

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146723.3 - phase: 0
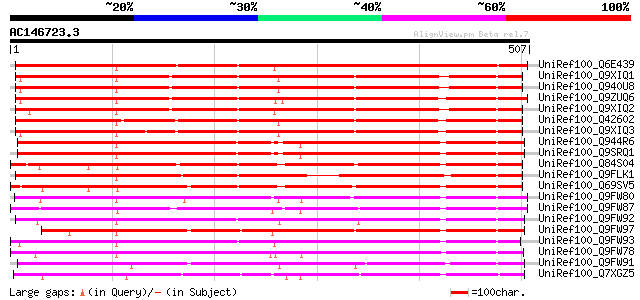
(507 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6E439 ACT11D09.3 [Cucumis melo] 513 e-144
UniRef100_Q9XIQ1 Putative cytochrome p450 protein [Arabidopsis t... 509 e-143
UniRef100_Q940U8 At1g64950/F13O11_25 [Arabidopsis thaliana] 508 e-142
UniRef100_Q9ZUQ6 Putative cytochrome p450 [Arabidopsis thaliana] 503 e-141
UniRef100_Q9XIQ2 Putative cytochrome P450 [Arabidopsis thaliana] 491 e-137
UniRef100_Q42602 Cytochrome P450 89A2 [Arabidopsis thaliana] 491 e-137
UniRef100_Q9XIQ3 F13O11.23 protein [Arabidopsis thaliana] 478 e-133
UniRef100_Q944R6 AT3g03470/T21P5_11 [Arabidopsis thaliana] 448 e-124
UniRef100_Q9SRQ1 Putative cytochrome P450 [Arabidopsis thaliana] 448 e-124
UniRef100_Q84S04 Putative cytochrome P450 [Oryza sativa] 419 e-116
UniRef100_Q9FLK1 Cytochrome P450-like protein [Arabidopsis thali... 408 e-112
UniRef100_Q69SV5 Putative cytochrome P450 [Oryza sativa] 407 e-112
UniRef100_Q9FW80 Putative cytochrome P450 [Oryza sativa] 385 e-105
UniRef100_Q9FW87 Putative cytochrome P450 [Oryza sativa] 384 e-105
UniRef100_Q9FW92 Putative cytochrome P450 [Oryza sativa] 377 e-103
UniRef100_Q9FW97 Putative cytochrome P450 [Oryza sativa] 377 e-103
UniRef100_Q9FW93 Putative cytochrome P450 [Oryza sativa] 374 e-102
UniRef100_Q9FW78 Putative cytochrome P450 [Oryza sativa] 370 e-101
UniRef100_Q9FW91 Putative cytochrome P450 [Oryza sativa] 368 e-100
UniRef100_Q7XGZ5 Putative cytochrome P450 [Oryza sativa] 364 4e-99
>UniRef100_Q6E439 ACT11D09.3 [Cucumis melo]
Length = 518
Score = 513 bits (1321), Expect = e-144
Identities = 263/513 (51%), Positives = 353/513 (68%), Gaps = 15/513 (2%)
Query: 6 WFIITALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHAK 65
WFI L + L + S LPPGP +PI+T+ L+ S +++ ++R AK
Sbjct: 4 WFIFLISLSICSLLTSIFTHFQTSTKLPPGPPSIPILTNFLWLRRSSLQIESLLRSFVAK 63
Query: 66 HGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSCYG 119
+GP++T S P +FI+DR +AH+ L+QN +FADRP K + ++Q NI+S+ YG
Sbjct: 64 YGPVLTLRIGSRPTVFIADRSIAHKILVQNGALFADRPPALSVGKVITSNQHNISSASYG 123
Query: 120 PTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAIF 179
P W +R NL S +L PSR +S+S+ARK VLD L+++L+ ++ N V +I ++A+F
Sbjct: 124 PLWRLLRRNLTSQILHPSRVRSYSEARKWVLDILLNRLQSQSES-GNPVSVIENFQYAMF 182
Query: 180 CLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQLR 239
CLL MCFG+ +D +I ++++++R +ILS + LNF+PK +++LFRKRW+ F QLR
Sbjct: 183 CLLVLMCFGDKLDESQIREVENVERAMILSFQRFNILNFWPKF-TKILFRKRWEAFFQLR 241
Query: 240 KLQNDLLVELIQARKKVI-----EKNNDDDEYVVCYVDTLLNLRLPEEKRKLNEGEVVAL 294
K Q +L LI+AR+K + N+++E VV YVDTLL L LP+EKRKLN+ E+V L
Sbjct: 242 KNQEKVLTRLIEARRKANGNRENKAQNEEEEIVVSYVDTLLELELPDEKRKLNDDELVTL 301
Query: 295 CSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPYL 354
CSEFLTAGTDTTST L+W+MANLVK +Q L E++ V+G EV+EE L KLPYL
Sbjct: 302 CSEFLTAGTDTTSTALQWIMANLVKHPEIQNKLFVEMKGVMGNGSREEVKEEVLGKLPYL 361
Query: 355 KAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVFK 414
KAV+LEGLRRHPP H+ +P V ED L Y++PKN TVNFMVA+IG DP VWEDPT F
Sbjct: 362 KAVVLEGLRRHPPAHFVLPHAVKEDAELGNYVIPKNVTVNFMVAEIGRDPKVWEDPTAFN 421
Query: 415 PERFLKDAKNRNEFDA-FDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFE 473
PERF+K K + E A FD+ G KEIKMMPFGAGRRICP + LA+LHLEYFVANLV FE
Sbjct: 422 PERFVKGGKEKEEQVAEFDITGSKEIKMMPFGAGRRICPGFGLAILHLEYFVANLVWRFE 481
Query: 474 WKTSLPGSNVDLTEKQEFTMVMKYPLEAQISLR 506
WK + G VD++EK E T+ MK PL+A+I R
Sbjct: 482 WKV-VDGDEVDMSEKVELTVAMKKPLKAKIHPR 513
>UniRef100_Q9XIQ1 Putative cytochrome p450 protein [Arabidopsis thaliana]
Length = 510
Score = 509 bits (1311), Expect = e-143
Identities = 263/512 (51%), Positives = 353/512 (68%), Gaps = 29/512 (5%)
Query: 6 WFII--TALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLH 63
W +I + L LLL L+ RLR S S+ LPP P Y P I I L+ L +R +H
Sbjct: 4 WLLILGSLFLSLLLNLLFFRLRDSSSLPLPPDPNYFPFIGTIQWLRQGLGGLNNYLRSVH 63
Query: 64 AKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSC 117
+ GPIIT S P IF++DR LAHQAL+ N VFADRP K + ++Q NI+S
Sbjct: 64 HRLGPIITLRITSRPSIFVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHNISSCL 123
Query: 118 YGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHA 177
YG TW +R NL S +L PSR +S+S AR+ VL+ L + F N + +++++ +A
Sbjct: 124 YGATWRLLRRNLTSEILHPSRVRSYSHARRWVLEILFDR--FGKNRGEEPIVVVDHLHYA 181
Query: 178 IFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQ 237
+F LL MCFG+ +D ++I++++ +QRR +L + LN +PK ++++ RKRW+EF Q
Sbjct: 182 MFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILNLWPKF-TKLILRKRWEEFFQ 240
Query: 238 LRKLQNDLLVELIQARKKVIEKNN--------DDDEYVVCYVDTLLNLRLPEEKRKLNEG 289
+R+ Q+D+L+ LI+AR+K++E+ D+ EYV YVDTLL L LP+EKRKLNE
Sbjct: 241 MRREQHDVLLPLIRARRKIVEERKNRSSEEEEDNKEYVQSYVDTLLELELPDEKRKLNED 300
Query: 290 EVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLE 349
E+V+LCSEFL GTDTT+T L+W+MANLVK+ +Q+ L EEI+ VVGE E EV EED +
Sbjct: 301 EIVSLCSEFLNGGTDTTATALQWIMANLVKNPDIQKRLYEEIKSVVGE-EANEVEEEDAQ 359
Query: 350 KLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWED 409
K+PYL+AV++EGLRRHPP H+ +P VTED VL GY VPKNGT+NFMVA+IG DP VWE+
Sbjct: 360 KMPYLEAVVMEGLRRHPPGHFVLPHSVTEDTVLGGYKVPKNGTINFMVAEIGRDPKVWEE 419
Query: 410 PTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLV 469
