

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146720.6 - phase: 0 /pseudo
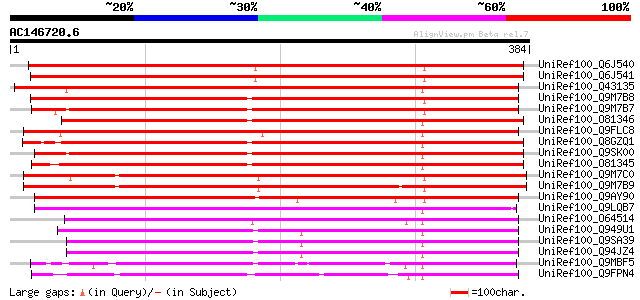
(384 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6J540 Cytochrome P450 [Lotus japonicus] 443 e-123
UniRef100_Q6J541 Cytochrome P450 [Lotus japonicus] 441 e-122
UniRef100_Q43135 Cytochrome P450 79A1 [Sorghum bicolor] 424 e-117
UniRef100_Q9M7B8 N-hydroxylating cytochrome P450 [Manihot escule... 422 e-116
UniRef100_Q9M7B7 N-hydroxylating cytochrome P450 [Manihot escule... 414 e-114
UniRef100_O81346 Cytochrome P450 79B2 [Arabidopsis thaliana] 391 e-107
UniRef100_Q9FLC8 Cytochrome P450 79A2 [Arabidopsis thaliana] 389 e-106
UniRef100_Q8GZQ1 Cytochrome P450 [Brassica napus] 387 e-106
UniRef100_Q9SK00 Putative cytochrome P450 [Arabidopsis thaliana] 386 e-106
UniRef100_O81345 Cytochrome P450 79B1 [Sinapis alba] 384 e-105
UniRef100_Q9M7C0 Cytochrome P450 CYP79E1 [Triglochin maritimum] 356 7e-97
UniRef100_Q9M7B9 Cytochrome P450 CYP79E2 [Triglochin maritimum] 355 2e-96
UniRef100_Q9AY90 Putative cytochrome p450tyr [Oryza sativa] 332 1e-89
UniRef100_Q9LQB7 F19C14.12 protein [Arabidopsis thaliana] 286 5e-76
UniRef100_O64514 YUP8H12R.1 protein [Arabidopsis thaliana] 273 6e-72
UniRef100_Q949U1 Putative cytochrome P450 protein [Arabidopsis t... 251 3e-65
UniRef100_Q9SA39 F3O9.20 protein [Arabidopsis thaliana] 233 9e-60
UniRef100_Q94JZ4 Cytochrome P450-like protein [Arabidopsis thali... 233 9e-60
UniRef100_Q9MBF5 Cytochrome P450 [Petunia hybrida] 182 1e-44
UniRef100_Q9FPN4 Flavonoid 3',5'-hydroxylase [Callistephus chine... 177 6e-43
>UniRef100_Q6J540 Cytochrome P450 [Lotus japonicus]
Length = 536
Score = 443 bits (1140), Expect = e-123
Identities = 219/390 (56%), Positives = 277/390 (70%), Gaps = 24/390 (6%)
Query: 15 WYLILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMM 74
W +L+++ + + K +S ++ K KLPPGP PWPIVGNLPEMLANRP WI K+M
Sbjct: 16 WTFLLVVIFFFIIFKVTKSHSVNKSKKYKLPPGPKPWPIVGNLPEMLANRPATIWIHKLM 75
Query: 75 NDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTP 134
++NT+IACIRL N VI ++ P IA E K DA FASRP S + ++G+LTT L P
Sbjct: 76 KEMNTEIACIRLANTIVIPVTCPTIACEFLKKHDASFASRPKIMSTDIASDGFLTTVLVP 135
Query: 135 FGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTK--IGGDGIVNVSVA 192
+GEQWKK+K+V+ N L+SP +H+WL KR EEADN++ Y+YNKC K G G+VN+ +A
Sbjct: 136 YGEQWKKMKRVLVNNLLSPQKHQWLLGKRNEEADNLMFYIYNKCCKDVNDGPGLVNIRIA 195
Query: 193 AQYYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRG 252
AQ+Y G+V R+L+ N RYFG ED GPG EE+E++ A FT+L+Y++AFS+SDF+P LR
Sbjct: 196 AQHYGGNVFRKLIFNTRYFGKVMEDGGPGFEEVEHINATFTILKYVYAFSISDFIPFLRR 255
Query: 253 LDLDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKDA------- 305
LDLDGH I KA IMKKYHDPII DRI+QW +G K +EDLLDVLI LKDA
Sbjct: 256 LDLDGHRSKIMKAMGIMKKYHDPIIHDRIKQWNDGLKTVEEDLLDVLIKLKDASNKPLLT 315
Query: 306 ---------------VDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFP 350
VDNPSNA EW LAE++NQPELLK+ATEELD+VVGK RLVQE D P
Sbjct: 316 LKEIKAQITELAIEMVDNPSNAFEWALAEMLNQPELLKRATEELDNVVGKERLVQESDIP 375
Query: 351 KLNYVKACAKEAFRRHPICDFNLPHVRCNE 380
KL +VKACA+EA R HP+ FN+PH+ N+
Sbjct: 376 KLQFVKACAREALRLHPMEYFNVPHLCMND 405
>UniRef100_Q6J541 Cytochrome P450 [Lotus japonicus]
Length = 535
Score = 441 bits (1135), Expect = e-122
Identities = 219/390 (56%), Positives = 278/390 (71%), Gaps = 25/390 (6%)
Query: 16 YLILMLLVYKFLIKHQRSTQKSEKPKP-KLPPGPTPWPIVGNLPEMLANRPTFRWIQKMM 74
+ L+++++ F+I T K K KLPPGP PWPIVGNLPEMLANRP WI K+M
Sbjct: 16 WTFLLVVIFSFMIFKVTKTHLVNKSKKYKLPPGPKPWPIVGNLPEMLANRPATIWIHKLM 75
Query: 75 NDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTP 134
++NT+IACIRL N VI ++ P IA E K DA FASRP S + ++G++TT L P
Sbjct: 76 KEMNTEIACIRLANTIVIPVTCPTIACEFLKKHDASFASRPKIMSTDIASDGFITTVLVP 135
Query: 135 FGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTK--IGGDGIVNVSVA 192
+GEQWKK+K+V+ N L+SP +H+WL KR EEADN++ Y+YNKC K G G+VN+ +A
Sbjct: 136 YGEQWKKMKRVLVNNLLSPQKHQWLLGKRNEEADNLMFYIYNKCCKDVNDGPGLVNIRIA 195
Query: 193 AQYYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRG 252
AQ+Y G+V R+L+ N RYFG ED GPG EE+E++ A FT+L+Y++AFS+SDF+P LR
Sbjct: 196 AQHYGGNVFRKLIFNSRYFGKVMEDGGPGFEEVEHINATFTILKYVYAFSISDFVPFLRR 255
