

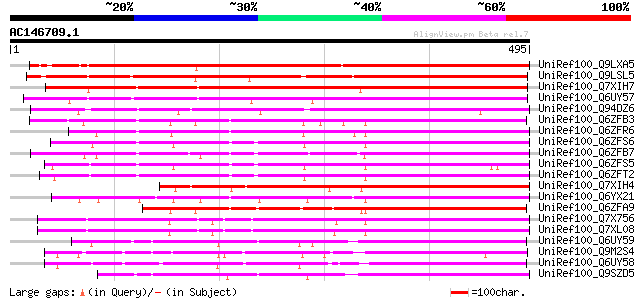
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146709.1 - phase: 0 /pseudo
(495 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LXA5 Lectin-like protein kinase-like [Arabidopsis th... 479 e-133
UniRef100_Q9LSL5 Receptor protein kinase-like protein [Arabidops... 445 e-123
UniRef100_Q7XIH7 Putative lectin-like protein kinase [Oryza sativa] 400 e-110
UniRef100_Q6UY57 Lectin-like receptor kinase 1;1 [Medicago trunc... 352 2e-95
UniRef100_Q94DZ6 Putative lectin-like receptor kinase 1;1 [Oryza... 337 4e-91
UniRef100_Q6ZFB3 Putative receptor-type protein kinase LRK1 [Ory... 332 1e-89
UniRef100_Q6ZFR6 Putative receptor-type protein kinase LRK1 [Ory... 327 6e-88
UniRef100_Q6ZFS6 Putative receptor-type protein kinase LRK1 [Ory... 321 3e-86
UniRef100_Q6ZFB7 Putative receptor-type protein kinase LRK1 [Ory... 316 8e-85
UniRef100_Q6ZFS5 Putative receptor-type protein kinase LRK1 [Ory... 315 2e-84
UniRef100_Q6ZFT2 Putative receptor-type protein kinase LRK1 [Ory... 313 6e-84
UniRef100_Q7XIH4 Putative lectin-like protein kinase [Oryza sativa] 299 1e-79
UniRef100_Q6YX21 Putative receptor kinase Lecrk [Oryza sativa] 297 4e-79
UniRef100_Q6ZFA9 Putative receptor-type protein kinase LRK1 [Ory... 286 1e-75
UniRef100_Q7X756 OSJNBb0096E05.1 protein [Oryza sativa] 274 5e-72
UniRef100_Q7XL08 OSJNBa0065J03.1 protein [Oryza sativa] 271 2e-71
UniRef100_Q6UY59 Lectin-like receptor kinase 7;2 [Medicago trunc... 268 3e-70
UniRef100_Q9M2S4 Probable serine/threonine-specific protein kina... 264 4e-69
UniRef100_Q6UY58 Lectin-like receptor kinase 7;3 [Medicago trunc... 264 5e-69
UniRef100_Q9SZD5 Serine/threonine-specific kinase like protein [... 260 6e-68
>UniRef100_Q9LXA5 Lectin-like protein kinase-like [Arabidopsis thaliana]
Length = 651
Score = 479 bits (1232), Expect = e-133
Identities = 249/478 (52%), Positives = 332/478 (69%), Gaps = 10/478 (2%)
Query: 20 MASSLYILTLFYMSLIFFNIPASSIHFKYPSFDPSDANIVYQGSAAPRDGEVNFNINENY 79
MA+S+ + + F + L F S+ F F + I YQG A +G V N +Y
Sbjct: 1 MANSILLFS-FVLVLPF----VCSVQFNISRFGSDVSEIAYQGDARA-NGAVELT-NIDY 53
Query: 80 SCQVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGFEIP 139
+C+ G Y ++V LW+ T K +DF+T ++F I+T+ YGHG AFFL P ++P
Sbjct: 54 TCRAGWATYGKQVPLWNPGTSKPSDFSTRFSFRIDTRNVGYGNYGHGFAFFLAPARIQLP 113
Query: 140 LNSDGGFMGLFNTTTMVSSSNQIVHVEFDSFANREFR--EARGHVGININSIISSATTPW 197
NS GGF+GLFN T SS+ +V+VEFD+F N E+ + + HVGIN NS++SS T W