P FKPERF++ +A D+ G + IKMMPFGAGRRICP LAMLHLEY+VAN+V
Sbjct: 420 PMAFKPERFME--------EAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMV 471
Query: 470 LNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEA 501
F+WK + G VDLTEK EFT+VMK+PL+A
Sbjct: 472 REFDWK-EVQGHEVDLTEKLEFTVVMKHPLKA 502
>UniRef100_Q940U8 At1g64950/F13O11_25 [Arabidopsis thaliana]
Length = 510
Score = 508 bits (1308), Expect = e-142
Identities = 262/512 (51%), Positives = 353/512 (68%), Gaps = 29/512 (5%)
Query: 6 WFII--TALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLH 63
W +I + L LLL L+ RLR S S+ LPP P Y P I I L+ L +R +H
Sbjct: 4 WLLILGSLFLSLLLNLLFFRLRDSSSLPLPPDPNYFPFIGTIQWLRQGLGGLNNYLRSVH 63
Query: 64 AKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSC 117
+ GPIIT S P IF++DR LAHQAL+ N VFADRP K + ++Q NI+S
Sbjct: 64 HRLGPIITLRITSRPSIFVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHNISSCL 123
Query: 118 YGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHA 177
YG TW +R NL S +L PSR +S+S AR+ VL+ L + F N + +++++ +A
Sbjct: 124 YGATWRLLRRNLTSEILHPSRVRSYSHARRWVLEILFDR--FGKNRGEEPIVVVDHLHYA 181
Query: 178 IFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQ 237
+F LL MCFG+ +D ++I++++ +QRR +L + LN +PK ++++ RKRW+EF Q
Sbjct: 182 MFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILNLWPKF-TKLILRKRWEEFFQ 240
Query: 238 LRKLQNDLLVELIQARKKVIEKNN--------DDDEYVVCYVDTLLNLRLPEEKRKLNEG 289
+R+ Q+D+L+ LI+AR+K++E+ D+ EYV YVDTLL L LP+EKRKLNE
Sbjct: 241 MRREQHDVLLPLIRARRKIVEERKNRSSEEEEDNKEYVQSYVDTLLELELPDEKRKLNED 300
Query: 290 EVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLE 349
E+V+LCSEFL GTDTT+T L+W+MANLVK+ +Q+ L EEI+ VVGE E EV EED +
Sbjct: 301 EIVSLCSEFLNGGTDTTATALQWIMANLVKNPDIQKRLYEEIKSVVGE-EANEVEEEDAQ 359
Query: 350 KLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWED 409
K+PYL+AV++EGLRRHPP H+ +P VTED VL GY VPKNGT+NFMVA+IG DP VWE+
Sbjct: 360 KMPYLEAVVMEGLRRHPPGHFVLPHSVTEDTVLGGYKVPKNGTINFMVAEIGRDPKVWEE 419
Query: 410 PTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLV 469
P FKPERF++ +A D+ G + IKMMPFGAG+RICP LAMLHLEY+VAN+V
Sbjct: 420 PMAFKPERFME--------EAVDITGSRGIKMMPFGAGKRICPGIGLAMLHLEYYVANMV 471
Query: 470 LNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEA 501
F+WK + G VDLTEK EFT+VMK+PL+A
Sbjct: 472 REFDWK-EVQGHEVDLTEKLEFTVVMKHPLKA 502
>UniRef100_Q9ZUQ6 Putative cytochrome p450 [Arabidopsis thaliana]
Length = 512
Score = 503 bits (1295), Expect = e-141
Identities = 263/517 (50%), Positives = 355/517 (67%), Gaps = 27/517 (5%)
Query: 6 WFII--TALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLH 63
W +I + L LLL L++ R R S S+ LPP P + P + + L+ L +R +H
Sbjct: 4 WLLILGSLFLSLLLNLLLFRRRDSSSLPLPPDPNFFPFLGTLQWLRQGLGGLNNYLRSVH 63
Query: 64 AKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSC 117
+ GPIIT S P IF++DR LAHQAL+ N VFADRP K + ++Q NI+SS
Sbjct: 64 HRLGPIITLRITSRPAIFVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHNISSSL 123
Query: 118 YGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHA 177
YG TW +R NL S +L PSR +S+S AR+ VL+ L + F + + +++++ +A
Sbjct: 124 YGATWRLLRRNLTSEILHPSRVRSYSHARRWVLEILFDR--FGKSRGEEPIVVVDHLHYA 181
Query: 178 IFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQ 237
+F LL MCFG+ +D ++I++++ +QRR +L + LN +PK ++++ RKRW+EF Q
Sbjct: 182 MFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILNLWPKF-TKLILRKRWEEFFQ 240
Query: 238 LRKLQNDLLVELIQARKKVIE--KNNDDDE------YVVCYVDTLLNLRLPEEKRKLNEG 289
+R+ Q+D+L+ LI+AR+K++E KN +E YV YVDTLL L LP+EKRKLNE
Sbjct: 241 MRREQHDVLLPLIRARRKIVEERKNRSSEEEEDNKVYVQSYVDTLLELELPDEKRKLNED 300
Query: 290 EVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLE 349
E+V+LCSEFL GTDTT+T L+W+MANLVK+ +Q+ L EEI+ VVGE E +EV EED +
Sbjct: 301 EIVSLCSEFLNGGTDTTATALQWIMANLVKNPEIQKRLYEEIKSVVGE-EAKEVEEEDAQ 359
Query: 350 KLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWED 409
K+PYLKAV++EGLRRHPP H+ +P VTED VL GY VPK GT+NFMVA+IG DP VWE+
Sbjct: 360 KMPYLKAVVMEGLRRHPPGHFVLPHSVTEDTVLGGYKVPKKGTINFMVAEIGRDPMVWEE 419
Query: 410 PTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLV 469
P FKPERF+ E +A D+ G + IKMMPFGAGRRICP LAMLHLEY+VAN+V
Sbjct: 420 PMAFKPERFM------GEEEAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMV 473
Query: 470 LNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEAQISLR 506
FEWK + G VDLTEK EFT+VMK+ L+A LR
Sbjct: 474 REFEWK-EVQGHEVDLTEKFEFTVVMKHSLKALAVLR 509
>UniRef100_Q9XIQ2 Putative cytochrome P450 [Arabidopsis thaliana]
Length = 511
Score = 491 bits (1265), Expect = e-137
Identities = 254/513 (49%), Positives = 348/513 (67%), Gaps = 30/513 (5%)
Query: 6 WFIITALLVLLLC---LIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKL 62
W +I L L L L+ R S S+ LPP P + P I + L+ L +R +
Sbjct: 4 WLLILGSLFLSLLVNHLLFRRRDSFSSLPLPPDPNFFPFIGTLKWLRKGLGGLDNYLRSV 63
Query: 63 HAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASS 116
H GPIIT S P IF++DR LAHQAL+ N VFADRP K + ++Q NI+S
Sbjct: 64 HHHLGPIITLRITSRPAIFVTDRSLAHQALVLNGAVFADRPPAESISKIISSNQHNISSC 123
Query: 117 CYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRH 176
YG TW +R NL S +L PSR +S+S AR+ VL+ L + F N + +++++ +
Sbjct: 124 LYGATWRLLRRNLTSEILHPSRLRSYSHARRWVLEILFGR--FGKNRGEEPIVVVDHLHY 181
Query: 177 AIFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFT 236
