQLRYAAEKSRRKFLYDTNVGAGLPVIENL 619
Query: 177 NRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDV 236
++++GD + + G LSG+L Y+ ++++G S+ A+ +GYTEPDPRDDL GMDV
Sbjct: 620 QNLLNAGDELMKFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDV 679
Query: 237 ARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAA 296
ARK LILAR GR + + I+IE + P E + D L LD RV KA
Sbjct: 680 ARKLLILARETGRELELADIEIEPVLPAEFNAE--GDVAAFMANLSQLDDLFAARVAKAR 737
Query: 297 SNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV I E C V I E+ N L +++ +N L Y+ Y PLV++G GA
Sbjct: 738 DEGKVLRYVGNIDEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGA 797
Query: 356 GNDTTAAGVLADIV 369
GND TAAGV AD++
Sbjct: 798 GNDVTAAGVFADLL 811
>UniRef100_P00561 Bifunctional aspartokinase/homoserine dehydrogenase I (AKI-HDI)
[Includes: Aspartokinase I (EC 2.7.2.4); Homoserine
dehydrogenase I (EC 1.1.1.3)] [Escherichia coli]
Length = 820
Score = 198 bits (504), Expect = 2e-49
Identities = 133/374 (35%), Positives = 198/374 (52%), Gaps = 32/374 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + +S ++ + LRV G+ +SK+L+ LN
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQSWLKNKHIDLRVCGVANSKALLTNVHGLN------ 517
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L++ E L++ + P + G+++ + + + VDCT+S
Sbjct: 518 -------LENWQEELAQAKE---------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP-----RRIRHESTVGAGLPVISSL 176
+ G VV NKK TS+M D+ L Y R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSM-DYYHQLRYAAEKSRRKFLYDTNVGAGLPVIENL 619
Query: 177 NRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDV 236
++++GD + + G LSG+L Y+ ++++G S+ A+ +GYTEPDPRDDL GMDV
Sbjct: 620 QNLLNAGDELMKFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDV 679
Query: 237 ARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAA 296
ARK LILAR GR + + I+IE + P E + D L LD RV KA
Sbjct: 680 ARKLLILARETGRELELADIEIEPVLPAEFNAE--GDVAAFMANLSQLDDLFAARVAKAR 737
Query: 297 SNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV I E C V I E+ N L +++ +N L Y+ Y PLV++G GA
Sbjct: 738 DEGKVLRYVGNIDEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGA 797
Query: 356 GNDTTAAGVLADIV 369
GND TAAGV AD++
Sbjct: 798 GNDVTAAGVFADLL 811
>UniRef100_Q65RV0 LysC protein [Mannheimia succiniciproducens]
Length = 816
Score = 197 bits (501), Expect = 4e-49
Identities = 125/372 (33%), Positives = 195/372 (51%), Gaps = 29/372 (7%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
K++ + L+G GGVG L++ I + + + + +RV + +S +++ ++ L+
Sbjct: 465 KSVDMFLVGVGGVGGELIEQIKQQKEYLAKKDIEIRVCALANSNKMLLNENGLS------ 518
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L + E LS S+ +L +L FVDCT +
Sbjct: 519 -------LDNWKEDLSNATQ----------PSDFDVLLSFIKLHHVVNPVFVDCTTAESV 561
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDF----EKLLTYPRRIRHESTVGAGLPVISSLN 177
+ + + G VV NKK T M + E R+ +++ VGAGLPVI +L
Sbjct: 562 SGLYARALSEGFHVVTPNKKANTREMAYYNLVRENARKNQRKFLYDTNVGAGLPVIENLQ 621
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD V R G LSG+L ++ ++E+G LSQ A+ G+TEPDPRDDL G DVA
Sbjct: 622 NLLAAGDEVERFNGILSGSLSFIFGKLEEGLTLSQATALAREKGFTEPDPRDDLSGQDVA 681