Sbjct: 114 PNSAGGFLGLFNGTNNQSSAFPLVYVEFDTFTNPEWDPLDVKSHVGINNNSLVSSNYTSW 173
Query: 198 NASKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEWITVGFS 257
NA+ H+ D V I Y+S +NL+VSW Y TS+P EN+SLS IDL KV+P +T+GFS
Sbjct: 174 NATSHNQDIGRVLIFYDSARRNLSVSWTYDLTSDPLENSSLSYIIDLSKVLPSEVTIGFS 233
Query: 258 AATSYVQELNYLLSWEFNSTLATSGDSKTKETRLVIILTVSCGVIVIGVGALVAYALFWR 317
A + V E N LLSWEF+S+L K++ + +I+ +S V+ + + +F
Sbjct: 234 ATSGGVTEGNRLLSWEFSSSLELIDIKKSQNDKKGMIIGISVSGFVLLTFFITSLIVFL- 292
Query: 318 KRKRSNKQKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKG 377
KRK+ K+ EE +LTS+N+DLERGAGPR+FTYK+L A NNF+ DRKLG+GGFGAVY+G
Sbjct: 293 KRKQQKKKAEETENLTSINEDLERGAGPRKFTYKDLASAANNFADDRKLGEGGFGAVYRG 352
Query: 378 YFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFM 437
Y LD+ VA+KK + GS+QGK+E+VTEVK+IS LRHRNLV+L+GWCH+K EFL++YEFM
Sbjct: 353 YLNSLDMMVAIKKFAGGSKQGKREFVTEVKIISSLRHRNLVQLIGWCHEKDEFLMIYEFM 412
Query: 438 PNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
PNGSLD+HLFGK+ L+W VR KI LGLAS LLYLHEEWE+CVVHRDIK+SNVMLDS+
Sbjct: 413 PNGSLDAHLFGKKPHLAWHVRCKITLGLASALLYLHEEWEQCVVHRDIKASNVMLDSN 470
>UniRef100_Q9LSL5 Receptor protein kinase-like protein [Arabidopsis thaliana]
Length = 675
Score = 445 bits (1145), Expect = e-123
Identities = 238/485 (49%), Positives = 327/485 (67%), Gaps = 19/485 (3%)
Query: 17 TQSMASSLYILTLFYMSLIFFNIPASSIHFKYPSFDPSD-ANIVYQGSAAP-RDGEVNFN 74
+ SM++S+ L+LF +F S++F + SF D +I Y G A P DG VNFN
Sbjct: 13 SSSMSNSILFLSLF----LFLPFVVDSLYFNFTSFRQGDPGDIFYHGDATPDEDGTVNFN 68
Query: 75 INENYSCQVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPY 134
N + QVG + YS+KV +W TGK +DF+T ++F I+ R+ S GHG+ FFL P
Sbjct: 69 -NAEQTSQVGWITYSKKVPIWSHKTGKASDFSTSFSFKIDA--RNLSADGHGICFFLAPM 125
Query: 135 GFEIPLNSDGGFMGLFNTTTMVSSSNQIVHVEFDSFANREF--REARGHVGININSIISS 192
G ++P S GGF+ LF SSS +VHVEFD+F N + + HVGIN NS++SS
Sbjct: 126 GAQLPAYSVGGFLNLFTRKNNYSSSFPLVHVEFDTFNNPGWDPNDVGSHVGINNNSLVSS 185
Query: 193 ATTPWNASKHSGDTAEVWIRYNSTTKNLTVSWKYQ--TTSNPQENTSLSISIDLMKVMPE 250
T WNAS HS D I Y+S TKNL+V+W Y+ TS+P+E++SLS IDL KV+P
Sbjct: 186 NYTSWNASSHSQDICHAKISYDSVTKNLSVTWAYELTATSDPKESSSLSYIIDLAKVLPS 245
Query: 251 WITVGFSAATSYVQELNYLLSWEFNSTLATSGDSKTKETRLVIILTVSCGVIVIGVGALV 310
+ GF AA E + LLSWE +S+L DS ++R+ +++ +S V ++
Sbjct: 246 DVMFGFIAAAGTNTEEHRLLSWELSSSL----DSDKADSRIGLVIGISASGFVFLTFMVI 301
Query: 311 AYALFWRKRKRSNKQKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGG 370
+ W +++R K+++ ++ S+N DLER AGPR+F+YK+L ATN FS RKLG+GG