A+F LL MCFG+ +D ++I++++ +QRR +L + L +PK +++++RKRW+EF
Sbjct: 182 AMFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILTLWPKF-TKLIYRKRWEEFF 240
Query: 237 QLRKLQNDLLVELIQARKKVI--------EKNNDDDEYVVCYVDTLLNLRLPEEKRKLNE 288
Q++ Q D+L+ LI+AR+K++ E+ D+ EYV YVDTLL++ LP+EKRKLNE
Sbjct: 241 QMQSEQQDVLLPLIRARRKIVDERKKRSSEEEKDNKEYVQSYVDTLLDVELPDEKRKLNE 300
Query: 289 GEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDL 348
E+V+LCSEFL AGTDTT+T L+W+MANLVK+ +Q+ L EEI+ +VGE E +EV E+D
Sbjct: 301 DEIVSLCSEFLNAGTDTTATALQWIMANLVKNPEIQRRLYEEIKSIVGE-EAKEVEEQDA 359
Query: 349 EKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWE 408
+K+PYLKAV++EGLRRHPP H+ +P VTED VL GY VPK GT+NFMVA+IG DP VWE
Sbjct: 360 QKMPYLKAVVMEGLRRHPPGHFVLPHSVTEDTVLGGYKVPKKGTINFMVAEIGRDPKVWE 419
Query: 409 DPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANL 468
+P FKPERF++ +A D+ G + IKMMPFGAGRRICP LAMLHLEY+VAN+
Sbjct: 420 EPMAFKPERFME--------EAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANM 471
Query: 469 VLNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEA 501
V FEW+ + G VDLTEK EFT+VMK+PL+A
Sbjct: 472 VREFEWQ-EVQGHEVDLTEKLEFTVVMKHPLKA 503
>UniRef100_Q42602 Cytochrome P450 89A2 [Arabidopsis thaliana]
Length = 506
Score = 491 bits (1264), Expect = e-137
Identities = 258/508 (50%), Positives = 347/508 (67%), Gaps = 25/508 (4%)
Query: 6 WFIITALLV-LLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHA 64
W +I A L LL ++LR R+S S LPP P +LP + + L+ L+ +R +H
Sbjct: 4 WLLILASLSGSLLLHLLLRRRNSSSPPLPPDPNFLPFLGTLQWLREGLGGLESYLRSVHH 63
Query: 65 KHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSCY 118
+ GPI+T S P IF++DR L H+AL+ N V+ADRP K + H NI+S Y
Sbjct: 64 RLGPIVTLRITSRPAIFVADRSLTHEALVLNGAVYADRPPPAVISKIVDEH--NISSGSY 121
Query: 119 GPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAI 178
G TW +R N+ S +L PSR +S+S AR VL+ L + + + LI+++ +A+
Sbjct: 122 GATWRLLRRNITSEILHPSRVRSYSHARHWVLEILFERFRNHGG--EEPIVLIHHLHYAM 179
Query: 179 FCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQL 238
F LL MCFG+ +D ++I++++ IQR +LS + N +PK ++++ RKRWQEF Q+
Sbjct: 180 FALLVLMCFGDKLDEKQIKEVEFIQRLQLLSLTKFNIFNIWPKF-TKLILRKRWQEFLQI 238
Query: 239 RKLQNDLLVELIQARKKVIEKNN-----DDDEYVVCYVDTLLNLRLPEEKRKLNEGEVVA 293
R+ Q D+L+ LI+AR+K++E+ D +YV YVDTLL+L LPEE RKLNE +++
Sbjct: 239 RRQQRDVLLPLIRARRKIVEERKRSEQEDKKDYVQSYVDTLLDLELPEENRKLNEEDIMN 298
Query: 294 LCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPY 353
LCSEFLTAGTDTT+T L+W+MANLVK +Q+ L EEI+ VVGE E +EV EED+EK+PY
Sbjct: 299 LCSEFLTAGTDTTATALQWIMANLVKYPEIQERLHEEIKSVVGE-EAKEVEEEDVEKMPY 357
Query: 354 LKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVF 413
LKAV+LEGLRRHPP H+ +P VTED VL GY VPKNGT+NFMVA+IG DP WE+P F
Sbjct: 358 LKAVVLEGLRRHPPGHFLLPHSVTEDTVLGGYKVPKNGTINFMVAEIGRDPVEWEEPMAF 417
Query: 414 KPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFE 473
KPERF+ E +A D+ G + IKMMPFGAGRRICP LAMLHLEY+VAN+V F+
Sbjct: 418 KPERFM------GEEEAVDLTGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMVREFQ 471
Query: 474 WKTSLPGSNVDLTEKQEFTMVMKYPLEA 501
WK + G VDLTEK EFT+VMK+PL+A
Sbjct: 472 WK-EVQGHEVDLTEKLEFTVVMKHPLKA 498
>UniRef100_Q9XIQ3 F13O11.23 protein [Arabidopsis thaliana]
Length = 511
Score = 478 bits (1229), Expect = e-133
Identities = 248/512 (48%), Positives = 342/512 (66%), Gaps = 28/512 (5%)
Query: 6 WFII--TALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLH 63
W +I + L LLL L+ RLR S S+ LPP P + P + + L+ +R +H
Sbjct: 4 WLLILGSLSLSLLLNLLFFRLRDSSSLPLPPAPNFFPFLGTLQWLRQGLGGFNNYVRSVH 63
Query: 64 AKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSC 117
+ GPIIT S P IF++D LAHQAL+ N VFADRP K L N+Q I S
Sbjct: 64 HRLGPIITLRITSRPAIFVADGSLAHQALVLNGAVFADRPPAAPISKILSNNQHTITSCL 123
Query: 118 YGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHA 177
YG TW +R N+ I L PSR KS+S R VL+ L +L+ + + +++ +A
Sbjct: 124 YGVTWRLLRRNITEI-LHPSRMKSYSHVRHWVLEILFDRLRKSGG--EEPIVVFDHLHYA 180
Query: 178 IFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQ 237
+F +L MCFG+ +D ++I++++ +QR+++L + LN PK ++++ RKRW+EF Q
Sbjct: 181 MFAVLVLMCFGDKLDEKQIKQVEYVQRQMLLGFARYSILNLCPKF-TKLILRKRWEEFFQ 239
Query: 238 LRKLQNDLLVELIQARKKVIEKNN--------DDDEYVVCYVDTLLNLRLPEEKRKLNEG 289
+R+ Q D+L+ LI AR+K++E+ ++ EYV YVDTLL++ LP+EKRKLNE
Sbjct: 240 MRREQQDVLLRLIYARRKIVEERKKRSSEEEEENKEYVQSYVDTLLDVELPDEKRKLNED 299
Query: 290 EVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLE 349
E+V+LCSEFL AG+DTT+T L+W+MANLVK++ +Q+ L EEI VVGE E + V E+D +
Sbjct: 300 EIVSLCSEFLIAGSDTTATVLQWIMANLVKNQEIQERLYEEITNVVGE-EAKVVEEKDTQ 358
Query: 350 KLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWED 409
K+PYLKAV++E LRRHPP + +P VTED VL GY VPK GT+NF+VA+IG DP VWE+
Sbjct: 359 KMPYLKAVVMEALRRHPPGNTVLPHSVTEDTVLGGYKVPKKGTINFLVAEIGRDPKVWEE 418
Query: 410 PTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLV 469
P FKPERF+ E +A D+ G + IKMMPFGAGRRICP LAMLHLEY+VAN+V
Sbjct: 419 PMAFKPERFM------GEEEAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMV 472
Query: 470 LNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEA 501
F+WK + G VDLTEK EFT++MK+PL+A
Sbjct: 473 REFQWK-EVEGHEVDLTEKVEFTVIMKHPLKA 503
>UniRef100_Q944R6 AT3g03470/T21P5_11 [Arabidopsis thaliana]
Length = 511
Score = 448 bits (1153), Expect = e-124
Identities = 247/513 (48%), Positives = 341/513 (66%), Gaps = 29/513 (5%)
