

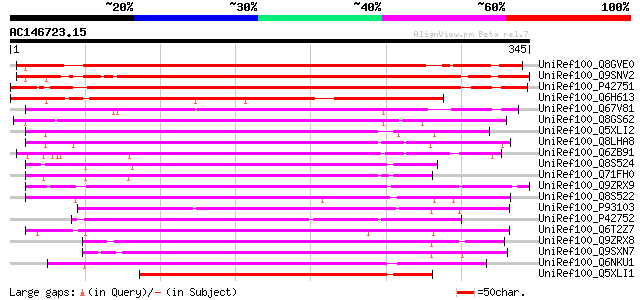
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146723.15 + phase: 0
(345 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8GVE0 Cyclin D1 [Helianthus tuberosus] 382 e-104
UniRef100_Q9SNV2 Cyclin D1 [Antirrhinum majus] 359 5e-98
UniRef100_P42751 Cyclin delta-1 [Arabidopsis thaliana] 358 2e-97
UniRef100_Q6H613 Putative cyclin D1 [Oryza sativa] 246 7e-64
UniRef100_Q67V81 Putative cyclin D1 [Oryza sativa] 231 2e-59
UniRef100_Q8GS62 Cyclin D [Physcomitrella patens] 214 4e-54
UniRef100_Q5XLI2 D-type cyclin [Saccharum officinarum] 199 7e-50
UniRef100_Q8LHA8 Putative D-type cyclin [Oryza sativa] 199 9e-50
UniRef100_Q6ZB91 Putative D-type cyclin [Oryza sativa] 194 4e-48
UniRef100_Q8S524 D-type cyclin [Zea mays] 192 1e-47
UniRef100_Q71FH0 Cyclin D2 [Triticum aestivum] 191 3e-47
UniRef100_Q9ZRX9 Cyclin D2.1 protein [Nicotiana tabacum] 184 3e-45
UniRef100_Q8S522 D-type cyclin [Zea mays] 183 7e-45
UniRef100_P93103 Cyclin-D like protein [Chenopodium rubrum] 182 1e-44
UniRef100_P42752 Cyclin delta-2 [Arabidopsis thaliana] 182 1e-44
UniRef100_Q6T2Z7 Cyclin d2 [Glycine max] 179 8e-44
UniRef100_Q9ZRX8 Cyclin D3.1 protein [Nicotiana tabacum] 176 8e-43
UniRef100_Q9SXN7 NtcycD3-1 protein [Nicotiana tabacum] 174 4e-42
UniRef100_Q6NKU1 At5g10440 [Arabidopsis thaliana] 172 9e-42
UniRef100_Q5XLI1 D-type cyclin [Saccharum officinarum] 172 9e-42
>UniRef100_Q8GVE0 Cyclin D1 [Helianthus tuberosus]
Length = 315
Score = 382 bits (980), Expect = e-104
Identities = 201/339 (59%), Positives = 246/339 (72%), Gaps = 30/339 (8%)
Query: 5 STSDC--ELLCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEHEF 62
S SDC +LLC EDS + D PECS D + S + ++SIA FIE E
Sbjct: 4 SCSDCFSDLLCCEDSGILSGDDRPECSYDFEYSG------------DFDDSIAEFIEQER 51
Query: 63 KFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLP 122
KFVPG DYV RFQS+ L++S REE++AWILKV +YGFQPLTAYLSVNY+DRF+ R P
Sbjct: 52 KFVPGIDYVERFQSQVLDASAREESVAWILKVQRFYGFQPLTAYLSVNYLDRFIYCRGFP 111
Query: 123 ESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWR 182
+NGWPLQLLSVACLSLAAKMEE L+PS+LD Q+EGAKYIF+P+TI RME LVL++LDWR
Sbjct: 112 VANGWPLQLLSVACLSLAAKMEETLIPSILDLQVEGAKYIFEPKTIRRMEFLVLSVLDWR 171
Query: 183 LRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSA 242
LRS+TP SF+ FF+ K+D +G +T F+ISRAT+IILSNIQ+AS L Y PSCIAAA IL A
Sbjct: 172 LRSVTPFSFIGFFSHKIDPSGMYTGFLISRATQIILSNIQEASLLEYWPSCIAAATILCA 231
Query: 243 ANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTARTR 302
A+++ +S +N +HAESWC+GLSKEKI CY L+Q +PK+LP + V T
Sbjct: 232 ASDLSKFSLINADHAESWCDGLSKEKITKCYRLVQ----------SPKILP-VHVRVMTA 280
Query: 303 RWSTVSSSSSSPSSSSSPSFSLSYKKRKLNSCFWVDVDK 341
R ST S SSS SSS SP YKKRKLN+ W++ DK
Sbjct: 281 RVSTESGDSSSSSSSPSP-----YKKRKLNNYSWIEEDK 314
>UniRef100_Q9SNV2 Cyclin D1 [Antirrhinum majus]
Length = 330
Score = 359 bits (922), Expect = 5e-98
Identities = 194/346 (56%), Positives = 255/346 (73%), Gaps = 27/346 (7%)
Query: 5 STSDC--ELLCGEDSSEVLTG---DLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIE 59
S SDC +LLCGEDS+ + +G DLPE +SD++S +P+ + +ESIA +E
Sbjct: 4 SCSDCFSDLLCGEDSNIIFSGGGDDLPEYTSDVES-----IPT------DVDESIAGLLE 52
Query: 60 HEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSR 119
E + + G + S ++S++SSTR E+ AWILKV YYGFQPLTAYL+V+Y DRFL++
Sbjct: 53 DE-RDLAGVN--SSSSNQSVDSSTRTESTAWILKVQRYYGFQPLTAYLAVSYFDRFLNAH 109