Query: 253 LDLDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKDA------- 305
LDLDGH I KA +IM+KYHDPII+DRI+QW +G K +EDLLDVLI LKDA
Sbjct: 256 LDLDGHRSKIMKAMRIMRKYHDPIIDDRIKQWNDGLKTVEEDLLDVLIKLKDANNKPLLT 315
Query: 306 ---------------VDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFP 350
VDNPSNA EW LAE+INQPELLK+ATEELD+VVGK RLVQE D P
Sbjct: 316 LKELKAQIIELAIEMVDNPSNAFEWALAEMINQPELLKRATEELDNVVGKERLVQESDIP 375
Query: 351 KLNYVKACAKEAFRRHPICDFNLPHVRCNE 380
KL +VKACA+EA R HP+ FN+PH+ N+
Sbjct: 376 KLQFVKACAREALRLHPMEYFNVPHLCMND 405
>UniRef100_Q43135 Cytochrome P450 79A1 [Sorghum bicolor]
Length = 557
Score = 424 bits (1091), Expect = e-117
Identities = 207/406 (50%), Positives = 281/406 (68%), Gaps = 33/406 (8%)
Query: 4 LVVSDQLSTTLWYLILMLLVYKFLIKHQRSTQKSEKP----------KPKLPPGPTPWPI 53
L+ S + L +++ + + + L + ++ST K P LPPGP PWP+
Sbjct: 17 LLSSSAILKLLLFVVTLSYLARALRRPRKSTTKCSSTTCASPPAGVGNPPLPPGPVPWPV 76
Query: 54 VGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFAS 113
VGNLPEML N+P FRWI +MM ++ TDIAC++LG VHV++I+ PEIARE+ KQDA F S
Sbjct: 77 VGNLPEMLLNKPAFRWIHQMMREMGTDIACVKLGGVHVVSITCPEIAREVLRKQDANFIS 136
Query: 114 RPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRY 173
RP ++++E + GY L+P+G+QWKK+++V+++E++ P RH WLHDKR +EADN+ RY
Sbjct: 137 RPLTFASETFSGGYRNAVLSPYGDQWKKMRRVLTSEIICPSRHAWLHDKRTDEADNLTRY 196
Query: 174 VYNKCTKIG-GDGIVNVSVAAQYYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIF 232
VYN TK GD V+V A++Y G+VIRRL+ N+RYFG D GPG E+ +++A+F
Sbjct: 197 VYNLATKAATGDVAVDVRHVARHYCGNVIRRLMFNRRYFGEPQADGGPGPMEVLHMDAVF 256
Query: 233 TVLQYLFAFSVSDFMPCLRGLDLDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEK 292
T L L+AF VSD++P LRGLDLDGHE+I+K+A + + HD +I+DR +QWK+G++ E
Sbjct: 257 TSLGLLYAFCVSDYLPWLRGLDLDGHEKIVKEANVAVNRLHDTVIDDRWRQWKSGERQEM 316
Query: 293 EDLLDVLISLKD----------------------AVDNPSNAVEWGLAELINQPELLKKA 330
ED LDVLI+LKD AVDNPSNAVEW LAE++N PE++ KA
Sbjct: 317 EDFLDVLITLKDAQGNPLLTIEEVKAQSQDITFAAVDNPSNAVEWALAEMVNNPEVMAKA 376
Query: 331 TEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPICDFNLPHV 376
EELD VVG+ RLVQE D PKLNYVKAC +EAFR HP+ FN+PHV
Sbjct: 377 MEELDRVVGRERLVQESDIPKLNYVKACIREAFRLHPVAPFNVPHV 422
>UniRef100_Q9M7B8 N-hydroxylating cytochrome P450 [Manihot esculenta]
Length = 542
Score = 422 bits (1084), Expect = e-116
Identities = 197/384 (51%), Positives = 275/384 (71%), Gaps = 26/384 (6%)
Query: 16 YLILMLLVYKFLIKHQRSTQKSEKPKP-KLPPGPTPWPIVGNLPEMLANRPTFRWIQKMM 74
++ L + + ++K Q+S E K LPPGPTPWP++GN+PEM+ RPTFRWI ++M
Sbjct: 30 FVTLFISIVSTIVKLQKSAANKEGSKKLPLPPGPTPWPLIGNIPEMIRYRPTFRWIHQLM 89
Query: 75 NDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTP 134
D+NTDI IR G + + IS P +ARE+ K DAIF++RP + S + ++ GYLTT + P
Sbjct: 90 KDMNTDICLIRFGRTNFVPISCPVLAREILKKNDAIFSNRPKTLSAKSMSGGYLTTIVVP 149
Query: 135 FGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQ 194
+ +QWKK++K++++E++SP RHKWLHDKR EEADN+V Y++N + + VN+ A +
Sbjct: 150 YNDQWKKMRKILTSEIISPARHKWLHDKRAEEADNLVFYIHN---QFKANKNVNLRTATR 206
Query: 195 YYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLD 254
+Y G+VIR+++ +KRYFG G D GPG EEIE+++A+FT L+YL+ F +SDF+P L GLD
Sbjct: 207 HYGGNVIRKMVFSKRYFGKGMPDGGPGPEEIEHIDAVFTALKYLYGFCISDFLPFLLGLD 266
Query: 255 LDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKDA--------- 305
LDG E+ + A K ++ Y +P+I++RIQQWK+G++ E EDLLDV I+LKD+
Sbjct: 267 LDGQEKFVLDANKTIRDYQNPLIDERIQQWKSGERKEMEDLLDVFITLKDSDGNPLLTPD 326
Query: 306 -------------VDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKL 352
VDNPSNA+EW + E++NQPE+LKKATEELD VVGK RLVQE D P L
Sbjct: 327 EIKNQIAEIMIATVDNPSNAIEWAMGEMLNQPEILKKATEELDRVVGKDRLVQESDIPNL 386
Query: 353 NYVKACAKEAFRRHPICDFNLPHV 376
+YVKACA+EAFR HP+ FN+PHV
Sbjct: 387 DYVKACAREAFRLHPVAHFNVPHV 410
>UniRef100_Q9M7B7 N-hydroxylating cytochrome P450 [Manihot esculenta]
Length = 541
Score = 414 bits (1063), Expect = e-114
Identities = 202/384 (52%), Positives = 274/384 (70%), Gaps = 28/384 (7%)
Query: 17 LILMLLVYKFLIKHQR--STQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMM 74
+ L + + +IK Q+ S +K+ K P LPPGPTPWP++GN+PEM+ RPTFRWI ++M