Query: 8 IITALLVLLLCLIILRLRSSQSIH-LPPGPLYLPIITDI-FLLQISFSKLKPIIRKLHAK 65
II ++ L I L+L S H LPPGP P+I +I +L + +FS + ++R L ++
Sbjct: 6 IIFLIISSLTFSIFLKLIFFFSTHKLPPGPPRFPVIGNIIWLKKNNFSDFQGVLRDLASR 65
Query: 66 HGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSCYG 119
HGPIIT H S P I+++DR LAHQAL+QN VF+DR K + ++Q +I SS YG
Sbjct: 66 HGPIITLHVGSKPSIWVTDRSLAHQALVQNGAVFSDRSLALPTTKVITSNQHDIHSSVYG 125
Query: 120 PTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAIF 179
W +R NL S +LQPSR K+ + +RK L+ L+ + E + + ++++RHA+F
Sbjct: 126 SLWRTLRRNLTSEILQPSRVKAHAPSRKWSLEILVDLFETEQREKGHISDALDHLRHAMF 185
Query: 180 CLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQLR 239
LL MCFGE + +I +I++ Q ++++S + LN FP V++ L R++W+EF +LR
Sbjct: 186 YLLALMCFGEKLRKEEIREIEEAQYQMLISYTKFSVLNIFPS-VTKFLLRRKWKEFLELR 244
Query: 240 KLQNDLLVELIQARKKVIEKNNDDDEYVVCYVDTLLNLRLPEE------KRKLNEGEVVA 293
K Q +++ + AR K E D V+CYVDTLLNL +P E KRKL++ E+V+
Sbjct: 245 KSQESVILRYVNARSK--ETTGD----VLCYVDTLLNLEIPTEEKEGGKKRKLSDSEIVS 298
Query: 294 LCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEV-VGEREERE-VREEDLEKL 351
LCSEFL A TD T+T+++W+MA +VK +Q+ + EE++ V GE EERE +REEDL KL
Sbjct: 299 LCSEFLNAATDPTATSMQWIMAIMVKYPEIQRKVYEEMKTVFAGEEEEREEIREEDLGKL 358
Query: 352 PYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPT 411
YLKAVILE LRRHPP HY VT D VL G+L+P+ GT+NFMV ++G DP +WEDP
Sbjct: 359 SYLKAVILECLRRHPPGHYLSYHKVTRDTVLGGFLIPRQGTINFMVGEMGRDPKIWEDPL 418
Query: 412 VFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLN 471
FKPERFL++ E FD+ G +EIKMMPFGAGRR+CP Y L++LHLEY+VANLV
Sbjct: 419 TFKPERFLEN----GEACDFDMTGTREIKMMPFGAGRRMCPGYALSLLHLEYYVANLVWK 474
Query: 472 FEWKTSLPGSNVDLTEKQEF-TMVMKYPLEAQI 503
FEWK + G VDL+EKQ+F TMVMK P +A I
Sbjct: 475 FEWK-CVEGEEVDLSEKQQFITMVMKNPFKANI 506
>UniRef100_Q9SRQ1 Putative cytochrome P450 [Arabidopsis thaliana]
Length = 511
Score = 448 bits (1153), Expect = e-124
Identities = 247/513 (48%), Positives = 341/513 (66%), Gaps = 29/513 (5%)
Query: 8 IITALLVLLLCLIILRLRSSQSIH-LPPGPLYLPIITDI-FLLQISFSKLKPIIRKLHAK 65
II ++ L I L+L S H LPPGP P+I +I +L + +FS + ++R L ++
Sbjct: 6 IIFLIISSLTFSIFLKLIFFFSTHKLPPGPPRFPVIGNIIWLKKNNFSDFQGVLRDLASR 65
Query: 66 HGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSCYG 119
HGPIIT H S P I+++DR LAHQAL+QN VF+DR K + ++Q +I SS YG
Sbjct: 66 HGPIITLHVGSKPSIWVTDRSLAHQALVQNGAVFSDRSLALPTTKVITSNQHDIHSSVYG 125
Query: 120 PTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAIF 179
W +R NL S +LQPSR K+ + +RK L+ L+ + E + + ++++RHA+F
Sbjct: 126 SLWRTLRRNLTSEILQPSRVKAHAPSRKWSLEILVDLFETEQREKGHISDALDHLRHAMF 185
Query: 180 CLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQLR 239
LL MCFGE + +I +I++ Q ++++S + LN FP V++ L R++W+EF +LR
Sbjct: 186 YLLALMCFGEKLRKEEIREIEEAQYQMLISYTKFSVLNIFPS-VTKFLLRRKWKEFLELR 244
Query: 240 KLQNDLLVELIQARKKVIEKNNDDDEYVVCYVDTLLNLRLPEE------KRKLNEGEVVA 293
K Q +++ + AR K E D V+CYVDTLLNL +P E KRKL++ E+V+
Sbjct: 245 KSQESVILRYVNARSK--ETTGD----VLCYVDTLLNLEIPTEEKEGGKKRKLSDSEIVS 298
Query: 294 LCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEV-VGEREERE-VREEDLEKL 351
LCSEFL A TD T+T+++W+MA +VK +Q+ + EE++ V GE EERE +REEDL KL
Sbjct: 299 LCSEFLNAATDPTATSMQWIMAIMVKYPEIQRKVYEEMKTVFAGEEEEREEIREEDLGKL 358
Query: 352 PYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPT 411
YLKAVILE LRRHPP HY VT D VL G+L+P+ GT+NFMV ++G DP +WEDP
Sbjct: 359 SYLKAVILECLRRHPPGHYLSYHKVTHDTVLGGFLIPRQGTINFMVGEMGRDPKIWEDPL 418
Query: 412 VFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLN 471
FKPERFL++ E FD+ G +EIKMMPFGAGRR+CP Y L++LHLEY+VANLV
Sbjct: 419 TFKPERFLEN----GEACDFDMTGTREIKMMPFGAGRRMCPGYALSLLHLEYYVANLVWK 474
Query: 472 FEWKTSLPGSNVDLTEKQEF-TMVMKYPLEAQI 503
FEWK + G VDL+EKQ+F TMVMK P +A I
Sbjct: 475 FEWK-CVEGEEVDLSEKQQFITMVMKNPFKANI 506
>UniRef100_Q84S04 Putative cytochrome P450 [Oryza sativa]
Length = 516
Score = 419 bits (1078), Expect = e-116
Identities = 233/521 (44%), Positives = 332/521 (63%), Gaps = 39/521 (7%)
Query: 1 MEAMAWFIITALLVLLLCLIILRLRSS---------QSIHLPPGPLYLPIITDIFLLQIS 51
ME ++ +T+LL L C ++LR R+S ++ LPPGP +P++ + L
Sbjct: 1 MEDWLFYSLTSLLCLA-CSLLLRARASAASPKAAAAEAAPLPPGPRTVPVLGPLLFLARR 59
Query: 52 FSKLKPIIRKLHAKHGPIITFHFW--STPHIFISDRLLAHQALIQNAVVFADRPK----- 104
++P +R++ A+HGP+ TF S P IF++ R AH+AL+Q FA RP+
Sbjct: 60 DFDVEPTLRRIAAEHGPVFTFAPLGPSRPTIFVAARGAAHRALVQRGAAFASRPRGGGGP 119
Query: 105 ---FLKNHQFNIASSCYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEA 161
L + N++S+ YGPTW A+R ++S +L P+R ++FS AR+ VLD LIS ++ E
Sbjct: 120 ASALLSSGGRNVSSAPYGPTWRALRRCISSGVLNPARLRAFSDARRWVLDVLISHVRGEG 179
Query: 162 NFLNNSVELINYIRHAIFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPK 221
V ++ ++A+FCLL MCFG+ ++ +I+ +QR L+ + F P
Sbjct: 180 GA---PVTVMEPFQYAMFCLLVYMCFGDRPGDARVREIEALQRDLLGNFLSFQVFAFLPP 236
Query: 222 IVSRVLFRKRWQEFTQLRKLQNDLLVELIQARKKVIEKNNDDDEYVVCYVDTLLNLRLPE 281
I ++++FR+RW + LR+ Q +L V LI+AR++ + CYVD+L+NL +PE
Sbjct: 237 I-TKLVFRERWNKLVSLRRRQEELFVPLIRARREAGAGGD-------CYVDSLVNLTIPE 288
Query: 282 EK-RKLNEGEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREE 340
+ R L +GE+V+LCSEF++AGTDTT+T L+W++ANLVK+ +Q L EEI VG +
Sbjct: 289 DGGRGLTDGEIVSLCSEFMSAGTDTTATALQWILANLVKNPAMQDRLREEIAAAVGS--D 346
Query: 341 REVREEDLEKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADI 400