Query: 120 PLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTIL 179
LP+ NGWP+QLLSVACLSLAAKMEE LVPSLLD Q+EGA +IF+PR I RMELLVL +L
Sbjct: 110 HLPKLNGWPMQLLSVACLSLAAKMEESLVPSLLDLQVEGANFIFEPRNIQRMELLVLRVL 169
Query: 180 DWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAI 239
DWRLRSI+P +LSFFA K+D TGT+T F+ SRA EIILS +Q+ S + YRPSCIAAA +
Sbjct: 170 DWRLRSISPFCYLSFFALKIDPTGTYTGFLTSRAKEIILSTVQETSLIEYRPSCIAAATM 229
Query: 240 LSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTA 299
LS+AN++P +SF+ +HAE+WC+GL K+ I C +LIQ + S+N + PKVLPQLRV
Sbjct: 230 LSSANDLPKFSFITAQHAEAWCDGLHKDNIASCIKLIQGVESNNRPKKQPKVLPQLRVMT 289
Query: 300 RTRRWSTVSSSSSSPSSSSSPSFSLSYKKRKLNSCFWVDVDKGNSE 345
R ++++SS SS S+SSSP SYK+RKLN+ D DK +S+
Sbjct: 290 R----ASLASSESSSSTSSSP----SYKRRKLNNSSRADDDKESSD 327
>UniRef100_P42751 Cyclin delta-1 [Arabidopsis thaliana]
Length = 335
Score = 358 bits (918), Expect = 2e-97
Identities = 198/347 (57%), Positives = 247/347 (71%), Gaps = 27/347 (7%)
Query: 1 MPLSSTSDCELLCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEH 60
M +S ++D +L CGEDS V +G E + D SS P +SIA FIE
Sbjct: 12 MSVSFSNDMDLFCGEDSG-VFSG---ESTVDFSSSEVDSWPG---------DSIACFIED 58
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E FVPG DY+SRFQ+RSL++S RE+++AWILKV YY FQPLTAYL+VNYMDRFL +R
Sbjct: 59 ERHFVPGHDYLSRFQTRSLDASAREDSVAWILKVQAYYNFQPLTAYLAVNYMDRFLYARR 118
Query: 121 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILD 180
LPE++GWP+QLL+VACLSLAAKMEE LVPSL DFQ+ G KY+F+ +TI RMELLVL++LD
Sbjct: 119 LPETSGWPMQLLAVACLSLAAKMEEILVPSLFDFQVAGVKYLFEAKTIKRMELLVLSVLD 178
Query: 181 WRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAIL 240
WRLRS+TP F+SFFA K+D +GTF F IS ATEIILSNI++ASFL Y PS IAAAAIL
Sbjct: 179 WRLRSVTPFDFISFFAYKIDPSGTFLGFFISHATEIILSNIKEASFLEYWPSSIAAAAIL 238
Query: 241 SAANEIPNW-SFVNP-EHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVT 298
ANE+P+ S VNP E E+WC+GLSKEKI+ CY L++ + NN+ N PKV+ +LRV+
Sbjct: 239 CVANELPSLSSVVNPHESPETWCDGLSKEKIVRCYRLMKAMAIENNRLNTPKVIAKLRVS 298
Query: 299 ARTRRWSTVSSSSSSPS-SSSSPSFSLSYKKRKLNSCFWVDVDKGNS 344
R SS+ + PS SSSP K+RKL+ WV + S
Sbjct: 299 VR------ASSTLTRPSDESSSPC-----KRRKLSGYSWVGDETSTS 334
>UniRef100_Q6H613 Putative cyclin D1 [Oryza sativa]
Length = 320
Score = 246 bits (628), Expect = 7e-64
Identities = 140/303 (46%), Positives = 187/303 (61%), Gaps = 32/303 (10%)
Query: 1 MPLSSTSDCELLCGEDSSEVLTG---DLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVF 57
MP LLC ED+ + D+ C+++ D S + L++ SIA
Sbjct: 1 MPADDDDASYLLCAEDAGAAVFDVAVDISTCTTEDDECCS--VGGEELYSAA---SIAEL 55
Query: 58 IEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLD 117
I E ++ P DY R +SRS++ + R E+++WILKV EY GF PLTAYL+VNYMDRFL
Sbjct: 56 IGGEAEYSPRSDYPDRLRSRSIDPAARAESVSWILKVQEYNGFLPLTAYLAVNYMDRFLS 115
Query: 118 SRPLP---------ESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQ---IEGAKYIFQP 165
R LP E GW +QLL+VACLSLAAKMEE LVPSLLD Q +E ++Y+F+P
Sbjct: 116 LRHLPVFVLFPSMQEGQGWAMQLLAVACLSLAAKMEETLVPSLLDLQASTVECSRYVFEP 175
Query: 166 RTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDAS 225
RTI RME L+LT L+WRLRS+TP +F+ FFACK S ++ + D
Sbjct: 176 RTICRMEFLILTALNWRLRSVTPFTFIDFFACKHISNA------------MVQNANSDIQ 223
Query: 226 FLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQ 285
FL + PS +AAAA+L A E P+ +FVNPE A +WC GL++E I CY+L+Q++V N Q
Sbjct: 224 FLDHCPSSMAAAAVLCATGETPSLAFVNPELAVNWCIGLAEEGISSCYQLMQQLVIGNVQ 283
Query: 286 RNA 288
R+A
Sbjct: 284 RSA 286