Sbjct: 30 ITLFISIVSTVIKLQKRASYKKASKNFP-LPPGPTPWPLIGNIPEMIRYRPTFRWIHQLM 88
Query: 75 NDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTP 134
D+NTDI IR G +V+ IS P IARE+ K DA+F++RP + ++ GYLTT + P
Sbjct: 89 KDMNTDICLIRFGKTNVVPISCPVIAREILKKHDAVFSNRPKILCAKTMSGGYLTTIVVP 148
Query: 135 FGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQ 194
+ +QWKK++KV+++E++SP RHKWLHDKR EEAD +V Y+ N + + VNV +AA+
Sbjct: 149 YNDQWKKMRKVLTSEIISPARHKWLHDKRAEEADQLVFYINN---QYKSNKNVNVRIAAR 205
Query: 195 YYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLD 254
+Y G+VIR+++ +KRYFG G D GPG EEI +V+AIFT L+YL+ F +SD++P L GLD
Sbjct: 206 HYGGNVIRKMMFSKRYFGKGMPDGGPGPEEIMHVDAIFTALKYLYGFCISDYLPFLEGLD 265
Query: 255 LDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKDA--------- 305
LDG E+I+ A K ++ +P+IE+RIQQW++G++ E EDLLDV I+L+D+
Sbjct: 266 LDGQEKIVLNANKTIRDLQNPLIEERIQQWRSGERKEMEDLLDVFITLQDSDGKPLLNPD 325
Query: 306 -------------VDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKL 352
+DNP+NAVEW + ELINQPELL KATEELD VVGK RLVQE D P L
Sbjct: 326 EIKNQIAEIMIATIDNPANAVEWAMGELINQPELLAKATEELDRVVGKDRLVQESDIPNL 385
Query: 353 NYVKACAKEAFRRHPICDFNLPHV 376
NYVKACA+EAFR HP+ FN+PHV
Sbjct: 386 NYVKACAREAFRLHPVAYFNVPHV 409
>UniRef100_O81346 Cytochrome P450 79B2 [Arabidopsis thaliana]
Length = 541
Score = 391 bits (1005), Expect = e-107
Identities = 184/365 (50%), Positives = 258/365 (70%), Gaps = 26/365 (7%)
Query: 39 KPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITISDPE 98
K KP LPPGPT WPI+G +P ML +RP FRW+ +M LNT+IAC++LGN HVIT++ P+
Sbjct: 51 KKKPYLPPGPTGWPIIGMIPTMLKSRPVFRWLHSIMKQLNTEIACVKLGNTHVITVTCPK 110
Query: 99 IARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLRHKW 158
IARE+ +QDA+FASRP +++ + ++NGY T +TPFG+Q+KK++KV+ ELV P RH+W
Sbjct: 111 IAREILKQQDALFASRPLTYAQKILSNGYKTCVITPFGDQFKKMRKVVMTELVCPARHRW 170
Query: 159 LHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFG-NGSED 217
LH KR EE D++ +VYN + G V+ ++Y G+ I++L+ R F N + D
Sbjct: 171 LHQKRSEENDHLTAWVYN---MVKNSGSVDFRFMTRHYCGNAIKKLMFGTRTFSKNTAPD 227
Query: 218 YGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLDLDGHERIIKKACKIMKKYHDPII 277
GP +E++E++EA+F L + FAF +SD++P L GLDL+GHE+I++++ IM KYHDPII
Sbjct: 228 GGPTVEDVEHMEAMFEALGFTFAFCISDYLPMLTGLDLNGHEKIMRESSAIMDKYHDPII 287
Query: 278 EDRIQQWKNGKKIEKEDLLDVLISLKD----------------------AVDNPSNAVEW 315
++RI+ W+ GK+ + ED LD+ IS+KD A DNPSNAVEW
Sbjct: 288 DERIKMWREGKRTQIEDFLDIFISIKDEQGNPLLTADEIKPTIKELVMAAPDNPSNAVEW 347
Query: 316 GLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPICDFNLPH 375
+AE++N+PE+L+KA EE+D VVGK RLVQE D PKLNYVKA +EAFR HP+ FNLPH
Sbjct: 348 AMAEMVNKPEILRKAMEEIDRVVGKERLVQESDIPKLNYVKAILREAFRLHPVAAFNLPH 407
Query: 376 VRCNE 380
V ++
Sbjct: 408 VALSD 412
>UniRef100_Q9FLC8 Cytochrome P450 79A2 [Arabidopsis thaliana]
Length = 529
Score = 389 bits (998), Expect = e-106
Identities = 190/395 (48%), Positives = 267/395 (67%), Gaps = 29/395 (7%)
Query: 11 STTLWYLILMLLVYKFLIKHQRSTQK---SEKPKPKLPPGPTPWPIVGNLPEMLA-NRPT 66
ST + I+ LL+ +K + + S LPPGP WP++GNLPE+L N+P
Sbjct: 4 STPMLAFIIGLLLLALTMKRKEKKKTMLISPTRNLSLPPGPKSWPLIGNLPEILGRNKPV 63
Query: 67 FRWIQKMMNDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNG 126
FRWI +M +LNTDIACIRL N HVI ++ P IARE+ KQD++FA+RP + EY + G
Sbjct: 64 FRWIHSLMKELNTDIACIRLANTHVIPVTSPRIAREILKKQDSVFATRPLTMGTEYCSRG 123
Query: 127 YLTTALTPFGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDG- 185
YLT A+ P GEQWKK+++V+++ + S + + KR EEADN+VRY+ N+ K G+
Sbjct: 124 YLTVAVEPQGEQWKKMRRVVASHVTSKKSFQMMLQKRTEEADNLVRYINNRSVKNRGNAF 183
Query: 186 -IVNVSVAAQYYSGDVIRRLLLNKRYFGNGSED-YGPGLEEIEYVEAIFTVLQYLFAFSV 243
++++ +A + YSG+V R+++ R+FG GSED GPGLEEIE+VE++FTVL +L+AF++
Sbjct: 184 VVIDLRLAVRQYSGNVARKMMFGIRHFGKGSEDGSGPGLEEIEHVESLFTVLTHLYAFAL 243
Query: 244 SDFMPCLRGLDLDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLK 303
SD++P LR LDL+GHE+++ A + + KY+DP +++R+ QW+NGK E +D LD+ I K
Sbjct: 244 SDYVPWLRFLDLEGHEKVVSNAMRNVSKYNDPFVDERLMQWRNGKMKEPQDFLDMFIIAK 303
Query: 304 D----------------------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKG 341
D VDNPSNA EWG+AE+IN+P +++KA EE+D VVGK
Sbjct: 304 DTDGKPTLSDEEIKAQVTELMLATVDNPSNAAEWGMAEMINEPSIMQKAVEEIDRVVGKD 363