EVREEDL+ +PYLKAV+LEGLRRHPP HY +P V ++ L+GY VP N +NF V +I
Sbjct: 347 GEVREEDLQAMPYLKAVVLEGLRRHPPGHYVLPHAVEDETTLDGYRVPANTPMNFAVGEI 406
Query: 401 GLDPTVWEDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLH 460
GLD VW P VF+PERFL E + D+ G KEIKMMPFGAGRR+CP LA+LH
Sbjct: 407 GLDGEVWASPEVFRPERFLPG----GEGEDVDLTGSKEIKMMPFGAGRRVCPGMALALLH 462
Query: 461 LEYFVANLVLNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEA 501
LEYFVANLV F+W+ + G VDLTEK EFT+VMK PL+A
Sbjct: 463 LEYFVANLVWEFDWR-EVAGDEVDLTEKLEFTVVMKRPLKA 502
>UniRef100_Q9FLK1 Cytochrome P450-like protein [Arabidopsis thaliana]
Length = 483
Score = 408 bits (1049), Expect = e-112
Identities = 223/504 (44%), Positives = 311/504 (61%), Gaps = 47/504 (9%)
Query: 6 WFIITALLVLLLCLIILRLR-SSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHA 64
WF I + + L+ L R ++ ++ LPP P + P+ L+ F +R +H
Sbjct: 4 WFHIFCVSFTVNFLLYLFFRRTNNNLPLPPNPNFFPMPGPFQWLRQGFDDFYSYLRSIHH 63
Query: 65 KHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSCY 118
+ GPII+ +S P IF+SDR LAH+AL+ N VF+DRP K + ++Q I+S Y
Sbjct: 64 RLGPIISLRIFSVPAIFVSDRSLAHKALVLNGAVFSDRPPALPTGKIITSNQHTISSGSY 123
Query: 119 GPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAI 178
G TW +R NL S +L PSR KS+S AR+ VL+ L S+++ V +++++R+A+
Sbjct: 124 GATWRLLRRNLTSEILHPSRVKSYSNARRSVLENLCSRIRNHGEEAKPIV-VVDHLRYAM 182
Query: 179 FCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQL 238
F LL MCFG+ +D +I++++ +QRR +++ + LN FP +++ RKRW+EF
Sbjct: 183 FSLLVLMCFGDKLDEEQIKQVEFVQRRELITLPRFNILNVFPSF-TKLFLRKRWEEFLTF 241
Query: 239 RKLQNDLLVELIQARKKV-IEKNNDDDEYVVCYVDTLLNLRLPEEKRKLNEGEVVALCSE 297
R+ ++L+ LI++R+K+ IE + EY+ YVDTLL+L LP+EKRKLNE E+
Sbjct: 242 RREHKNVLLPLIRSRRKIMIESKDSGKEYIQSYVDTLLDLELPDEKRKLNEDEI------ 295
Query: 298 FLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPYLKAV 357
+Q+ L EEI+ V+GE EE+E+ EE+++K+PYLKAV
Sbjct: 296 ------------------------EIQKRLYEEIKSVIGEEEEKEIEEEEMKKMPYLKAV 331
Query: 358 ILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVFKPER 417
+LEGLR HPP H +P V+ED L GY VPK GT N VA IG DPTVWE+P FKPER
Sbjct: 332 VLEGLRLHPPGHLLLPHRVSEDTELGGYRVPKKGTFNINVAMIGRDPTVWEEPMEFKPER 391
Query: 418 FLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFEWKTS 477
F+ + K DV G + IKMMPFGAGRRICP AMLHLEYFV NLV FEWK
Sbjct: 392 FIGEDKE------VDVTGSRGIKMMPFGAGRRICPGIGSAMLHLEYFVVNLVKEFEWK-E 444
Query: 478 LPGSNVDLTEKQEFTMVMKYPLEA 501
+ G VDL+EK EFT+VMKYPL+A
Sbjct: 445 VEGYEVDLSEKWEFTVVMKYPLKA 468
>UniRef100_Q69SV5 Putative cytochrome P450 [Oryza sativa]
Length = 514
Score = 407 bits (1046), Expect = e-112
Identities = 229/515 (44%), Positives = 324/515 (62%), Gaps = 34/515 (6%)
Query: 1 MEAMAWFIITALLVLLLCLIILRLRSSQSI----HLPPGPLYLPIITDIFLLQISFSKLK 56
ME ++ +T LL+ L C ++LR R+S + LPPGP +P++ + L ++
Sbjct: 1 MEDWIFYSLT-LLLCLACSLLLRARASAAAVEVAPLPPGPRTVPVLGPLLFLARRDFDVE 59
Query: 57 PIIRKLHAKHGPIITFHFW--STPHIFISDRLLAHQALIQNAVVFADRPK-------FLK 107
P +R++ A+HGP+ TF S P IF++ R AH+AL+Q FA RP+ L
Sbjct: 60 PTLRRIAAEHGPVFTFAPLGPSRPTIFVAARGPAHRALVQRGAAFASRPRGVSPASVLLT 119
Query: 108 NHQFNIASSCYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNS 167
+ N++S+ +GP W A+R ++S +L P+R ++FS AR+ VLDAL+S ++ E
Sbjct: 120 SGGRNVSSAQHGPIWRALRRCISSGVLNPARLRAFSDARRWVLDALVSHIRGEGGAPLTV 179
Query: 168 VELINYIRHAIFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVL 227
+E Y A+FCLL MCFG+ ++ +I+ +QR L+ + F P I +R++
Sbjct: 180 MEPFQY---AMFCLLVYMCFGDRPGDARVREIEALQRELLSNFLSFEVFAFLPPI-TRLV 235
Query: 228 FRKRWQEFTQLRKLQNDLLVELIQARKKVIEKNNDDDEYVVCYVDTLLNLRLPEEK-RKL 286
FR+RW + LR+ Q +L LI+AR++ + CYVD+L+ L +PE+ R L
Sbjct: 236 FRRRWNKLVSLRRRQEELFAPLIRARREAGAGGD-------CYVDSLVKLTIPEDGGRGL 288
Query: 287 NEGEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREE 346
+ E+V+LCSEF++AGTDTT+T L+W++ANLVK+ +Q L EEI + EVREE
Sbjct: 289 TDVEIVSLCSEFMSAGTDTTATALQWILANLVKNPAMQDKLREEITAAA---VDGEVREE 345
Query: 347 DLEKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTV 406
DL+ +PYLKAV+LEGLRRHPP H+ +P V E+ L+GY VP N VNF V +IGLD V
Sbjct: 346 DLQAMPYLKAVVLEGLRRHPPDHFLLPHTVEEETTLDGYRVPANTPVNFAVGEIGLDSEV 405
Query: 407 WEDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVA 466
W P VF+PERFL E + D+ G KEIKMMPFGAGRRICP LA+LHLEYFVA
Sbjct: 406 WTSPEVFRPERFLAG----GEGEDVDLTGSKEIKMMPFGAGRRICPGMALALLHLEYFVA 461
Query: 467 NLVLNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEA 501
NLV FEW+ + G VDLT+K +FT+VMK PL+A
Sbjct: 462 NLVREFEWR-EVAGDEVDLTQKLQFTVVMKRPLKA 495
>UniRef100_Q9FW80 Putative cytochrome P450 [Oryza sativa]
Length = 520
Score = 385 bits (990), Expect = e-105
Identities = 221/524 (42%), Positives = 306/524 (58%), Gaps = 32/524 (6%)
Query: 5 AWFIITALLVLLLCLIILRLRSSQ--SIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKL 62
+W ++ LVL L +++ R R S S +PPGPL +P++ + L+ S + L+P++++L
Sbjct: 6 SWLLLLVALVLPLVVLLARRRRSGGGSRRIPPGPLAVPVLGSLLWLRHSSANLEPLLQRL 65
Query: 63 HAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP----KFLKNHQFNIASSCY 118
A++GP+++ S IF++DR +AH AL++ ADRP L I S Y
Sbjct: 66 IARYGPVVSLRVGSRLSIFVADRRVAHAALVERGAALADRPDVTRSLLGETGNTITRSSY 125
Query: 119 GPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVE---LINYIR 175
GP W +R NL + L PSR + F+ AR V L+ KL A + ++ R
Sbjct: 126 GPVWRVLRRNLVAETLHPSRVRLFAPARSWVRRVLVDKLADGARPESEPPRPRVVVETFR 185