>UniRef100_Q67V81 Putative cyclin D1 [Oryza sativa]
Length = 363
Score = 231 bits (590), Expect = 2e-59
Identities = 149/354 (42%), Positives = 201/354 (56%), Gaps = 47/354 (13%)
Query: 11 LLCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEHEF-KFVPG-- 67
LLCGED+ E+ P S SS S +++ +++E+ + E + PG
Sbjct: 31 LLCGEDAGELEREGEPAQGSSPSSSLSCAAAAAAAADDDDEDEDEHGVHGEVVQVTPGGE 90
Query: 68 -----FDY-------------VSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSV 109
+DY ++ E+++WILKV +GFQP TAYL+V
Sbjct: 91 EHCYDYDYDVDVPVGAELVMPACSPPRTAVHRPGWSESVSWILKVRSVHGFQPATAYLAV 150
Query: 110 NYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTIL 169
+YMDRF+ SR LP+ +GW QLL VACLSLAAKMEE P LLD QIEG ++IF+PRTI
Sbjct: 151 SYMDRFMSSRSLPD-HGWASQLLCVACLSLAAKMEESSAPPLLDLQIEGTRFIFEPRTIQ 209
Query: 170 RMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTY 229
RMEL+VL LDWRLRS+TP +F+ FFACK+ S+G + + RA +IILS I + FL +
Sbjct: 210 RMELIVLVELDWRLRSVTPFAFVDFFACKVGSSGRSSRILALRACQIILSAIHELEFLNH 269
Query: 230 RPSCIAAAAILSAANEIP----NWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQ 285
S +AAAA+L A NE P + S V+ E A SWC GL++E+I CY+L+Q
Sbjct: 270 CASSMAAAAVLFAVNESPAAMSHRSSVSSESAASWCIGLTEERISSCYQLLQR------- 322
Query: 286 RNAPKVLPQLRVTARTR-RWSTVSSSSSSPSSSSSPSFSLSYKKRKLNSCFWVD 338
L TAR R R + ++ SS +SSSS S K+RKL+ F D
Sbjct: 323 --------ALNATARKRKRHPMILAACSSVTSSSSRS-----KRRKLDGHFGED 363
>UniRef100_Q8GS62 Cyclin D [Physcomitrella patens]
Length = 360
Score = 214 bits (544), Expect = 4e-54
Identities = 136/345 (39%), Positives = 191/345 (54%), Gaps = 19/345 (5%)
Query: 3 LSSTSDC--ELLCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEH 60
+S + DC L C ED S + C + D SQ F E++E+IA +
Sbjct: 1 MSPSVDCLASLYCAEDVSGTAWNESEMCGA-ADRVFESQPAVFMDFPVEDDEAIATLLMK 59
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E +F+P DY+ R+QSR L R AI WILKVH +Y + PLT L+VNYMDRFL
Sbjct: 60 EAQFMPEADYLERYQSRKLSLEARLAAIEWILKVHSFYNYSPLTVALAVNYMDRFLSRYY 119
Query: 121 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILD 180
PE W LQLLSVAC+SLAAKMEE VP LLDFQ+E ++IF+ TI RMELLVL+ L+
Sbjct: 120 FPEGKEWMLQLLSVACISLAAKMEESDVPILLDFQVEQEEHIFEAHTIQRMELLVLSTLE 179
Query: 181 WRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAIL 240
WR+ +TP S++ +F KL + ++SR +EIIL +I+ + L Y PS +AAA+I+
Sbjct: 180 WRMSGVTPFSYVDYFFHKLGVSDLLLRALLSRVSEIILKSIRVTTSLQYLPSVVAAASII 239
Query: 241 SAANEIP-----------NWSFVNPEHAESWCEGLSKEKIIGCY----ELIQEIVSSNNQ 285
A E+ N VN E + C ++ IG Y L ++I+ ++
Sbjct: 240 CALEEVTTIRTGDLLRTFNELLVNVESVKD-CYIDMRQSEIGPYCVRMGLKRKILHASEP 298
Query: 286 RNAPKVLPQLRVTARTRRWSTVSSSSSSPSSSSSPSFSLSYKKRK 330
++ VL V++ + SS SSP + SP + S +KR+
Sbjct: 299 QSPVGVLEAADVSSPSGTVLGFSSRESSPDVTDSPPSTNSQRKRR 343
>UniRef100_Q5XLI2 D-type cyclin [Saccharum officinarum]
Length = 343
Score = 199 bits (507), Expect = 7e-50
Identities = 127/319 (39%), Positives = 180/319 (55%), Gaps = 19/319 (5%)
Query: 11 LLCGEDSSEVLT--GDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEHEFKFVPGF 68
LLC EDSS +L + E L SS ++ S +F EE +A F+E E +P
Sbjct: 13 LLCAEDSSSILDLEAEAEEEEVMLARSSRTRGDPSVVFPVPSEECVAGFVEAEAAHMPRE 72
Query: 69 DYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWP 128
DY R + ++ R +A+ WI KVH YYGF PLTA L+VNY+DRFL LPE W
Sbjct: 73 DYAERLRGGGMDLRVRMDAVDWIWKVHAYYGFGPLTACLAVNYLDRFLSLYQLPEGKAWT 132
Query: 129 LQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITP 188