Query: 342 RLVQEYDFPKLNYVKACAKEAFRRHPICDFNLPHV 376
RLV E D P LNYVKAC KEAFR HP+ FNLPH+
Sbjct: 364 RLVIESDLPNLNYVKACVKEAFRLHPVAPFNLPHM 398
>UniRef100_Q8GZQ1 Cytochrome P450 [Brassica napus]
Length = 540
Score = 387 bits (994), Expect = e-106
Identities = 193/394 (48%), Positives = 265/394 (66%), Gaps = 31/394 (7%)
Query: 10 LSTTLWYLILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRW 69
L TTL + LV L+K +T +K K LPPGPT WPI+G +P ML +RP FRW
Sbjct: 26 LLTTLQAFAAITLV--MLLKKVFTT---DKKKLSLPPGPTGWPIIGMVPTMLKSRPVFRW 80
Query: 70 IQKMMNDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLT 129
+ +M LNT+IAC+RLGN HVIT++ P+IARE+ +QDA+FASRP +++ ++NGY T
Sbjct: 81 LHSIMKQLNTEIACVRLGNTHVITVTCPKIAREILKQQDALFASRPMTYAQNVLSNGYKT 140
Query: 130 TALTPFGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNV 189
+TPFGEQ+KK++KV+ ELV P RH+WLH KR EE D++ +VYN + G V+
Sbjct: 141 CVITPFGEQFKKMRKVVMTELVCPARHRWLHQKRAEENDHLTAWVYN---LVKNSGSVDF 197
Query: 190 SVAAQYYSGDVIRRLLLNKRYFG-NGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMP 248
++Y G+ I++L+ R F N + D GP E+IE++EA+F L + F+F +SD++P
Sbjct: 198 RFVTRHYCGNAIKKLMFGTRTFSENTAPDGGPTAEDIEHMEAMFEALGFTFSFCISDYLP 257
Query: 249 CLRGLDLDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKD---- 304
L GLDL+GHE+I++ + IM KYHDPI++ RI+ W+ GK+ + ED LD+ IS+KD
Sbjct: 258 MLTGLDLNGHEKIMRDSSAIMDKYHDPIVDARIKMWREGKRTQIEDFLDIFISIKDEQGN 317
Query: 305 ------------------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQE 346
A DNPSNAVEW +AE++N+PE+L KA EE+D VVGK RLVQE
Sbjct: 318 PLLTADEIKPTIKELVMAAPDNPSNAVEWAMAEMVNKPEILHKAMEEIDRVVGKERLVQE 377
Query: 347 YDFPKLNYVKACAKEAFRRHPICDFNLPHVRCNE 380
D PKLNYVKA +EAFR HP+ FNLPHV ++
Sbjct: 378 SDIPKLNYVKAILREAFRLHPVAAFNLPHVALSD 411
>UniRef100_Q9SK00 Putative cytochrome P450 [Arabidopsis thaliana]
Length = 566
Score = 386 bits (992), Expect = e-106
Identities = 188/385 (48%), Positives = 266/385 (68%), Gaps = 27/385 (7%)
Query: 19 LMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLN 78
L L + ++ +S+ +++K P LPPGPT +PIVG +P ML NRP FRW+ +M +LN
Sbjct: 57 LAALCFLMILNKIKSSSRNKKLHP-LPPGPTGFPIVGMIPAMLKNRPVFRWLHSLMKELN 115
Query: 79 TDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQ 138
T+IAC+RLGN HVI ++ P+IARE+ +QDA+FASRP +++ + ++NGY T +TPFGEQ
Sbjct: 116 TEIACVRLGNTHVIPVTCPKIAREIFKQQDALFASRPLTYAQKILSNGYKTCVITPFGEQ 175
Query: 139 WKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSG 198
+KK++KVI E+V P RH+WLHD R EE D++ ++YN + V++ ++Y G
Sbjct: 176 FKKMRKVIMTEIVCPARHRWLHDNRAEETDHLTAWLYN---MVKNSEPVDLRFVTRHYCG 232
Query: 199 DVIRRLLLNKRYFGNGSE-DYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLDLDG 257
+ I+RL+ R F +E D GP LE+IE+++A+F L + FAF +SD++P L GLDL+G
Sbjct: 233 NAIKRLMFGTRTFSEKTEADGGPTLEDIEHMDAMFEGLGFTFAFCISDYLPMLTGLDLNG 292
Query: 258 HERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKD------------- 304
HE+I++++ IM KYHDPII++RI+ W+ GK+ + ED LD+ IS+KD
Sbjct: 293 HEKIMRESSAIMDKYHDPIIDERIKMWREGKRTQIEDFLDIFISIKDEAGQPLLTADEIK 352
Query: 305 ---------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYV 355
A DNPSNAVEW +AE+IN+PE+L KA EE+D VVGK R VQE D PKLNYV
Sbjct: 353 PTIKELVMAAPDNPSNAVEWAIAEMINKPEILHKAMEEIDRVVGKERFVQESDIPKLNYV 412
Query: 356 KACAKEAFRRHPICDFNLPHVRCNE 380
KA +EAFR HP+ FNLPHV ++
Sbjct: 413 KAIIREAFRLHPVAAFNLPHVALSD 437
>UniRef100_O81345 Cytochrome P450 79B1 [Sinapis alba]
Length = 542
Score = 384 bits (987), Expect = e-105
Identities = 187/387 (48%), Positives = 262/387 (67%), Gaps = 32/387 (8%)
Query: 17 LILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMND 76
+ L++L+ K L+ + K K LPPGPT WPI+G +P ML +RP FRW+ +M
Sbjct: 36 ITLVMLLKKVLVND------TNKKKLSLPPGPTGWPIIGMVPTMLKSRPVFRWLHSIMKQ 89
Query: 77 LNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFG 136
LNT+IAC+RLG+ HVIT++ P+IARE+ +QDA+FASRP +++ ++NGY T +TPFG
Sbjct: 90 LNTEIACVRLGSTHVITVTCPKIAREVLKQQDALFASRPMTYAQNVLSNGYKTCVITPFG 149
Query: 137 EQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYY 196
EQ+KK++KV+ ELV P RH+WLH KR EE D++ +VYN + V+ ++Y
Sbjct: 150 EQFKKMRKVVMTELVCPARHRWLHQKRAEENDHLTAWVYN---MVNNSDSVDFRFVTRHY 206
Query: 197 SGDVIRRLLLNKRYFG-NGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLDL 255