Query: 176 HAIFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEF 235
+A+FCLL MCFGE +D + I QR +L + ++ F V++ +FR+R Q
Sbjct: 186 YAMFCLLVLMCFGERLDEAAVRAIGAAQRDWLLYVARKTSVFAFYPAVTKHIFRRRLQIG 245
Query: 236 TQLRKLQNDLLVELIQARKKVIEKNN-----------DDDEYVVCYVDTLLNLRLPEEK- 283
LR+ Q +L V LI AR+ KN+ + + YVDTLL++ LP+
Sbjct: 246 LALRRQQKELFVPLIDARRA--RKNHIQQAGGPPVPEKETTFEHSYVDTLLDVSLPDTDG 303
Query: 284 -RKLNEGEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREERE 342
L E E+V LCSEFL AGTDTT+T L+W+MA LVK+ +Q L +EI+ + + E
Sbjct: 304 DSALTEDELVMLCSEFLNAGTDTTATALQWIMAELVKNPSIQSKLHDEIKS---KTSDDE 360
Query: 343 VREEDLEKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGL 402
+ EED +PYLKAVILEGLR+HPP H+ +P ED+ + GYL+PK TVNFMVA++G
Sbjct: 361 ITEEDTHDMPYLKAVILEGLRKHPPAHFVLPHKAAEDVEVGGYLIPKGATVNFMVAEMGR 420
Query: 403 DPTVWEDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLE 462
D WE P F PERFL + + DV G +E++MMPFG GRRIC +AMLHLE
Sbjct: 421 DEREWEKPMEFIPERFLAG----GDGEGVDVTGSREVRMMPFGVGRRICAGLGVAMLHLE 476
Query: 463 YFVANLVLNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEAQISLR 506
YFVANLV FEWK + G VDLTEK EFT VM PL AQ+ R
Sbjct: 477 YFVANLVKEFEWK-EVAGDEVDLTEKNEFTTVMAKPLRAQLVKR 519
>UniRef100_Q9FW87 Putative cytochrome P450 [Oryza sativa]
Length = 514
Score = 384 bits (985), Expect = e-105
Identities = 224/523 (42%), Positives = 308/523 (58%), Gaps = 32/523 (6%)
Query: 1 MEAMAWF-IITALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPII 59
++A W ++ A LV L L+ LR +S +PPGPL +P++ ++ L S + L+P++
Sbjct: 4 VDASQWLMLLLAFLVALFILLSLRKCGGRS-RVPPGPLAVPVLGNLLWLSHSSADLEPLL 62
Query: 60 RKLHAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRPK----FLKNHQFNIAS 115
R+L A +GP+++ S IF++DR +AH AL++ ADRP+ L + I+
Sbjct: 63 RRLIAVYGPVVSLRVGSHLSIFVADRRVAHAALVERGAALADRPEVTRALLGENGNTISR 122
Query: 116 SCYGPTWHAIRNNLASIMLQPSRFKS-FSKARKCVLDALISKLKFEANFLNNSVELINYI 174
YGPTW +R NL + L PSR ++ F+ AR AL+ L + L +
Sbjct: 123 GNYGPTWRLLRRNLVAETLHPSRARAAFAPARSWARRALVDGL------VGGGAVLADAF 176
Query: 175 RHAIFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQE 234
RHA+FCLL MCFGE +D + I D Q +L + F V++ +FR R Q
Sbjct: 177 RHAMFCLLVLMCFGEWLDEAAVRAIGDAQHGWLLHYATKMKVFAFCPAVTKHIFRGRIQT 236
Query: 235 FTQLRKLQNDLLVELIQARKK---------VIEKNNDDDEYVVCYVDTLLNLRLPEE--K 283
LR+ Q +L + LI AR++ V EK E+ YVDTLL+++LPE+
Sbjct: 237 SLALRRRQKELFMPLISARRERKNQLAERAVPEKETTTFEH--SYVDTLLDIKLPEDGGD 294
Query: 284 RKLNEGEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREV 343
R L + E+V LCSEFL AGTDT STTL+W+MA LVK+ +Q L +EI+ + + E+
Sbjct: 295 RALTDDEMVRLCSEFLDAGTDTMSTTLQWIMAELVKNPSIQSKLHDEIKSKTSDDHD-EI 353
Query: 344 REEDLEKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLD 403
EED +K+PYLKAVILEGLR+HPP H+ +P ED+ + GYL+PK TVNFMVA++G D
Sbjct: 354 TEEDTQKMPYLKAVILEGLRKHPPGHFALPHKAAEDMEVGGYLIPKGATVNFMVAEMGRD 413
Query: 404 PTVWEDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEY 463
WE+P F PERFL + + DV G K I+MMPFG GRRIC AMLHLEY
Sbjct: 414 EKEWENPMEFMPERFLPG----GDGEGVDVTGSKGIRMMPFGVGRRICAGLNTAMLHLEY 469
Query: 464 FVANLVLNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEAQISLR 506
FVAN+V FEW+ + G VD EK EFT VM PL AQ+ R
Sbjct: 470 FVANMVREFEWR-EIAGEEVDFAEKLEFTTVMAKPLRAQLVRR 511
>UniRef100_Q9FW92 Putative cytochrome P450 [Oryza sativa]
Length = 523
Score = 377 bits (968), Expect = e-103
Identities = 218/521 (41%), Positives = 305/521 (57%), Gaps = 29/521 (5%)
Query: 6 WFI-ITALLVLLLCLIILRLR-----SSQSIHLPPGPLYLPIITDIFLLQISFSKLKPII 59
W + + A+LVL+ L ++ R + LPPGP +P++ L S + +P++
Sbjct: 4 WHVAVAAILVLIPFLRLILSRRGGRGGGKRGRLPPGPPAVPLLGSTVWLTNSLADAEPLL 63
Query: 60 RKLHAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP-----KFLKNHQFNIA 114
R+L A+HGP+++ S +F++DR LAH AL++ ADRP + L I+
Sbjct: 64 RRLIARHGPVVSLRVASRLLVFVADRRLAHAALVEKGASLADRPAMASTRLLGESDNLIS 123
Query: 115 SSCYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYI 174
+ YGP W +R NL + L PSR + F+ AR V L+ KL+ E ++
Sbjct: 124 RASYGPVWRLLRRNLVAETLHPSRVRLFAPARAWVRRVLVEKLRDENGDAAAPHAVVETF 183
Query: 175 RHAIFCLLFSMCFGENVDSRKIEKIKDIQR-RLILSSGQLGALNFFPKIVSRVLFRKRWQ 233
++A+FCLL MCFGE +D + I QR L+ S ++ FFP V++ LFR R Q
Sbjct: 184 QYAMFCLLVLMCFGERLDEDAVRAIAVAQRDALLYLSSKMPVFAFFPA-VTKHLFRGRLQ 242
Query: 234 EFTQLRKLQNDLLVELIQARKKVIEKNND-------DDEYVVCYVDTLLNLRLPEE-KRK 285
+ LR+ Q +L V LI AR++ ++ + + YVDTLL+++LP++ R
Sbjct: 243 KAHALRRRQTELFVPLINARREYKKRQGGANGEPKKETTFEHSYVDTLLDIKLPDDGNRP 302
Query: 286 LNEGEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGERE---ERE 342
L + E+V LCSEFL AGTDTTST L+W+MA LVK+ +Q L +EI G +RE
Sbjct: 303 LTDDEMVNLCSEFLNAGTDTTSTALQWIMAELVKNPSIQSKLHDEIMAKTGGGGGGGQRE 362
Query: 343 VREEDLEKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGL 402
V EED+ +PYLKAV+LEGLR+HPP H +P ED+ + GYL+PK TVNFMVA++G
Sbjct: 363 VSEEDIHDMPYLKAVVLEGLRKHPPGHMVLPHRAAEDMEIGGYLIPKGTTVNFMVAEMGR 422
Query: 403 DPTVWEDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLE 462
D WE P F PERFL + + DV G +EI+MMPFG GRRIC +AMLH+E
Sbjct: 423 DEKEWEKPMEFMPERFLAG----GDGEGVDVTGSREIRMMPFGVGRRICAGLGVAMLHVE 478
Query: 463 YFVANLVLNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEAQI 503
YFVAN+V FEWK + G VD EK EFT VM PL A++
Sbjct: 479 YFVANMVSEFEWK-EVAGDEVDFAEKIEFTTVMAKPLRARL 518
>UniRef100_Q9FW97 Putative cytochrome P450 [Oryza sativa]
Length = 516
Score = 377 bits (967), Expect = e-103