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR G + + +++ES+ PK + + +F+ L LD + RVEKA +
Sbjct: 682 RKLLILARESGLELELSDVEVESVLPKGFS-EGKSAVEFMEI-LPQLDAEFAARVEKAGA 739
Query: 298 NGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGN 357
VLRYV I +C+V I E+ + L +++ +N L YTR Y PL+++G GAGN
Sbjct: 740 QNKVLRYVGQINDGKCKVSIVEVDADDPLYKVKNGENALAFYTRYYQPIPLLLRGYGAGN 799
Query: 358 DTTAAGVLADIV 369
TAAG+ ADI+
Sbjct: 800 AVTAAGIFADIL 811
>UniRef100_P44505 Bifunctional aspartokinase/homoserine dehydrogenase (AK-HD)
[Includes: Aspartokinase (EC 2.7.2.4); Homoserine
dehydrogenase (EC 1.1.1.3)] [Haemophilus influenzae]
Length = 815
Score = 196 bits (497), Expect = 1e-48
Identities = 123/372 (33%), Positives = 199/372 (53%), Gaps = 29/372 (7%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
K + + L+G GGVG L++ + + + + + +RV I +S +++ ++ LN
Sbjct: 464 KVVDMFLVGVGGVGGELIEQVKRQKEYLAKKNVEIRVCAIANSNRMLLDENGLN------ 517
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L K+ E+ ++ D F+ +L FVDCT++
Sbjct: 518 ----LEDWKNDLENATQPSDFDVLLSFI-------------KLHHVVNPVFVDCTSAESV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIRH----ESTVGAGLPVISSLN 177
+ + + G VV NKK T + + +L + +H E+ VGAGLPVI +L
Sbjct: 561 AGLYARALKEGFHVVTPNKKANTRELVYYNELRQNAQASQHKFLYETNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + G LSG+L ++ ++E+G LS+V A+ G+TEPDPRDDL G DVA
Sbjct: 621 NLLAAGDELEYFEGILSGSLSFIFGKLEEGLSLSEVTALAREKGFTEPDPRDDLSGQDVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR G + + +++E + PK D + D+F++ L LD++ + RV A +
Sbjct: 681 RKLLILAREAGIELELSDVEVEGVLPKGFS-DGKSADEFMAM-LPQLDEEFKTRVATAKA 738
Query: 298 NGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGN 357
G VLRYV I +C+V I + N+ L +++ +N L YTR Y PL+++G GAGN
Sbjct: 739 EGKVLRYVGKISEGKCKVSIVAVDLNNPLYKVKDGENALAFYTRYYQPIPLLLRGYGAGN 798
Query: 358 DTTAAGVLADIV 369
TAAG+ ADI+
Sbjct: 799 AVTAAGIFADIL 810
>UniRef100_UPI0000254EC0 UPI0000254EC0 UniRef100 entry
Length = 293
Score = 193 bits (491), Expect = 6e-48
Identities = 109/281 (38%), Positives = 160/281 (56%), Gaps = 8/281 (2%)
Query: 96 KILEIASQLGKKTGLAFVDCTASSDTIVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLT 155
K ++ S+ G +DCT++ D Q ++ G V+ NKK + ++K+
Sbjct: 11 KFVKHVSEDGANQNTLIIDCTSNEDISSKYSQWLESGLHVITPNKKAFSGNFDAYKKIKD 70
Query: 156 YPRRIR----HESTVGAGLPVISSLNRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLS 211
+E+TV AGLPVIS++ +I +GD ++ I G SGTL Y+ + + KP S
Sbjct: 71 LSNSSGGYCFYETTVAAGLPVISTVQSLIDTGDTIHSIEGIFSGTLAYLFNVYDGTKPFS 130
Query: 212 QVVRAAKSLGYTEPDPRDDLGGMDVARKALILARILGRRINMDSIQIESLYPKEMGPDVM 271
Q+V AKS GYTEPDPRDDL GMDVARK I+ R GR + MD+ IE+L P+E+ +
Sbjct: 131 QIVSEAKSSGYTEPDPRDDLTGMDVARKLAIIGREAGRALKMDTFNIENLVPEEL---MN 187
Query: 272 TDDDFLSCGLLLLDKDIQERVEKAASNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLR 330
D D D D+QER A N VLRY + + +G+ E PK+ A ++
Sbjct: 188 VDSDQFLDLFKSYDNDMQERFLNAKKNSMVLRYTAKLNKDNEVFIGLSEYPKDHAFSNIQ 247
Query: 331 GSDNVLEVYTRCYSNQPLVIQGAGAGNDTTAAGVLADIVDI 371
+DN++++++ YS P+VIQG GAG + TAAG+ ADIV +
Sbjct: 248 LTDNIIQIHSDRYSKNPMVIQGPGAGPEVTAAGIFADIVTL 288