QLLSVACLSLAAKMEE VP LD QI A+Y+F+ +TI RMELLVL+ L WR++++TP
Sbjct: 133 TQLLSVACLSLAAKMEETYVPPSLDLQIGDARYVFEAKTIQRMELLVLSTLKWRMQAVTP 192
Query: 189 LSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPN 248
S++ +F +L+ + + R+ E+IL + L +RPS IAAA + A E
Sbjct: 193 FSYIDYFLHRLNGGDAPSRRAVLRSAELILCTARGTHCLDFRPSEIAAAVAAAVAGE--- 249
Query: 249 WSFVNPEHA----ESWC-EGLSKEKIIGCYELIQEIVS--SNNQRNAPKVLPQLR-VTAR 300
EHA ++ C + KE++ C E IQ V+ R+ VL ++ R
Sbjct: 250 ------EHAVDIDKACCTHRVHKERVSRCLEAIQATVALPGTVPRSPTGVLDAAGCLSYR 303
Query: 301 TRRWSTVSSSSSSPSSSSS 319
+ + +++++S +SSSS
Sbjct: 304 SDDTAVAAAAAASHASSSS 322
>UniRef100_Q8LHA8 Putative D-type cyclin [Oryza sativa]
Length = 356
Score = 199 bits (506), Expect = 9e-50
Identities = 124/344 (36%), Positives = 187/344 (54%), Gaps = 23/344 (6%)
Query: 11 LLCGEDSSEVLT-----GDLPECSSDLDSSSSSQLP-----SSSLFAEEEEESIAVFIEH 60
LLC EDSS VL G E +++L ++F + +E +A+ +E
Sbjct: 12 LLCAEDSSSVLGLGGFGGGGGEVAAELGCGGGGGFDFFGFGGGAVFPIDSDEFVALLVEK 71
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E P Y+ + + LE S R++AI WI KVH YY F PL+ YL+VNY+DRFL S
Sbjct: 72 EMDHQPQRGYLEKLELGGLECSWRKDAIDWICKVHSYYNFGPLSLYLAVNYLDRFLSSFN 131
Query: 121 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILD 180
LP W QLLSV+CLSLA KMEE +VP +D Q+ A+Y+F+ R I RMEL+V+ L
Sbjct: 132 LPHDESWMQQLLSVSCLSLATKMEETVVPLPMDLQVFDAEYVFEARHIKRMELIVMKTLK 191
Query: 181 WRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAIL 240
WRL+++TP SF+ +F K + ++ + S +++ + ++D+ FL++RPS IAAA +L
Sbjct: 192 WRLQAVTPFSFIGYFLDKFNEGKPPSYTLASWCSDLTVGTLKDSRFLSFRPSEIAAAVVL 251
Query: 241 SAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQE--IVSSNNQRNAPKVLPQLRVT 298
+ E + N ES ++KE ++ CYEL+ E +V NA +P +T
Sbjct: 252 AVLAE-NQFLVFNSALGESEIP-VNKEMVMRCYELMVEKALVKKIRNSNASSSVPHSPIT 309
Query: 299 ARTRRWSTVSSSSSSPSSSSSPSFSLSY---------KKRKLNS 333
+ S ++ SS S S + Y K+R+LN+
Sbjct: 310 VLDAACFSFRSDDTTLGSSQSNSNNKDYNSQDSAPASKRRRLNT 353
>UniRef100_Q6ZB91 Putative D-type cyclin [Oryza sativa]
Length = 383
Score = 194 bits (492), Expect = 4e-48
Identities = 135/352 (38%), Positives = 186/352 (52%), Gaps = 37/352 (10%)
Query: 5 STSDCE------LLCGEDSSEVL-----TGDLPE-----CSS---DL------DSSSSSQ 39
S+S C LLC ED+S +L G++ E C S DL S
Sbjct: 4 SSSSCHDAAASMLLCAEDNSSILWLEDEEGEVGERRSGGCRSMVGDLAAGGGGGSGGGGV 63
Query: 40 LPSSSLFAEEEEESIAVFIEHEFKFVPGFDYVSRFQSRS--LESSTREEAIAWILKVHEY 97
+F + EE +A +E E +P DY R + ++ R EAI WI +V+ Y
Sbjct: 64 EEEEDMFPRQSEECVASLVEREQAHMPRADYGERLRGGGGDVDLRVRSEAIGWIWEVYTY 123
Query: 98 YGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIE 157
Y F +TAYL+VNY+DRFL LPE W QLLSVACLS+AAKMEE +VP LD QI
Sbjct: 124 YNFSSVTAYLAVNYLDRFLSQYELPEGRDWMTQLLSVACLSIAAKMEETVVPQCLDLQIG 183
Query: 158 GAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEII 217
+++F+ TI RMELLVLT L+WR++++TP S++ +F KL+S + R++E+I
Sbjct: 184 EPRFLFEVETIHRMELLVLTNLNWRMQAVTPFSYIDYFLRKLNSGNAAPRSWLLRSSELI 243
Query: 218 LSNIQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQ 277
L FL +RPS IAAA + A E V + AE++ + K +++ C E IQ
Sbjct: 244 LRIAAGTGFLEFRPSEIAAAVAATVAGEAT--GVVEEDIAEAFTH-VDKGRVLQCQEAIQ 300
Query: 278 EIVSSNNQRNAPKVLPQLRVTARTRRWSTVSSSSSSPSSSSS--PSFSLSYK 327
+ S N + P A TRR S +SSSS P S + + LSYK
Sbjct: 301 DHHYSMATINTVQPKP-----ASTRRGSASASSSSVPESPVAVLDAGCLSYK 347
>UniRef100_Q8S524 D-type cyclin [Zea mays]
Length = 358
Score = 192 bits (488), Expect = 1e-47
Identities = 110/293 (37%), Positives = 172/293 (58%), Gaps = 24/293 (8%)