G+ I++L+ R F N + + GP E+IE++EA+F L + F+F +SD++P L GLDL
Sbjct: 207 CGNAIKKLMFGTRTFSQNTAPNGGPTAEDIEHMEAMFEALGFTFSFCISDYLPILTGLDL 266
Query: 256 DGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKD----------- 304
+GHE+I++ + IM KYHDPII+ RI+ W+ GKK + ED LD+ IS+KD
Sbjct: 267 NGHEKIMRDSSAIMDKYHDPIIDARIKMWREGKKTQIEDFLDIFISIKDEEGNPLLTADE 326
Query: 305 -----------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLN 353
A DNPSNAVEW +AE++N+PE+L+KA EE+D VVGK RLVQE D PKLN
Sbjct: 327 IKPTIKELVMAAPDNPSNAVEWAMAEMVNKPEILRKAMEEIDRVVGKERLVQESDIPKLN 386
Query: 354 YVKACAKEAFRRHPICDFNLPHVRCNE 380
YVKA +EAFR HP+ FNLPHV ++
Sbjct: 387 YVKAILREAFRLHPVAAFNLPHVALSD 413
>UniRef100_Q9M7C0 Cytochrome P450 CYP79E1 [Triglochin maritimum]
Length = 540
Score = 356 bits (913), Expect = 7e-97
Identities = 185/400 (46%), Positives = 256/400 (63%), Gaps = 30/400 (7%)
Query: 11 STTLWYLILMLLVYKFLIKHQRSTQKSEKPKPK---LPPGPTPWPIVGNLPEMLANRPTF 67
+ T+ +L+L+ FL ++ K K K K LPPGP PWPIVG+L M NRP+F
Sbjct: 17 TATVLFLLLLTTALSFLFLFKQHLTKLTKSKSKSTTLPPGPRPWPIVGSLVSMYMNRPSF 76
Query: 68 RWIQKMMNDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGY 127
RWI M I CIRLG VHV+ ++ PEIARE DA FASRP + Y + G+
Sbjct: 77 RWILAQMEGRR--IGCIRLGGVHVVPVNCPEIAREFLKVHDADFASRPVTVVTRYSSRGF 134
Query: 128 LTTALTPFGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIG--GDG 185
+ A+ P GEQWKK+++V+++E+++ R +W R EEADNI+RY+ +C G
Sbjct: 135 RSIAVVPLGEQWKKMRRVVASEIINAKRLQWQLGLRTEEADNIMRYITYQCNTSGDTNGA 194
Query: 186 IVNVSVAAQYYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSD 245
I++V A ++Y +VIRR+L KRYFG+G E GPG EEIE+V+A F VL ++AF+ +D
Sbjct: 195 IIDVRFALRHYCANVIRRMLFGKRYFGSGGEGGGPGKEEIEHVDATFDVLGLIYAFNAAD 254
Query: 246 FMPCLRGLDLDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKI-EKEDLLDVLISLKD 304
++ L+ LDL G E+ +KKA ++ KYHD +IE R ++ G++ + EDLLDVL+SLKD
Sbjct: 255 YVSWLKFLDLHGQEKKVKKAIDVVNKYHDSVIESRRERKVEGREDKDPEDLLDVLLSLKD 314
Query: 305 A----------------------VDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGR 342
+ VDNPSNAVEW LAE++N P++L+KAT+E+D VVG+ R
Sbjct: 315 SNGKPLLDVEEIKAQIADLTYATVDNPSNAVEWALAEMLNNPDILQKATDEVDQVVGRHR 374
Query: 343 LVQEYDFPKLNYVKACAKEAFRRHPICDFNLPHVRCNERH 382
LVQE DFP L Y++ACA+EA R HP+ FNLPHV + H
Sbjct: 375 LVQESDFPNLPYIRACAREALRLHPVAAFNLPHVSLRDTH 414
>UniRef100_Q9M7B9 Cytochrome P450 CYP79E2 [Triglochin maritimum]
Length = 533
Score = 355 bits (910), Expect = 2e-96
Identities = 187/397 (47%), Positives = 251/397 (63%), Gaps = 29/397 (7%)
Query: 11 STTLWYLILMLLVYKFLIKHQRSTQKSEKPKPK-LPPGPTPWPIVGNLPEMLANRPTFRW 69
S TL+ L+LM FL ++ K KPK LPPGP PWPIVG+L M NRP+FRW
Sbjct: 15 SATLFLLLLMTTALSFLFLFKQHLAKLTKPKSTTLPPGPRPWPIVGSLVSMYMNRPSFRW 74
Query: 70 IQKMMNDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLT 129
I M I CIRLG VHV+ ++ PEIARE D+ FASRP + Y + G+ +
Sbjct: 75 ILAQMEGRR--IGCIRLGGVHVVPVNCPEIAREFLKVHDSDFASRPVTVVTRYSSRGFRS 132
Query: 130 TALTPFGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIG--GDGIV 187
A+ P GEQWKK+++V+++E+++ R +W R EEADNIVRY+ +C G I+
Sbjct: 133 IAVVPLGEQWKKMRRVVASEIINAKRLQWQLGLRTEEADNIVRYITYQCNTSGDTSGAII 192
Query: 188 NVSVAAQYYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFM 247
+V A ++Y +VIRR+L KRYFG+G GPG EEIE+V+A F VL ++AF+ +D++
Sbjct: 193 DVRFALRHYCANVIRRMLFGKRYFGSGGVGGGPGKEEIEHVDATFDVLGLIYAFNAADYV 252
Query: 248 PCLRGLDLDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKDA-- 305
L+ LDL G E+ +KKA ++ KYHD +I+ R ++ K + EDLLDVL SLKD+
Sbjct: 253 SWLKFLDLHGQEKKVKKAIDVVNKYHDSVIDARTERKVEDK--DPEDLLDVLFSLKDSNG 310
Query: 306 --------------------VDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQ 345
VDNPSNAVEW LAE++N P +L+KAT+ELD VVG+ RLVQ
Sbjct: 311 KPLLDVEEIKAQIADLTYATVDNPSNAVEWALAEMLNNPAILQKATDELDQVVGRHRLVQ 370
Query: 346 EYDFPKLNYVKACAKEAFRRHPICDFNLPHVRCNERH 382
E DFP L Y++ACA+EA R HP+ FNLPHV + H
Sbjct: 371 ESDFPNLPYIRACAREALRLHPVAAFNLPHVSLRDTH 407
>UniRef100_Q9AY90 Putative cytochrome p450tyr [Oryza sativa]
Length = 543
Score = 332 bits (850), Expect = 1e-89
Identities = 166/389 (42%), Positives = 252/389 (64%), Gaps = 33/389 (8%)