Sbjct: 302 TTVVVWSRKQRKKKERD-IENMISINKDLEREAGPRKFSYKDLVSATNRFSSHRKLGEGG 360
Query: 371 FGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEF 430
FGAVY+G +++ VAVKK+S SRQGK E++ EVK+IS+LRHRNLV+L+GWC++K EF
Sbjct: 361 FGAVYEGNLKEINTMVAVKKLSGDSRQGKNEFLNEVKIISKLRHRNLVQLIGWCNEKNEF 420
Query: 431 LLVYEFMPNGSLDSHLFGKR-TPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSN 489
LL+YE +PNGSL+SHLFGKR LSW +R+KI LGLAS LLYLHEEW++CV+HRDIK+SN
Sbjct: 421 LLIYELVPNGSLNSHLFGKRPNLLSWDIRYKIGLGLASALLYLHEEWDQCVLHRDIKASN 480
Query: 490 VMLDS 494
+MLDS
Sbjct: 481 IMLDS 485
>UniRef100_Q7XIH7 Putative lectin-like protein kinase [Oryza sativa]
Length = 689
Score = 400 bits (1029), Expect = e-110
Identities = 214/474 (45%), Positives = 300/474 (63%), Gaps = 16/474 (3%)
Query: 35 IFFNIPASSIHFKYPSFDPSDANIVYQGSAAPRDGEVNF----------NINENYSCQVG 84
++F++ H SF + + GSA + E F +N + VG
Sbjct: 13 LYFSLSLKIAHVNPLSFKLNFTESNHNGSATIQLQEDAFYNKAVKLTKDELNGKITQSVG 72
Query: 85 RVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGFEIPLNSDG 144
R Y++ V LWDS TG+L FTT +TF I S YG GLAFFL Y +P NS
Sbjct: 73 RAIYTDPVPLWDSTTGQLASFTTRFTFKIYAPTNDSS-YGEGLAFFLSSYPSVVPNNSMD 131
Query: 145 GFMGLF-NTTTMVSSSNQIVHVEFDSFANREFREARGHVGININSIISSATTPWNASKHS 203
G++GLF N+ NQIV VEFDS N + HVGINI+SI+S A W +S +
Sbjct: 132 GYLGLFSNSNDQSDPLNQIVAVEFDSHKNTWDPDGN-HVGINIHSIVSVANVTWRSSIND 190
Query: 204 GDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEWITVGFSAATSYV 263
G A W+ Y + ++NL+V YQ N+SLS S+DL K +P+ +++GFSA+T
Sbjct: 191 GRIANAWVTYQANSRNLSVFLSYQDNPQFSGNSSLSYSVDLSKYLPDKVSIGFSASTGKF 250
Query: 264 QELNYLLSWEFNSTLATSGDSKTKETRLVIILTVSCGVIVIGVGALVAYALFWRKRKRSN 323
EL+ +L WEF+ST ++ + LVI L+ S V+V +G + + F R R+ +
Sbjct: 251 VELHQILYWEFDSTDVHLMKTEKTKGILVISLSTSGSVVVCSIGLVCFFLCFRRIRRTTR 310
Query: 324 KQKEEAMHLT---SMNDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFA 380
+++E L S++ + E+G GPRRF Y EL +AT+NF+ +RKLG+GGFGAVY+G+
Sbjct: 311 SREKEKEKLDCDESIDSEFEKGKGPRRFQYNELVVATDNFAAERKLGEGGFGAVYQGFLK 370
Query: 381 DLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNG 440
D ++++A+K++++GS QG+KEY++EVK+IS+LRHRNLV+L+GWCH+ GEFLLVYEFMPN
Sbjct: 371 DQNIEIAIKRVAKGSTQGRKEYISEVKIISRLRHRNLVQLVGWCHEHGEFLLVYEFMPNR 430
Query: 441 SLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDS 494
SLD HL+ L+W +R KI +G+AS LLYLHEEWE+CVVHRD+K SNVMLDS
Sbjct: 431 SLDKHLYDGGNLLAWPLRFKITIGVASALLYLHEEWEQCVVHRDVKPSNVMLDS 484
>UniRef100_Q6UY57 Lectin-like receptor kinase 1;1 [Medicago truncatula]
Length = 678