Query: 11 LLCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEE----------------EEESI 54
LLC ED++ +L L + + ++++ P ++ A ++ +
Sbjct: 12 LLCAEDNAAIL--GLDDDGEESSWAAAATPPRDTVAAAAATGVAVDGILTEFPLLSDDCV 69
Query: 55 AVFIEHEFKFVPGFDYVSRFQSRSLE---SSTREEAIAWILKVHEYYGFQPLTAYLSVNY 111
A +E E + +P Y+ + Q R + ++ R++AI WI KV E+Y F PLTA LSVNY
Sbjct: 70 ATLVEKEVEHMPAEGYLQKLQRRHGDLDLAAVRKDAIDWIWKVIEHYNFAPLTAVLSVNY 129
Query: 112 MDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRM 171
+DRFL + PE W QLL+VACLSLA+K+EE VP LD Q+ AK++F+ RTI RM
Sbjct: 130 LDRFLSTYEFPEGRAWMTQLLAVACLSLASKIEETFVPLPLDLQVAEAKFVFEGRTIKRM 189
Query: 172 ELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRP 231
ELLVL+ L WR+ ++T SF+ +F KL G + SR+++++LS + A F+ +RP
Sbjct: 190 ELLVLSTLKWRMHAVTACSFVEYFLHKLSDHGAPSLLARSRSSDLVLSTAKGAEFVVFRP 249
Query: 232 SCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNN 284
S IAA+ L+A E + E A S C+ L KE+++ C+E+IQE +++ +
Sbjct: 250 SEIAASVALAAIGECRSSVI---ERAASSCKYLDKERVLRCHEMIQEKITAGS 299
>UniRef100_Q71FH0 Cyclin D2 [Triticum aestivum]
Length = 353
Score = 191 bits (485), Expect = 3e-47
Identities = 116/285 (40%), Positives = 171/285 (59%), Gaps = 17/285 (5%)
Query: 11 LLCGEDSSEVL--TGDLPECSSDLDSSSSSQLPSSSLFAEE---------EEESIAVFIE 59
LLC ED++ +L D +CS +++ ++ + + A E +E +A +E
Sbjct: 13 LLCAEDNAAILGLDDDEEDCSWAAAAATPPRIAADAAAAAEGFLVDHPVQSDECVAALVE 72
Query: 60 HEFKFVPGFDYVSRFQSR--SLE-SSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFL 116
E + +P Y R +L+ ++ R +AI WI +V E++ F PLTA LSVNY+DRFL
Sbjct: 73 TEKEHMPADGYPQMLLRRPGALDLAAVRRDAIDWIWEVIEHFNFAPLTAVLSVNYLDRFL 132
Query: 117 DSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVL 176
PLPE W QLL+VACLSLA+KMEE VP +D Q+ A F+ RTI RMELLVL
Sbjct: 133 SVYPLPEGKAWVTQLLAVACLSLASKMEETYVPLPVDLQVVEANSAFEGRTIKRMELLVL 192
Query: 177 TILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAA 236
+ L WR++++T SF+ +F K + + SR+T++ILS + A FL +RPS IAA
Sbjct: 193 STLKWRMQAVTACSFIDYFLRKFNDHDAPSMLAFSRSTDLILSTAKGADFLVFRPSEIAA 252
Query: 237 AAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVS 281
+ L+A E N S V E A + C+ ++KE+++ CYELIQ+ V+
Sbjct: 253 SVALAAFGE-RNTSVV--ERATTTCKFINKERVLRCYELIQDKVA 294
>UniRef100_Q9ZRX9 Cyclin D2.1 protein [Nicotiana tabacum]
Length = 354
Score = 184 bits (467), Expect = 3e-45
Identities = 120/336 (35%), Positives = 187/336 (54%), Gaps = 14/336 (4%)
Query: 11 LLCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEHEFKFVPGFDY 70
L + + E + DL ++ + S LPS S EE ++ ++ E +F+P DY
Sbjct: 30 LTISQQNIETKSKDL-SFNNGIRSEPLIDLPSLS------EECLSFMVQREMEFLPKDDY 82
Query: 71 VSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQ 130
V R +S L+ S R+EA+ WILK H +YGF L+ LS+NY+DRFL LP S W +Q
Sbjct: 83 VERLRSGDLDLSVRKEALDWILKAHMHYGFGELSFCLSINYLDRFLSLYELPRSKTWTVQ 142
Query: 131 LLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLS 190
LL+VACLSLAAKMEE VP +D Q+ K++F+ +TI RMELLVL+ L WR+++ TP +
Sbjct: 143 LLAVACLSLAAKMEEINVPLTVDLQVGDPKFVFEGKTIQRMELLVLSTLKWRMQAYTPYT 202
Query: 191 FLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPNWS 250
F+ +F K++ + +IS + ++ILS I+ FL +R S IAA+ +S + EI
Sbjct: 203 FIDYFMRKMNGDQIPSRPLISGSMQLILSIIRSIDFLEFRSSEIAASVAMSVSGEIQAKD 262
Query: 251 FVNPEHAESWCEGLSKEKIIGCYELIQEIVSSN-NQRNAPKVLPQLRVTARTRRWSTVSS 309
+ + L K ++ C ELIQ++ ++ A ++PQ + + +S
Sbjct: 263 I--DKAMPCFFIHLDKGRVQKCVELIQDLTTATITTAAAASLVPQSPIGV-LEAAACLSY 319