Query: 19 LMLLVYKFLIKHQRSTQKSEKPK-PKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDL 77
++++V L++ +R K + LPPGP P++GN+ +ML N+P FRW+ +++ D
Sbjct: 16 MLVVVLVALVRRRRHRSKGAGGRLESLPPGPVGLPVIGNMHQMLVNKPVFRWVHRLLADA 75
Query: 78 NTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGE 137
+I C+RLG VHV+ ++ PE+ARE+ K DA+FA RP++++ E + GY + +++P G+
Sbjct: 76 GGEIVCVRLGPVHVVAVTSPEMAREVLRKNDAVFADRPTTFAAESFSVGYRSASISPHGD 135
Query: 138 QWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYS 197
QW+K+++V++ E++SP L R EEAD++VRYV +C + G V+V A+++
Sbjct: 136 QWRKMRRVLTAEILSPATEHRLRGARGEEADHLVRYVLVRCGRDG--AAVDVRHVARHFC 193
Query: 198 GDVIRRLLLNKRYF----GNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGL 253
G+VIRRL L +R+F + + PG +E E+V+A+F L YL AF VSD+ P L GL
Sbjct: 194 GNVIRRLTLGRRHFREPRADDEDAAAPGRDEAEHVDALFATLNYLDAFCVSDYFPALVGL 253
Query: 254 DLDGHERIIKKACKIMKKYHDPIIEDRIQQW----KNGKKIEKEDLLDVLISLKDA---- 305
DLDG E++IKK + + + HDP++E+R+++W K G++ + D LDVL SL DA
Sbjct: 254 DLDGQEKVIKKVMRTLNRLHDPVVEERVEEWRLLRKAGERRDVADFLDVLASLDDAAGRP 313
Query: 306 ------------------VDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEY 347
VDNPSNAVEW LAE++N+PE+++KA +ELD+VVG+ RLVQE
Sbjct: 314 LLTVEEIKAQTIDIMIATVDNPSNAVEWALAEMMNKPEVMRKAMDELDTVVGRDRLVQES 373
Query: 348 DFPKLNYVKACAKEAFRRHPICDFNLPHV 376
D LNY+KAC +EAFR HP FN P V
Sbjct: 374 DVRDLNYLKACIREAFRLHPYHPFNPPRV 402
>UniRef100_Q9LQB7 F19C14.12 protein [Arabidopsis thaliana]
Length = 530
Score = 286 bits (733), Expect = 5e-76
Identities = 154/380 (40%), Positives = 229/380 (59%), Gaps = 24/380 (6%)
Query: 19 LMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLN 78
L+L+ ++ R + KPK +LPPGP WPIVGN+ +M+ NRP WI ++M +L
Sbjct: 12 LVLISITLVLALARRFSRFMKPKGQLPPGPRGWPIVGNMLQMIINRPAHLWIHRVMEELQ 71
Query: 79 TDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQ 138
TDIAC R HVIT++ +IARE+ ++D + A R S+++ +++GY + + +GE
Sbjct: 72 TDIACYRFARFHVITVTSSKIAREVLREKDEVLADRSESYASHLISHGYKNISFSSYGEN 131
Query: 139 WKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSG 198
WK VKKV++ +L+SP R EADNIV YVYN C +NV Y
Sbjct: 132 WKLVKKVMTTKLMSPTTLSKTLGYRNIEADNIVTYVYNLCQLGSVRKPINVRDTILTYCH 191
Query: 199 DVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLDLDGH 258
V+ R++ +R+F E+ G G +E E+++AI+ L F+F++++++P LRG ++D
Sbjct: 192 AVMMRMMFGQRHFDEVVENGGLGPKEKEHMDAIYLALDCFFSFNLTNYIPFLRGWNVDKA 251
Query: 259 ERIIKKACKIMKKYHDPIIEDRIQQW-KNGKKIEKEDLLDVLISLKD------------- 304
E +++A I+ +DPII++RI W K G K +ED LD+LI+LKD
Sbjct: 252 ETEVREAVHIINICNDPIIQERIHLWRKKGGKQMEEDWLDILITLKDDQGMHLFTFDEIR 311
Query: 305 ---------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYV 355
+DN N VEW +AE++N PE+L+KAT ELD +VGK RLVQE D +LNY+
Sbjct: 312 AQCKEINLATIDNTMNNVEWTIAEMLNHPEILEKATNELDIIVGKDRLVQESDISQLNYI 371
Query: 356 KACAKEAFRRHPICDFNLPH 375
KAC+KE+FR HP F +PH
Sbjct: 372 KACSKESFRLHPANVF-MPH 390
>UniRef100_O64514 YUP8H12R.1 protein [Arabidopsis thaliana]
Length = 437
Score = 273 bits (698), Expect = 6e-72
Identities = 153/373 (41%), Positives = 217/373 (58%), Gaps = 37/373 (9%)
Query: 41 KPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITISDPEIA 100
+ KLPP P +PI+GNL ML NRPT +WI ++MND+ TDIAC R G VHVI I+ IA
Sbjct: 37 RKKLPPCPRGFPIIGNLVGMLKNRPTSKWIVRVMNDMKTDIACFRFGRVHVIVITSDVIA 96
Query: 101 RELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLRHKWLH 160
RE+ ++DA+FA RP S+S EY++ GY +GE+ K+KKV+S+EL+S L
Sbjct: 97 REVVREKDAVFADRPDSYSAEYISGGYNGVVFDEYGERQMKMKKVMSSELMSTKALNLLL 156
Query: 161 DKRVEEADNIVRYVYNKC----TKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFGNGSE 216
R E+DN++ YV+N +K +VNV ++ +V RLL +R+F +
Sbjct: 157 KVRNLESDNLLAYVHNLYNKDESKTKHGAVVNVRDIVCTHTHNVKMRLLFGRRHFKETTM 216
Query: 217 DYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLDLDGHERIIKKACKIMKKYHDPI 276
D GL E E+ +AIF L F+F V+D+ P LRG +L G E +++A ++ +Y+ I
Sbjct: 217 DGSLGLMEKEHFDAIFAALDCFFSFYVADYYPFLRGWNLQGEEAELREAVDVIARYNKMI 276
Query: 277 IEDRIQQWKNGKKIEK-----------EDLLDVLISLKD--------------------- 304
I+++I+ W+ K +D LD+L +LKD
Sbjct: 277 IDEKIELWRGQNKDYNRAETKNDVPMIKDWLDILFTLKDENGKPLLTPQEITHLSVDLDV 336
Query: 305 -AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAF 363
+DN N +EW LAE++NQ E+L+KA EE+D VVGK RLVQE D P LNYVKAC +E