Score = 352 bits (902), Expect = 2e-95
Identities = 199/499 (39%), Positives = 295/499 (58%), Gaps = 22/499 (4%)
Query: 14 SVETQSMASSLYILTLFYMSLIFFNIP-ASSIHFKYPSFDPSD--ANIVYQGSAAPRDGE 70
S + + S IL LF + L+ +P +S+ F +FD +N+ YQG +G
Sbjct: 7 SCSSNIINQSKNILVLFSLLLVLSYLPKTNSLLFNITNFDDPTVASNMSYQGDGKSTNGS 66
Query: 71 VNFNINENYSCQVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFF 130
++ N +Y +VGR FYS+ + LWD T LT FTT ++F I+ + + YG G F+
Sbjct: 67 IDLN-KVSYLFRVGRAFYSQPLHLWDKKTNTLTSFTTRFSFTIDKL--NDTTYGDGFVFY 123
Query: 131 LVPYGFEIPLNSDGGFMGLFNTTTMVSS-SNQIVHVEFDSFANREFREARGHVGININSI 189
L P G++IP NS GG GLFN TT + N +V VEFD+F + HVGI+ N++
Sbjct: 124 LAPLGYQIPPNSAGGVYGLFNATTNSNFVMNYVVGVEFDTFVGPTDPPMK-HVGIDDNAL 182
Query: 190 ISSATTPWNASKHSGDTAEVWIRYNSTTKNLTVSWKYQTT----SNPQENTSLSISIDLM 245
S A ++ K+ G V I YNS K L V W ++ N+S+S IDLM
Sbjct: 183 TSVAFGKFDIDKNLGRVCYVLIDYNSDEKMLEVFWSFKGRFVKGDGSYGNSSISYQIDLM 242
Query: 246 KVMPEWITVGFSAATSYVQELNYLLSWEFNSTLATSGDSKTK----------ETRLVIIL 295
K +PE++ +GFSA+T E N + SWEF+S L S + + +T +V++
Sbjct: 243 KKLPEFVNIGFSASTGLSTESNVIHSWEFSSNLEDSNSTTSLVEGNDGKGSLKTVIVVVA 302
Query: 296 TVSCGVIVIGVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDDLERGAGPRRFTYKELDL 355
+ ++V + ++V + + RKRK ++ +E DL++ PRRF Y EL
Sbjct: 303 VIVPVILVFLIASIVGWVIVKRKRKNCDEGLDEYGIPFPKKFDLDKATIPRRFEYSELVA 362
Query: 356 ATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHR 415
ATN F DR LG+GG+G VYKG + L VAVK+I ++ ++ EV++IS+L HR
Sbjct: 363 ATNGFDDDRMLGRGGYGQVYKGALSYLGKIVAVKRIFADFENSERVFINEVRIISRLIHR 422
Query: 416 NLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEE 475
NLV+ +GWCH++ E LLV+E+MPNGSLD+HLFG + L+W VR+K+ALG+A+ L YLH++
Sbjct: 423 NLVQFIGWCHEQDELLLVFEYMPNGSLDTHLFGDKKSLAWEVRYKVALGVANALRYLHDD 482
Query: 476 WERCVVHRDIKSSNVMLDS 494
E+CV+HRDIKS+NV+LD+
Sbjct: 483 AEQCVLHRDIKSANVLLDT 501
>UniRef100_Q94DZ6 Putative lectin-like receptor kinase 1;1 [Oryza sativa]
Length = 696
Score = 337 bits (865), Expect = 4e-91
Identities = 202/502 (40%), Positives = 298/502 (59%), Gaps = 39/502 (7%)
Query: 21 ASSLYILTLFYMSLIFFNIPASSIHFKYPSFDPSDANIVYQGSA-------APRDGEVNF 73
A++ ++L + + L+ A S + + S D + + I +QG A RD +
Sbjct: 8 AAAPFLLLVALLLLLPSPAAAFSFTYNFTSADTAPSGIAFQGDAFFNKFIRLTRDERIG- 66
Query: 74 NINENYSCQVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVP 133
+ GR F+S V L D + + F+T ++F I S + G GLAFFL P
Sbjct: 67 ----PITSSAGRAFFSRPVPLCDPVSRRRASFSTAFSFSIAAPDPSAAS-GDGLAFFLSP 121
Query: 134 YGFEIPLNSDGGFMGLFNTTTMVSSS----NQIVHVEFDSFANREFREARGHVGININSI 189