Query: 310 SSSSPSSSSSPSFSLSYKKRKLNSCFWVDVDKGNSE 345
S + S + S K+RKL++ ++ G SE
Sbjct: 320 KSGDERTVGSCTTSSHTKRRKLDTS---SLEHGTSE 352
>UniRef100_Q8S522 D-type cyclin [Zea mays]
Length = 390
Score = 183 bits (464), Expect = 7e-45
Identities = 121/362 (33%), Positives = 184/362 (50%), Gaps = 43/362 (11%)
Query: 11 LLCGEDSSEVLTGDLPECSSDLDSSSSSQLPS------SSLFAEEEEESIAVFIEHEFKF 64
LLC E+ S +L D E + + P + LF + EE +A +E E
Sbjct: 13 LLCAEEHSSILWYDEEEEELEAVGRRRGRSPGYGDDFGADLFPPQSEECVAGLVERERDH 72
Query: 65 VPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPES 124
+PG Y R + R EA+ WI K + ++ F+PLTAYL+VNY+DRFL +P+
Sbjct: 73 MPGPCYGDRLRGGGGCLCVRREAVDWIWKAYTHHRFRPLTAYLAVNYLDRFLSLSEVPDG 132
Query: 125 NGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLR 184
W QLL+VAC+SLAAKMEE VP LD Q+ A+Y+F+ +T+ RMELLVLT L+WR+
Sbjct: 133 KDWMTQLLAVACVSLAAKMEETAVPQCLDLQVGDARYVFEAKTVQRMELLVLTTLNWRMH 192
Query: 185 SITPLSFLSFFACKLDSTGTFT--HFIISRATEIILSNIQDASFLTYRPSCIAAAAILSA 242
++TP S++ +F KL + G+ + ++ E+IL + + +RPS IAAA +
Sbjct: 193 AVTPFSYVDYFLNKLSNGGSTAPRSCWLLQSAELILRAARGTGCVGFRPSEIAAAVAAAV 252
Query: 243 ANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVS-------------SNNQRNAP 289
A ++ + V + C + KE+++ C E I + S S +R++P
Sbjct: 253 AGDVDDADGVE----NACCAHVDKERVLRCQEAIGSMASSAAIDGDATVPPKSARRRSSP 308
Query: 290 KVLP-----------------QLRVTARTRRWSTVSSSSSSPSSSSSPSFS-LSYKKRKL 331
+P R +T ++S P SSSS S S ++ K+RKL
Sbjct: 309 VPVPVPVPQSPVGVLDAAACLSYRSEEAATATATSAASHGPPGSSSSSSTSPVTSKRRKL 368
Query: 332 NS 333
S
Sbjct: 369 AS 370
>UniRef100_P93103 Cyclin-D like protein [Chenopodium rubrum]
Length = 372
Score = 182 bits (462), Expect = 1e-44
Identities = 109/298 (36%), Positives = 168/298 (55%), Gaps = 13/298 (4%)
Query: 46 FAEEEEESIAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTA 105
F E +A ++E + G DY+ RF++ L+ R I WI KV +Y F PL
Sbjct: 71 FFVANHECLASLFDNERQHFLGLDYLKRFRNGDLDLGARNLVIDWIHKVQSHYNFGPLCV 130
Query: 106 YLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQP 165
YLSVNY+DRFL + LP W +QLL VACLSLAAK++E VP +LD Q+ +K++F+
Sbjct: 131 YLSVNYLDRFLSAYELP-GKAWMMQLLGVACLSLAAKVDETDVPLILDLQVSESKFVFEA 189
Query: 166 RTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDAS 225
+TI RMELLVL+ L WR++S+TP SF+ +F KL + +I +A ++ILS I+
Sbjct: 190 KTIQRMELLVLSTLKWRMQSVTPFSFIDYFLYKLSGDKMPSKSLIFQAIQLILSTIKGID 249
Query: 226 FLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEI--VSSN 283
+ +RPS IAAA +S + F + + + + KE+++ C E++ ++ S +
Sbjct: 250 LMEFRPSEIAAAVAISVTQQTQIVEFTDKAFS-FLTDHVEKERLMKCVEIMHDLRMSSRS 308
Query: 284 NQRNAPKVLPQLRV---------TARTRRWSTVSSSSSSPSSSSSPSFSLSYKKRKLN 332
N A +PQ + + ++ ST S S S+ SSP+ + K+RKL+
Sbjct: 309 NGALASTSVPQSPIGVLDASACLSYKSDDTSTTPSGSCGNSAHSSPASAPPPKRRKLD 366
>UniRef100_P42752 Cyclin delta-2 [Arabidopsis thaliana]
Length = 361
Score = 182 bits (462), Expect = 1e-44
Identities = 106/259 (40%), Positives = 153/259 (58%), Gaps = 7/259 (2%)
Query: 42 SSSLFAEEEEESIAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQ 101
SSSL E+ I + E +F PG DYV R S L+ S R +A+ WILKV +Y F
Sbjct: 59 SSSL----SEDRIKEMLVREIEFCPGTDYVKRLLSGDLDLSVRNQALDWILKVCAHYHFG 114
Query: 102 PLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKY 161
L LS+NY+DRFL S LP+ W QLL+V+CLSLA+KMEE VP ++D Q+E K+
Sbjct: 115 HLCICLSMNYLDRFLTSYELPKDKDWAAQLLAVSCLSLASKMEETDVPHIVDLQVEDPKF 174
Query: 162 IFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNI 221