Identities = 205/488 (42%), Positives = 295/488 (60%), Gaps = 26/488 (5%)
Query: 32 LPPGPLYLPIITDIFLLQISFSKLKP--IIRKLHAKHGPIITFHFWSTPHIFISDRLLAH 89
LPPGP +P++ + LL + + ++P ++++L A++GPI++ + +F++DR LAH
Sbjct: 33 LPPGPPAVPLLGSVVLLTKALTDVEPELLLQRLIARYGPIVSLRMGTRVSVFVADRRLAH 92
Query: 90 QALIQNAVVFADRP-----KFLKNHQFNIASSCYGPTWHAIRNNLASIMLQPSRFKSFSK 144
AL++ ADRP + L + I + YGP W +R NL S L PSR + F+
Sbjct: 93 AALVEGGAALADRPGVPASRLLGENDNIITRAGYGPVWRLLRRNLVSETLHPSRARLFAP 152
Query: 145 ARKCVLDALISKLKFEANFLNNSVELINYIRHAIFCLLFSMCFGENVDSRKIEKIKDIQR 204
AR V ++ KL ++ V+ + Y A+FCLL +MCFGE +D + ++D QR
Sbjct: 153 ARYWVHRVIVDKLAASGQAPHDVVDTLQY---AMFCLLVNMCFGERLDEATVRAVEDAQR 209
Query: 205 RLILS-SGQLGALNFFPKIVSRVLFRKRWQEFTQLRKLQNDLLVELIQARKK-------V 256
L++ + Q+ +FP I LFR R ++ LR+ Q +L + LI AR++
Sbjct: 210 DLLIYITSQMAVFAYFPAITKH-LFRGRLEKIYALRRRQRELFMPLINARREYKKHGGET 268
Query: 257 IEKNNDDDEYVVCYVDTLLNLRLPEE-KRKLNEGEVVALCSEFLTAGTDTTSTTLEWVMA 315
+ N + YVDTLL+++LPE+ R L + E++ LCSEFL AGTDTTST L+W+MA
Sbjct: 269 TKTTNKETTLEHSYVDTLLDIKLPEDGNRALTDDEIIKLCSEFLNAGTDTTSTALQWIMA 328
Query: 316 NLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPYLKAVILEGLRRHPPTHYTIPRV 375
LVK+ +Q L +EI+ G+ ++ EV EED+ +PYL+AV+LEGLR+HPP H+ +P
Sbjct: 329 ELVKNPSIQSKLHDEIKSKTGD-DQPEVTEEDVHGMPYLRAVVLEGLRKHPPGHFVLPHR 387
Query: 376 VTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVFKPERFLKDAKNRNEFDAFDVRG 435
ED+ + GYL+PK TVNFMVA+IG D W P F PERFL + + DV G
Sbjct: 388 AAEDVEVGGYLIPKGATVNFMVAEIGRDEREWAKPMEFIPERFLPG----GDGEGVDVTG 443
Query: 436 IKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFEWKTSLPGSNVDLTEKQEFTMVM 495
K I+MMPFG GRRIC AM HLEYFVAN+V FEWK + G V+ EK+EFT VM
Sbjct: 444 SKGIRMMPFGVGRRICAGLSFAMHHLEYFVANMVREFEWK-EVAGDEVEFAEKREFTTVM 502
Query: 496 KYPLEAQI 503
PL A++
Sbjct: 503 AKPLRARL 510
>UniRef100_Q9FW93 Putative cytochrome P450 [Oryza sativa]
Length = 643
Score = 374 bits (961), Expect = e-102
Identities = 211/514 (41%), Positives = 298/514 (57%), Gaps = 21/514 (4%)
Query: 1 MEAMAWFI--ITALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPI 58
M+A F+ I L+ LL ++ R + LPPGP +P++ L S +P+
Sbjct: 1 MDAWQLFVPAIAILIPLLRLILFRRGDDGRRGRLPPGPPAVPLLGSTVWLTNSLYDAEPV 60
Query: 59 IRKLHAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFN 112
+++L ++HGP+++ F S +F++DR LAH AL+++ ADRP + +
Sbjct: 61 VKRLMSRHGPVVSLRFGSQLLVFVADRRLAHAALVESGASLADRPSQAASARLVGEGDTM 120
Query: 113 IASSCYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVE-LI 171
I+ + YGP W +R NL + L PSR F+ R V L+ +L+ E + ++
Sbjct: 121 ISRASYGPVWRLLRRNLVADTLHPSRAHLFAPVRARVRRLLVDRLREEHGEAEAAPRAVV 180
Query: 172 NYIRHAIFCLLFSMCFGENVDSRKIEKIKDIQRRLIL-SSGQLGALNFFPKIVSRVLFRK 230
++A+FCLL MCFGE +D + I +R +L S ++G NFFP I +R LFR
Sbjct: 181 ETFQYAMFCLLVLMCFGEQLDEDAVRAIGAAERDTMLYMSSEMGVFNFFPAI-TRHLFRG 239
Query: 231 RWQEFTQLRKLQNDLLVELIQARKKVI----EKNNDDDEYVVCYVDTLLNLRLPEE-KRK 285
R Q+ LR+ + +L V LI +R++ E + + YVD LL++ LPE+ R
Sbjct: 240 RLQKAHALRRRKEELFVPLINSRREYKKNGGEPKKETTTFTHSYVDNLLDINLPEDGNRA 299
Query: 286 LNEGEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVRE 345
L + E+V LCSEFL AGTD+TS L+W+MA LV++ +Q L EEI+ G + EV E
Sbjct: 300 LTDDELVMLCSEFLVAGTDSTSAALQWIMAELVRNPSIQSKLYEEIKSKTGGGGQHEVSE 359
Query: 346 EDLEKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPT 405
ED+ +PYLKAV+LEGLR+HPP H +P ED+ + GYL+PK VNFMVA++G D
Sbjct: 360 EDVHDMPYLKAVVLEGLRKHPPAHMLLPHKAAEDMDVGGYLIPKGTIVNFMVAEMGRDEK 419
Query: 406 VWEDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFV 465
WE P F PERFL + + DV K I+MMPFG GRRICP +AMLHLEYFV
Sbjct: 420 EWEKPMEFMPERFLPG----GDGEGVDVTSSKGIRMMPFGVGRRICPGLGIAMLHLEYFV 475
Query: 466 ANLVLNFEWKTSLPGSNVDLTEKQEFTMVMKYPL 499
AN+V FEWK + G VD EK+EF VM PL
Sbjct: 476 ANMVREFEWK-EVAGDEVDFAEKREFNTVMAKPL 508
>UniRef100_Q9FW78 Putative cytochrome P450 [Oryza sativa]
Length = 537
Score = 370 bits (950), Expect = e-101
Identities = 211/530 (39%), Positives = 300/530 (55%), Gaps = 32/530 (6%)
Query: 1 MEAMAWFIITALLVLLLCLIILR----LRSSQSIHLPPGPLYLPIITD-IFLLQISFSKL 55
++A W ++ + ++ +L ++ + +PPGPL +P++ ++L S + L
Sbjct: 3 IDATQWLLLLVVFLVAFLFTLLAKHGAVKRKHGVRVPPGPLAVPVLGSLVWLTHSSSANL 62
Query: 56 KPIIRKLHAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP-----KFLKNHQ 110
+P++R+L A+HGP+++ S IF++DR +AH AL+ ADRP L +
Sbjct: 63 EPLLRRLIARHGPVVSLRVGSRLSIFVADRRVAHAALVGRGAALADRPPDVTHSLLGESR 122
Query: 111 FNIASSCYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVEL 170
I S YGP W +R NL PSR + F+ AR V L+ KL + +
Sbjct: 123 NTITRSGYGPVWRLLRRNLVVETTHPSRVRLFAPARSWVRRVLVDKLADAGAHPASPPRV 182
Query: 171 INYIRHAIFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRK 230
+ R+A+F LL MCFGE +D + I Q +L G+ ++ F +++ LFR
Sbjct: 183 LEVFRYAMFSLLVLMCFGERLDEAAVRAIGAAQHDFLLYLGRKTSVFMFYPAITKHLFRG 242
Query: 231 RWQEFTQLRKLQNDLLVELIQAR---KKVIE----------KNNDDDEYVVCYVDTLLNL 277
R +R+ Q +L + LI AR KK I+ K +D+ + YVDTLL +
Sbjct: 243 RVHLGLAVRRRQKELFMPLIDARRERKKQIQQSGDSAASEKKKDDNTTFNHSYVDTLLTI 302
Query: 278 RLPEEK----RKLNEGEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEE 333
RL + R L + E+V+LCSEFL+AGTDTT+T L+W+MA LVK+ +Q L EEI+
Sbjct: 303 RLQDVDGDGDRALTDDEMVSLCSEFLSAGTDTTATALQWIMAELVKNPSIQSKLYEEIKA 362
Query: 334 VVGEREEREVREEDLEK-LPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGT 392