+ +P +S GG +GLFN+ + ++ +V VEFD++ N E+ + HVG+++ I
Sbjct: 122 FPSVLPNSSAGGLLGLFNSFSRGGAAAAHPRPLVAVEFDTYKN-EWDPSDDHVGVDLGGI 180
Query: 190 ISSATTPWNASKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTS--LSISIDLMKV 247
+S+AT W S G A + Y+ KNLTV+ Y + T L ++DLM+
Sbjct: 181 VSAATVDWPTSMKDGRRAHARVAYDGQAKNLTVALSYGDAAAAAALTDPVLWYAVDLMEY 240
Query: 248 MPEWITVGFSAATSYVQELNYLLSWEFNSTLATSGDSKTKETRLVIILTVSCGVIVIGVG 307
+P+ + VGFSAAT EL+ +L WEF S++ T +ET ++ ++ CG++++ V
Sbjct: 241 LPDAVAVGFSAATGEAAELHQVLYWEFTSSIDTK-----EETVILWVVLGLCGLLLVLVA 295
Query: 308 ALVAYALF-WRKR-KRSNKQKEEAMHLTSMNDD-LERGAGPRRFTYKELDLATNNFSKDR 364
A V + + WRK + ++ +E M + D+ +GPRRF Y +L AT NFS +R
Sbjct: 296 AGVLWFVSQWRKAGELADGDIDEEMGYDELADEEFFVESGPRRFRYSDLAAATKNFSDER 355
Query: 365 KLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWC 424
KLGQGGFGAVY+G+ +L L VA+K++S+GS QG+KEY EV++ISQLRHR+LV+L+GWC
Sbjct: 356 KLGQGGFGAVYRGFLKELGLAVAIKRVSKGSTQGRKEYAAEVRIISQLRHRHLVRLVGWC 415
Query: 425 HD-KGEFLLVYEFMPNGSLDSHLF----------GKRTPLSWGVRHKIALGLASGLLYLH 473
H+ +G+FLLVYE MPNGS+D HL+ G PLSW R+ +ALGLAS LLYLH
Sbjct: 416 HEHRGDFLLVYELMPNGSVDRHLYGGGGGSKKAGGAAPPLSWPTRYNVALGLASALLYLH 475
Query: 474 EEWERCVVHRDIKSSNVMLDSS 495
EE +CVVHRDIK SNVMLD++
Sbjct: 476 EECPQCVVHRDIKPSNVMLDAT 497
>UniRef100_Q6ZFB3 Putative receptor-type protein kinase LRK1 [Oryza sativa]
Length = 512
Score = 332 bits (851), Expect = 1e-89
Identities = 205/504 (40%), Positives = 290/504 (56%), Gaps = 34/504 (6%)
Query: 20 MASSLYILTLFYMSLIFFNIPASSIHFKYPSFDPSDANIVYQGSAAPRDG---EVNFNIN 76
MA++ + L M ++ A+++ F Y +F + NI QGSAA G E+ N
Sbjct: 12 MAAASALACLLVMLRCLPSVVATTVSFNYSTFSNAK-NITLQGSAAFAGGGCIEITTGSN 70
Query: 77 ENYSCQVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGF 136
S +GRV Y+ V LWD+ TG++ FTT ++F I G G+AFFLV Y
Sbjct: 71 LPSSGTMGRVAYTPPVQLWDAATGEVASFTTRFSFNITPTNLDNK--GDGMAFFLVGYPS 128
Query: 137 EIPLNSDGGFMGLFNT--TTMVSSSNQIVHVEFDSFANREF--REARGHVGININSIISS 192
+P DGG +GL + T+ N+ V VEFD++ NR+F H+GI++NSI S
Sbjct: 129 RMPDKGDGGALGLTSRYFDTVQPGENRFVAVEFDTYLNRDFDPNATYDHIGIDVNSIRSV 188
Query: 193 ATTPWNASKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEWI 252
T + +G+ + + YNS++ L+ T+ +LS ++DL +PE +
Sbjct: 189 QTESLPSFSLTGNMTAI-VDYNSSSSILSAQLVKTWTNGSTTLYNLSTTVDLKSALPEKV 247
Query: 253 TVGFSAATSYVQELNYLLSWEFNSTL------ATSGDSKTKETRLVIIL---TVSCGVIV 303
+VG AAT EL+ L SW FNS+ A T + L ++ TV + V
Sbjct: 248 SVGILAATGLSLELHQLHSWYFNSSFQQNPPPAVQHSPTTSGSGLAGVIAGATVGAALFV 307