+F+ +TI RMELLV+T L+WRL+++TP SF+ +F K+ +G + +I R++ IL+
Sbjct: 175 VFEAKTIKRMELLVVTTLNWRLQALTPFSFIDYFVDKI--SGHVSENLIYRSSRFILNTT 232
Query: 222 QDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVS 281
+ FL +RPS IAAAA +S + ++ E A S + +E++ C L++ +
Sbjct: 233 KAIEFLDFRPSEIAAAAAVSVSIS-GETECIDEEKALSSLIYVKQERVKRCLNLMRSLTG 291
Query: 282 SNNQRNAPKVLPQLRVTAR 300
N R Q RV R
Sbjct: 292 EENVRGTSLSQEQARVAVR 310
>UniRef100_Q6T2Z7 Cyclin d2 [Glycine max]
Length = 361
Score = 179 bits (455), Expect = 8e-44
Identities = 122/346 (35%), Positives = 186/346 (53%), Gaps = 27/346 (7%)
Query: 11 LLCGEDS-----SEVLTGDLPECSSDLDSSSSSQLPSSSLFAEE-------EEESIAVFI 58
LLC EDS S G + + + S + P F EE +ES+ + +
Sbjct: 11 LLCTEDSTVFDESHNNGGTIMTAMGVYEDTRSPRRPH---FDEEPDDLPLLSDESLVMMV 67
Query: 59 EHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDS 118
E E + G Y+++FQ+ L+ R EAI WI KV ++GF PL YLS+NY+DRFL +
Sbjct: 68 EKECQHWSGLRYLNKFQTGDLDFGARMEAIDWIHKVRSHFGFGPLCGYLSINYLDRFLFA 127
Query: 119 RPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTI 178
LP+ W +QLL+VAC+SLAAK++E VP LD Q+ +K++F+ +TI RMELLVL+
Sbjct: 128 YELPKGRVWTMQLLAVACVSLAAKLDETEVPLSLDLQVGESKFLFEAKTIQRMELLVLST 187
Query: 179 LDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAA- 237
L WR+++ITP +FL +F CK++ + I R+ ++I S + FL ++PS IAAA
Sbjct: 188 LKWRMQAITPFTFLDYFLCKINDDQSPLRSSIMRSIQLISSTARGIDFLEFKPSEIAAAV 247
Query: 238 -----AILSAANEIPNWSFVNPEHAES-WCEGLSKEKIIGCYELIQEIV----SSNNQRN 287
A A + V+ A S + + KE+++ C ++IQE+ S+ +
Sbjct: 248 KPSEIAAAVAMYVMGETQTVDTGKAISVLIQHVEKERLLKCVQMIQELSCNSGSAKDSSA 307
Query: 288 APKVLPQLRVTARTRRWSTVSSSSSSPSSSSSPSF-SLSYKKRKLN 332
+ LPQ + S ++ SS + S S K+RKLN
Sbjct: 308 SVTCLPQSPIGVLDALCFNYKSDDTNASSCVNSSHNSPVAKRRKLN 353
>UniRef100_Q9ZRX8 Cyclin D3.1 protein [Nicotiana tabacum]
Length = 373
Score = 176 bits (446), Expect = 8e-43
Identities = 106/285 (37%), Positives = 159/285 (55%), Gaps = 11/285 (3%)
Query: 49 EEEESIAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLS 108
E+EE +++F + + + + FQ SL S R +++ WILKV+ YYGF LTA L+
Sbjct: 72 EDEELLSLFSKEKETHC----WFNSFQDDSLLCSARVDSVEWILKVNGYYGFSALTAVLA 127
Query: 109 VNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTI 168
+NY DRFL S + W +QL +V CLSLAAK+EE VP LLDFQ+E AKY+F+ +TI
Sbjct: 128 INYFDRFLTSLHYQKDKPWMIQLAAVTCLSLAAKVEETQVPLLLDFQVEDAKYVFEAKTI 187
Query: 169 LRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLT 228
RMELLVL+ L WR+ +TPLSFL +L + R ++LS + D F+
Sbjct: 188 QRMELLVLSSLKWRMNPVTPLSFLDHIIRRLGLRNNIHWEFLRRCENLLLSIMADCRFVR 247
Query: 229 YRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEI----VSSNN 284
Y PS +A A +L +++ + V+ ++ ++KEK+ C+ELI E+ +S
Sbjct: 248 YMPSVLATAIMLHVIHQVEPCNSVDYQNQLLGVLKINKEKVNNCFELISEVCSKPISHKR 307
Query: 285 QRNAPKVLPQLRVTARTRRWSTVSSSSSSPSSSSSPSFSLSYKKR 329
+ P P + +S+ SS+ S S+S F + K R
Sbjct: 308 KYENPSHSPSGVIDP---IYSSESSNDSWDLESTSSYFPVFKKSR 349
>UniRef100_Q9SXN7 NtcycD3-1 protein [Nicotiana tabacum]
Length = 368
Score = 174 bits (440), Expect = 4e-42
Identities = 104/293 (35%), Positives = 161/293 (54%), Gaps = 14/293 (4%)
Query: 49 EEEESIAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLS 108
E+EE +++F + E + + F+ + ++ L +R+EA+ WILKV+ +YGF TA L+
Sbjct: 57 EDEELLSLFTK-EKETISNFETI---KTDPLLCLSRKEAVKWILKVNAHYGFSTFTAILA 112
Query: 109 VNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTI 168
+NY DRFL S + W +QL++V CLSLAAK+EE VP LLDFQ+E AKY+F+ +TI