>UniRef100_UPI00002A3B42 UPI00002A3B42 UniRef100 entry
Length = 321
Score = 192 bits (487), Expect = 2e-47
Identities = 110/279 (39%), Positives = 160/279 (56%), Gaps = 9/279 (3%)
Query: 98 LEIASQLGKKTGLA---FVDCTASSDTIVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLL 154
L++ Q K+ L VDCT+S ++ G VV ANKK TS+ ++ L+
Sbjct: 40 LKLVEQFVKQNHLVNPVLVDCTSSDALAAQYVDFLNAGFHVVAANKKATTSSYTYYQDLI 99
Query: 155 TYP----RRIRHESTVGAGLPVISSLNRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPL 210
R+ +E+ VGAGLPVI +L + +GD + G LSG+L Y+ ++DG L
Sbjct: 100 AATQKNNRKFLYETNVGAGLPVIDNLQSLFGAGDELLEFSGILSGSLSYMFGALQDGLSL 159
Query: 211 SQVVRAAKSLGYTEPDPRDDLGGMDVARKALILARILGRRINMDSIQIESLYPKEMGPDV 270
S+ AK G+TEPDPRDDL G DVARK LI+AR G ++ + I++ES+ P+
Sbjct: 160 SEATIKAKESGFTEPDPRDDLSGTDVARKLLIIAREAGMKLELSDIEVESVLPQGFAEGD 219
Query: 271 MTDDDFLSCGLLLLDKDIQERVEKAASNGNVLRYVCVIEGPRCEVGIQELPKNSALGRLR 330
D+ L LD D R+E+AAS G VLRYV I+ C+VGI+ + + AL +R
Sbjct: 220 SIDE--FMAKLPSLDADFNARIEQAASEGKVLRYVGTIKEGHCKVGIEAVDSSHALNVIR 277
Query: 331 GSDNVLEVYTRCYSNQPLVIQGAGAGNDTTAAGVLADIV 369
+N L + ++ Y +P VI+G GAG++ TAAGV AD++
Sbjct: 278 DGENALAILSQYYQPRPFVIRGYGAGSEVTAAGVFADVL 316
>UniRef100_Q8PLH8 Aspartokinase [Xanthomonas axonopodis]
Length = 835
Score = 189 bits (481), Expect = 8e-47
Identities = 108/264 (40%), Positives = 162/264 (60%), Gaps = 8/264 (3%)
Query: 113 VDCTASSDTIVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP----RRIRHESTVGA 168
+DC+ S++ + G VV NK+ + + +E + R R+E+TVGA
Sbjct: 567 IDCSGSAEVADRYAGWLAAGIHVVTPNKQAGSGPLARYEAIRAAADASGARFRYEATVGA 626
Query: 169 GLPVISSLNRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPR 228
GLPVI++L ++ +GD V I G SGTL ++ ++ E P +++V A+ +GYTEPDPR
Sbjct: 627 GLPVITTLRDLVDTGDAVTSIEGIFSGTLAWLFNKYEGSVPFAELVTQARGMGYTEPDPR 686
Query: 229 DDLGGMDVARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDI 288
DDL G+DVARK +ILAR GR I+++ +Q+ESL P+ + + DDF++ L +D
Sbjct: 687 DDLSGVDVARKLVILAREAGRSISLEDVQVESLVPEALRQ--ASVDDFMA-RLPEVDASF 743
Query: 289 QERVEKAASNGNVLRYVCVIEGPRC-EVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQP 347
+R+ +A + GNVLRYV + R VG+ ELP + A LR +DNV++ TR Y P
Sbjct: 744 AQRLAQAHARGNVLRYVAQLPPDRAPSVGLVELPADHAFANLRLTDNVVQFTTRRYCENP 803
Query: 348 LVIQGAGAGNDTTAAGVLADIVDI 371
LV+QG GAG + TAAGV AD++ +
Sbjct: 804 LVVQGPGAGPEVTAAGVFADLLRV 827
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.139 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 609,742,034
Number of Sequences: 2790947
Number of extensions: 25586709
Number of successful extensions: 62162
Number of sequences better than 10.0: 568
Number of HSP's better than 10.0 without gapping: 339
Number of HSP's successfully gapped in prelim test: 229
Number of HSP's that attempted gapping in prelim test: 60949
Number of HSP's gapped (non-prelim): 782
length of query: 376
length of database: 848,049,833
effective HSP length: 129
effective length of query: 247
effective length of database: 488,017,670
effective search space: 120540364490
effective search space used: 120540364490
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146743.7