Sbjct: 337 VGIDNAVNVIEWTLAEMLNQREILEKAVEEIDMVVGKERLVQESDVPNLNYVKACCRETL 396
Query: 364 RRHPICDFNLPHV 376
R HP F +PH+
Sbjct: 397 RLHPTNPFLVPHM 409
>UniRef100_Q949U1 Putative cytochrome P450 protein [Arabidopsis thaliana]
Length = 538
Score = 251 bits (640), Expect = 3e-65
Identities = 140/368 (38%), Positives = 211/368 (57%), Gaps = 30/368 (8%)
Query: 36 KSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITIS 95
K++ +LPPGP WPI+GNLPE+ RP ++ + M +L TDIAC + ITI+
Sbjct: 37 KTKDRSCQLPPGPPGWPILGNLPELFMTRPRSKYFRLAMKELKTDIACFNFAGIRAITIN 96
Query: 96 DPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLR 155
EIARE ++DA A RP + E + + Y + ++P+GEQ+ K+K+VI+ E++S
Sbjct: 97 SDEIAREAFRERDADLADRPQLFIMETIGDNYKSMGISPYGEQFMKMKRVITTEIMSVKT 156
Query: 156 HKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFGNG- 214
K L R EADN++ YV++ + V+V ++ Y V R+L +R+
Sbjct: 157 LKMLEAARTIEADNLIAYVHSMYQR---SETVDVRELSRVYGYAVTMRMLFGRRHVTKEN 213
Query: 215 --SEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMP-CLRGLDLDGHERIIKKACKIMKK 271
S+D G E ++E IF L L +FS +D++ LRG ++DG E+ + + C I++
Sbjct: 214 VFSDDGRLGNAEKHHLEVIFNTLNCLPSFSPADYVERWLRGWNVDGQEKRVTENCNIVRS 273
Query: 272 YHDPIIEDRIQQWK-NGKKIEKEDLLDVLISLKD----------------------AVDN 308
Y++PII++R+Q W+ G K ED LD I+LKD A+DN
Sbjct: 274 YNNPIIDERVQLWREEGGKAAVEDWLDTFITLKDQNGKYLVTPDEIKAQCVEFCIAAIDN 333
Query: 309 PSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPI 368
P+N +EW L E++ PE+L+KA +ELD VVG+ RLVQE D P LNY+KAC +E FR HP
Sbjct: 334 PANNMEWTLGEMLKNPEILRKALKELDEVVGRDRLVQESDIPNLNYLKACCRETFRIHPS 393
Query: 369 CDFNLPHV 376
+ H+
Sbjct: 394 AHYVPSHL 401
>UniRef100_Q9SA39 F3O9.20 protein [Arabidopsis thaliana]
Length = 524
Score = 233 bits (593), Expect = 9e-60
Identities = 136/361 (37%), Positives = 196/361 (53%), Gaps = 30/361 (8%)
Query: 43 KLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITISDPEIARE 102
+LPPG WPI+GNLPE++ RP ++ M +L TDIAC H ITI+ EIARE
Sbjct: 41 QLPPGRPGWPILGNLPELIMTRPRSKYFHLAMKELKTDIACFNFAGTHTITINSDEIARE 100
Query: 103 LCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLRHKWLHDK 162
++DA A RP E + + Y T + +GE + K+KKVI+ E++S L
Sbjct: 101 AFRERDADLADRPQLSIVESIGDNYKTMGTSSYGEHFMKMKKVITTEIMSVKTLNMLEAA 160
Query: 163 RVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFGNG---SEDYG 219
R EADN++ Y+++ + V+V ++ Y V R+L +R+ S+D
Sbjct: 161 RTIEADNLIAYIHSMYQR---SETVDVRELSRVYGYAVTMRMLFGRRHVTKENMFSDDGR 217
Query: 220 PGLEEIEYVEAIFTVLQYLFAFSVSDFMP-CLRGLDLDGHERIIKKACKIMKKYHDPIIE 278
G E ++E IF L L FS D++ L G ++DG E K +++ Y++PII+
Sbjct: 218 LGKAEKHHLEVIFNTLNCLPGFSPVDYVDRWLGGWNIDGEEERAKVNVNLVRSYNNPIID 277
Query: 279 DRIQQWK-NGKKIEKEDLLDVLISLKD----------------------AVDNPSNAVEW 315
+R++ W+ G K ED LD I+LKD A+DNP+N +EW
Sbjct: 278 ERVEIWREKGGKAAVEDWLDTFITLKDQNGNYLVTPDEIKAQCVEFCIAAIDNPANNMEW 337
Query: 316 GLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPICDFNLPH 375
L E++ PE+L+KA +ELD VVGK RLVQE D LNY+KAC +E FR HP + PH
Sbjct: 338 TLGEMLKNPEILRKALKELDEVVGKDRLVQESDIRNLNYLKACCRETFRIHPSAHYVPPH 397
Query: 376 V 376
V
Sbjct: 398 V 398
>UniRef100_Q94JZ4 Cytochrome P450-like protein [Arabidopsis thaliana]
Length = 537
Score = 233 bits (593), Expect = 9e-60
Identities = 136/361 (37%), Positives = 196/361 (53%), Gaps = 30/361 (8%)
Query: 43 KLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITISDPEIARE 102
+LPPG WPI+GNLPE++ RP ++ M +L TDIAC H ITI+ EIARE
Sbjct: 43 QLPPGRPGWPILGNLPELIMTRPRSKYFHLAMKELKTDIACFNFAGTHTITINSDEIARE 102
Query: 103 LCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLRHKWLHDK 162
++DA A RP E + + Y T + +GE + K+KKVI+ E++S L
Sbjct: 103 AFRERDADLADRPQLSIVESIGDNYKTMGTSSYGEHFMKMKKVITTEIMSVKTLNMLEAA 162
Query: 163 RVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFGNG---SEDYG 219
R EADN++ Y+++ + V+V ++ Y V R+L +R+ S+D
Sbjct: 163 RTIEADNLIAYIHSMYQR---SETVDVRELSRVYGYAVTMRMLFGRRHVTKENMFSDDGR 219
Query: 220 PGLEEIEYVEAIFTVLQYLFAFSVSDFMP-CLRGLDLDGHERIIKKACKIMKKYHDPIIE 278
G E ++E IF L L FS D++ L G ++DG E K +++ Y++PII+
Sbjct: 220 LGKAEKHHLEVIFNTLNCLPGFSPVDYVDRWLGGWNIDGEEERAKVNVNLVRSYNNPIID 279
Query: 279 DRIQQWK-NGKKIEKEDLLDVLISLKD----------------------AVDNPSNAVEW 315