+ + E+ E+D LPYLKAVILEGLR+HPP H +P ED+ + GYL+PK T
Sbjct: 363 TMSGDNDDEINEDDARNNLPYLKAVILEGLRKHPPMHLLLPHKAAEDVEVGGYLIPKGAT 422
Query: 393 VNFMVADIGLDPTVWEDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICP 452
VNFMVA+IG D WE PT F PERF+ + + DV G +EI+MMPFGAGRRIC
Sbjct: 423 VNFMVAEIGRDEKEWEKPTEFIPERFMAGG---GDGEGVDVTGSREIRMMPFGAGRRICA 479
Query: 453 AYKLAMLHLEYFVANLVLNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEAQ 502
A +AMLHLEYFVAN+V FEWK + G VD E+ EFT VM L +
Sbjct: 480 ALSVAMLHLEYFVANMVKEFEWK-EVAGDEVDFAERLEFTTVMAKSLRGK 528
>UniRef100_Q9FW91 Putative cytochrome P450 [Oryza sativa]
Length = 512
Score = 368 bits (945), Expect = e-100
Identities = 210/511 (41%), Positives = 296/511 (57%), Gaps = 24/511 (4%)
Query: 8 IITALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHAKHG 67
I+ +L+L L ++ SS+ LPPGP +P+ ++ L+ S + ++P++ L K+G
Sbjct: 6 ILAGVLLLPLLFLLRNAASSRRRRLPPGPPAVPLFGNLLWLRHSAADVEPLLLTLFKKYG 65
Query: 68 PIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRPKFLKNHQFNIASSC-----YGPTW 122
P++T S IF++DR LAH ALI ADRP+ + ++ + YG W
Sbjct: 66 PVVTLRIGSRLSIFVADRHLAHAALIAAGAKLADRPQAATSTLLGVSDNIITRANYGAMW 125
Query: 123 HAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAIFCLL 182
+R NL S LQ SR F+ AR V L+ KL+ N +E Y +FCLL
Sbjct: 126 RLLRRNLVSQTLQQSRVDQFAPARVWVRRVLMEKLRGSGEEAPNVMEAFQY---TMFCLL 182
Query: 183 FSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQLRKLQ 242
MCFGE +D + +++ +R +L + ++ FF +++ LFR R + LR+ Q
Sbjct: 183 VLMCFGERLDEPAVRDVEEAERAWLLYISRRMSVFFFFPWITKHLFRGRLEAAHALRRRQ 242
Query: 243 NDLLVELIQARKKVIEKNND------DDEYVVCYVDTLLNLRLPEE-KRKLNEGEVVALC 295
+L V LI+AR++ + + + YVDTLL++++PEE R L + E+V LC
Sbjct: 243 KELFVPLIEARREYKRLASQGLPPARETTFQHSYVDTLLDVKIPEEGNRALTDDEIVTLC 302
Query: 296 SEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVG---EREEREVREEDLEKLP 352
SEFL AGTDTTST L+W+MA LVK+ VQ+ L EI G E ER VR++D K+P
Sbjct: 303 SEFLNAGTDTTSTGLQWIMAELVKNPAVQEKLYAEINATCGGDDELLERNVRDKD-NKMP 361
Query: 353 YLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTV 412
YL AV+ EGLR+HPP H+ +P ED+ + GYL+PK TVNFMVA+IG D WE+P
Sbjct: 362 YLNAVVKEGLRKHPPGHFVLPHKAAEDMDVGGYLIPKGATVNFMVAEIGRDEREWENPMQ 421
Query: 413 FKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNF 472
F PERFL D+ G K IKMMPFG GRRIC +AMLHLEYFV ++V+ F
Sbjct: 422 FMPERFLDGGHGA----GVDMHGTKGIKMMPFGVGRRICAGLNIAMLHLEYFVGSMVMEF 477
Query: 473 EWKTSLPGSNVDLTEKQEFTMVMKYPLEAQI 503
EWK + G V+ EK+EFT VM PL ++
Sbjct: 478 EWK-EVEGHEVEFAEKREFTTVMAKPLRPRL 507
>UniRef100_Q7XGZ5 Putative cytochrome P450 [Oryza sativa]
Length = 530
Score = 364 bits (934), Expect = 4e-99
Identities = 211/531 (39%), Positives = 299/531 (55%), Gaps = 41/531 (7%)
Query: 4 MAWFIITALLVLLLCLIILRLRSSQSI-------HLPPGPLYLPIITDIFLLQISFSKLK 56
+ W ++ A L+L L++ S LPPGP +P++ ++ S +
Sbjct: 3 LTWLLLLATLLLSTTLLVFLFHGGSSATGGEKRRRLPPGPATVPVLGNLLWATNSGMDIM 62
Query: 57 PIIRKLHAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRPKFLKNHQFN---- 112
+R+LHA+HGP++ S + ++DR LAH AL+++ ADRP+F
Sbjct: 63 RAVRRLHARHGPMLGLRMGSRLEVIVADRRLAHAALVESGAAMADRPEFASRALLGLDTA 122
Query: 113 -IASSCYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELI 171
I++S YGP W R N + + P+R + F+ AR VL+ L KL+ ++
Sbjct: 123 TISNSSYGPLWRLFRRNFVAEVAHPARLRQFAPARAAVLEELTDKLRRRQEDAGAGT-IL 181
Query: 172 NYIRHAIFCLLFSMCFGENVDSRKIEKIKDIQRRLIL-SSGQLGALNFFPKIVSRVLFRK 230
++A+F LL +MCFGE +D R + I QR L+L SS +L F P I +R LF
Sbjct: 182 ETFQYAMFFLLVAMCFGELLDERAVRDIAAAQRDLLLHSSKKLRVFAFLPAITTR-LFAG 240
Query: 231 RWQEFTQLRKLQNDLLVELIQARKKVIEKNNDDDEYVVC---------------YVDTLL 275
R + +R+ + + LI AR+ KN DD YVDTLL
Sbjct: 241 RMKAMIAMRQRLKGMFMPLIDARRA--RKNLVDDHGDATAPPPPAASATTLPHSYVDTLL 298
Query: 276 NLRLPEE--KRKLNEGEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEE 333
NLR+ + +R L + E+VALCSEFL GTDTTST LEW+MA LVK+ +Q L EI+
Sbjct: 299 NLRINDNGGERALTDDEMVALCSEFLNGGTDTTSTALEWIMAELVKNPTIQDKLHGEIKG 358
Query: 334 VVGEREEREVREEDLEKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTV 393
+ + V EED++K+PYLKAV++EGLRRHPP H+ +P ED+ L GY +PK V
Sbjct: 359 AITSNSGK-VSEEDVQKMPYLKAVVMEGLRRHPPGHFVLPHAPAEDMELGGYTIPKGTLV 417
Query: 394 NFMVADIGLDPTVWEDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPA 453
NF VAD+G+D W+ P F PERF+ + + D+ G +EI+MMPFGAGRRICP
Sbjct: 418 NFTVADMGMDGAAWDRPREFLPERFMAG----GDGEGVDITGTREIRMMPFGAGRRICPG 473
Query: 454 YKLAMLHLEYFVANLVLNFEWKTSLPGSNVDLT-EKQEFTMVMKYPLEAQI 503
+A LHLEYFVAN+V FEW+ + G VD+ EK EFT+VM+ PL A++
Sbjct: 474 LGVATLHLEYFVANMVAAFEWRAA-EGEAVDVDGEKLEFTVVMEKPLRARL 523
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.140 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 837,220,195
Number of Sequences: 2790947
Number of extensions: 35699573
Number of successful extensions: 153956
Number of sequences better than 10.0: 4524
Number of HSP's better than 10.0 without gapping: 2797
Number of HSP's successfully gapped in prelim test: 1728
Number of HSP's that attempted gapping in prelim test: 140509
Number of HSP's gapped (non-prelim): 5592
length of query: 507
length of database: 848,049,833
effective HSP length: 132
effective length of query: 375
effective length of database: 479,644,829
effective search space: 179866810875
effective search space used: 179866810875
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146723.3