Sbjct: 113 INYFDRFLSSLHFQKDKPWMIQLVAVTCLSLAAKVEETQVPLLLDFQVEDAKYVFEAKTI 172
Query: 169 LRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLT 228
RMELLVL+ L WR+ +TPLSF+ +L + + I+L I D FL+
Sbjct: 173 QRMELLVLSSLKWRMNPVTPLSFVDHIIRRLGLKSHIHWEFLKQCERILLLVIADCRFLS 232
Query: 229 YRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEI----VSSNN 284
Y PS +A A +L +++ + + ++ +SKEK+ CYELI E+ +S
Sbjct: 233 YMPSVLATATMLHVIHQVEPCNAADYQNQLLEVLNISKEKVNDCYELITEVSYNSISHKR 292
Query: 285 QRNAPKVLPQLRVTA------RTRRWSTVSSSSSSPSSSSSPSFSLSYKKRKL 331
+ +P P + W +SSS + S F +KK ++
Sbjct: 293 KYESPINSPSAVIDTFYSSENSNESWDLQTSSSIPSTYSPRDQFLPLFKKSRV 345
>UniRef100_Q6NKU1 At5g10440 [Arabidopsis thaliana]
Length = 298
Score = 172 bits (437), Expect = 9e-42
Identities = 106/298 (35%), Positives = 162/298 (53%), Gaps = 12/298 (4%)
Query: 26 PECSSDLDSSSSSQLPSSSLFAE----EEEESIAVFIEHEFKFVPGFDYVSRFQSRSLES 81
P S+ D S+ + + S+F E EE + IE E + P DY+ R ++ L+
Sbjct: 7 PNLVSNFDDEKSNSVDTRSIFQMGFPLESEEIVREMIEKERQHSPRDDYLKRLRNGDLDF 66
Query: 82 STREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAA 141
+ R +A+ WI K E F PL L++NY+DRFL LP W +QLL+VACLSLAA
Sbjct: 67 NVRIQALGWIWKACEELQFGPLCICLAMNYLDRFLSVHDLPSGKAWTVQLLAVACLSLAA 126
Query: 142 KMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDS 201
K+EE VP L+ Q+ ++F+ +++ RMELLVL +L WRLR++TP S++ +F K++
Sbjct: 127 KIEETNVPELMQLQVGAPMFVFEAKSVQRMELLVLNVLRWRLRAVTPCSYVRYFLSKING 186
Query: 202 TGTFTHF-IISRATEIILSNIQDASFLTYRPSCIAAAAILSAANE-IPNWSFVNPEHAES 259
H +++R+ ++I S + FL +R S IAAA LS + E +SF S
Sbjct: 187 YDQEPHSRLVTRSLQVIASTTKGIDFLEFRASEIAAAVALSVSGEHFDKFSF------SS 240
Query: 260 WCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTARTRRWSTVSSSSSSPSSS 317
L KE++ E+I+ SS++ + + Q + + ST S SSS S S
Sbjct: 241 SFSSLEKERVKKIGEMIERDGSSSSSQTPNNTVLQFKSRRYSHSLSTASVSSSLTSLS 298
>UniRef100_Q5XLI1 D-type cyclin [Saccharum officinarum]
Length = 254
Score = 172 bits (437), Expect = 9e-42
Identities = 91/195 (46%), Positives = 127/195 (64%), Gaps = 3/195 (1%)
Query: 87 AIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEP 146
AI WI KV E+Y F PLTA LSVNY+DRFL + PE W QLL+VACLSLA+KMEE
Sbjct: 1 AIDWIWKVIEHYNFAPLTAVLSVNYLDRFLSTYEFPEDRAWMTQLLAVACLSLASKMEET 60
Query: 147 LVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFT 206
VP LD Q+ K++F+ RTI RMEL VL L WR+ ++T S++ +F KL G +
Sbjct: 61 FVPLPLDLQVAETKFVFEGRTIRRMELHVLNTLKWRMHAVTACSYVKYFLHKLSDHGAPS 120
Query: 207 HFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSK 266
SR+++++LS + A F+ +RPS IAA+ L+A E + E A S C+ L+K
Sbjct: 121 LLARSRSSDLVLSTAKGAEFVVFRPSEIAASVALAAMGECRSSVI---ERAASSCKYLNK 177
Query: 267 EKIIGCYELIQEIVS 281
E+++ C+E+IQE ++
Sbjct: 178 ERVLRCHEMIQEKIT 192
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 558,041,338
Number of Sequences: 2790947
Number of extensions: 22234130
Number of successful extensions: 121715
Number of sequences better than 10.0: 724
Number of HSP's better than 10.0 without gapping: 309
Number of HSP's successfully gapped in prelim test: 415
Number of HSP's that attempted gapping in prelim test: 120192
Number of HSP's gapped (non-prelim): 944
length of query: 345
length of database: 848,049,833
effective HSP length: 128
effective length of query: 217
effective length of database: 490,808,617
effective search space: 106505469889
effective search space used: 106505469889
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146723.15