+R++ W+ G K ED LD I+LKD A+DNP+N +EW
Sbjct: 280 ERVEIWREKGGKAAVEDWLDTFITLKDQNGNYLVTPDEIKAQCVEFCIAAIDNPANNMEW 339
Query: 316 GLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPICDFNLPH 375
L E++ PE+L+KA +ELD VVGK RLVQE D LNY+KAC +E FR HP + PH
Sbjct: 340 TLGEMLKNPEILRKALKELDEVVGKDRLVQESDIRNLNYLKACCRETFRIHPSAHYVPPH 399
Query: 376 V 376
V
Sbjct: 400 V 400
>UniRef100_Q9MBF5 Cytochrome P450 [Petunia hybrida]
Length = 539
Score = 182 bits (462), Expect = 1e-44
Identities = 128/391 (32%), Positives = 198/391 (49%), Gaps = 52/391 (13%)
Query: 16 YLILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEM--LANRPTFRWIQKM 73
YL+ L+ + F+ +RS QK + LPPGP WPIVGNL ++ L +R + K
Sbjct: 30 YLVSKLVHFSFI---ERSKQKINR----LPPGPKQWPIVGNLFQLGQLPHRDMASFCDKY 82
Query: 74 MNDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALT 133
+ +RLGNV IT +DPEI RE+ ++QD IFASRP + + ++ G AL
Sbjct: 83 -----GPLVYLRLGNVDAITTNDPEIIREILVQQDDIFASRPRTLAAIHLAYGCGDVALA 137
Query: 134 PFGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAA 193
P G +WK+++++ L++ R + R +EA ++V V+ K K VN+
Sbjct: 138 PLGPKWKRMRRICMEHLLTTKRLESFGKHRADEAQSLVEDVWAKTQK---GETVNLRDLL 194
Query: 194 QYYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGL 253
+S + + R+LL K++F G+E GP E +E++ + L + D++P R +
Sbjct: 195 GAFSMNNVTRMLLGKQFF--GAESAGP-QEAMEFMHITHELFWLLGVIYLGDYLPLWRWI 251
Query: 254 DLDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIE--KEDLLDVLISLKD------- 304
D G E+ +++ K + +H IIE + KNGK ++ + D +DVL+SL
Sbjct: 252 DPHGCEKKMREVEKRVDDFHMRIIE---EHRKNGKNVDEGEMDFVDVLLSLPGEDEGDGN 308
Query: 305 --------------------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLV 344
A D + EW +AE+I P +LKK EELD VVG R+V
Sbjct: 309 GKQHMDDTEIKALIQDMIAAATDTSAVTNEWPMAEVIKHPNVLKKIQEELDIVVGSDRMV 368
Query: 345 QEYDFPKLNYVKACAKEAFRRHPICDFNLPH 375
E D L Y++ +E FR HP F +PH
Sbjct: 369 TESDLVHLKYLRCVVRETFRMHPAGPFLIPH 399
>UniRef100_Q9FPN4 Flavonoid 3',5'-hydroxylase [Callistephus chinensis]
Length = 510
Score = 177 bits (448), Expect = 6e-43
Identities = 126/388 (32%), Positives = 180/388 (45%), Gaps = 57/388 (14%)
Query: 17 LILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLA-NRPTFRWIQKMMN 75
L+++ LV F+ +H +LPPGP PWP+VGNLP + A T +
Sbjct: 15 LVIIALVNMFITRHTN----------RLPPGPAPWPVVGNLPHLGAIPHHTLAALATKYG 64
Query: 76 DLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPF 135
L +RLG VHV+ S P +A + D FASRP + +++ Y P+
Sbjct: 65 PL----VYLRLGFVHVVVASSPSVAAQFLKVHDLKFASRPPNSGAKHIAYNYQDMVFAPY 120
Query: 136 GEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQY 195
G QW +K+ + L S R EE + R + G VN+
Sbjct: 121 GPQWTMFRKICKDHLFSSKALDDFRHVRQEEVAILARGLAG-----AGRSKVNLGQQLNM 175
Query: 196 YSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFA-FSVSDFMPCLRGLD 254
+ + + R++L+KR FGN S P E + + T L +L F++ D++P L LD
Sbjct: 176 CTANTLARMMLDKRVFGNESGGDDPKANEFKEMA---TELMFLAGQFNIGDYIPVLDWLD 232
Query: 255 LDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKE--DLLDVLISLKD-------- 304
L G + +KK K+ D I+++ K I D+L LISLKD
Sbjct: 233 LQGIVKKMKKLHTRFDKFLDVILDEH-------KVIASGHIDMLSTLISLKDDTSVDGRK 285
Query: 305 ----------------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYD 348
D SN VEW +AELI QP LLK+A EE+DSVVG+ RLV E D
Sbjct: 286 PSDIEIKALLLELFVAGTDTSSNTVEWAIAELIRQPHLLKRAQEEMDSVVGQNRLVTEMD 345
Query: 349 FPKLNYVKACAKEAFRRHPICDFNLPHV 376
+L +++A KEAFR HP +LP +
Sbjct: 346 LSQLTFLQAIVKEAFRLHPSTPLSLPRI 373
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 701,250,561
Number of Sequences: 2790947
Number of extensions: 31290724
Number of successful extensions: 101362
Number of sequences better than 10.0: 2306
Number of HSP's better than 10.0 without gapping: 1471
Number of HSP's successfully gapped in prelim test: 835
Number of HSP's that attempted gapping in prelim test: 96936
Number of HSP's gapped (non-prelim): 3232
length of query: 384
length of database: 848,049,833
effective HSP length: 129
effective length of query: 255
effective length of database: 488,017,670
effective search space: 124444505850
effective search space used: 124444505850
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146720.6