Query: 304 IGVGALVAYALFWR--KRKRSNKQKEEAMHLTSMNDD-----------LERGAGPRRFTY 350
+ + A+V + R K++R ++ EEA H+ DD +E G GPR+ Y
Sbjct: 308 VLLFAMVVVLVRRRRSKKRREVEEAEEARHVGLARDDDDDDDGEPIVEIEMGMGPRQIPY 367
Query: 351 KELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISR-GSRQGKKEYVTEVKVI 409
++L ATN+F+ + KLGQGGFGAVY+GY + L VA+K+ ++ S+QGKKEY +E+KVI
Sbjct: 368 QDLIEATNSFAAEEKLGQGGFGAVYRGYLREQGLAVAIKRFAKDSSKQGKKEYRSEIKVI 427
Query: 410 SQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGL 469
S+LRHRNLV+L+GWCH + E LL+YE +PN SLD HL G T L+W +R KI LGL S L
Sbjct: 428 SRLRHRNLVQLIGWCHGRDELLLIYELVPNRSLDIHLHGNGTFLTWPMRVKIVLGLGSAL 487
Query: 470 LYLHEEWERCVVHRDIKSSNVMLD 493
YLHEEWE+CVVHRDIK SNVMLD
Sbjct: 488 FYLHEEWEQCVVHRDIKPSNVMLD 511
>UniRef100_Q6ZFR6 Putative receptor-type protein kinase LRK1 [Oryza sativa]
Length = 705
Score = 327 bits (837), Expect = 6e-88
Identities = 201/468 (42%), Positives = 272/468 (57%), Gaps = 32/468 (6%)
Query: 57 NIVYQGSAA-PRDGEVNFNINENYSC--QVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVI 113
NI QGSAA DG + N +GRV YS V LW++ TG++ FTT ++F I
Sbjct: 26 NITLQGSAAIAGDGWIEITTGSNLPSGGTMGRVAYSPPVQLWEAATGEVASFTTRFSFNI 85
Query: 114 NTQGRSPSLYGHGLAFFLVPYGFEIPLNSDGGFMGLFNTT--TMVSSSNQIVHVEFDSFA 171
G G+AFFLV Y +P +DGG +GL + T ++S N+ V VEFD+F
Sbjct: 86 TPTNLDNK--GDGMAFFLVGYPSRMPDTADGGALGLTSRTFDAVMSGDNRFVAVEFDTFN 143
Query: 172 NREFREAR-GHVGININSIISSATTPWNASKHSGDTAEVWIRYNSTTKNLTVSWKYQTTS 230
N A H+G+++NSI+S T + +G+ A + + YNS++ L+V T+
Sbjct: 144 NSFDPSATYDHIGVDVNSIVSVQTESLPSFSLTGNMAAI-VDYNSSSSILSVQLVKTWTN 202
Query: 231 NPQENTSLSISIDLMKVMPEWITVGFSAATSYVQELNYLLSWEFNSTL------ATSGDS 284
+LS ++DL +PE ++VGFSAAT EL+ L SW FNS+ A
Sbjct: 203 GSTTLYNLSTTVDLKTALPEKVSVGFSAATGSSLELHQLHSWYFNSSFQQNPPPAAQPSP 262
Query: 285 KTKETRLVIILTVSCGVIVIGVGALVAYALFWRKRKRSNKQKE-----EAMHLTSMNDD- 338
T L ++ + + V L A + +R+RS K++E EA H+ DD
Sbjct: 263 TTSGPGLAGVIAGATAGGALFVVLLFAMIVVLVRRRRSKKRREAEEAEEARHVGLAGDDD 322
Query: 339 ----------LERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAV 388
+E G GPR+ Y EL AT +F+ + KLGQGGFGAVY+GY + L VA+
Sbjct: 323 DDDDGEPIVEIEMGMGPRQIPYHELVEATKSFAAEEKLGQGGFGAVYRGYLREQGLAVAI 382
Query: 389 KKISR-GSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLF 447
K+ ++ S+QGKKEY +E+KVIS+LRHRNLV+L+GWCH + E LLVYE +PN SLD HL
Sbjct: 383 KRFAKDSSKQGKKEYRSEIKVISRLRHRNLVQLIGWCHGRDELLLVYELVPNRSLDIHLH 442
Query: 448 GKRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
G T L+W +R KI LGL S L YLHEEWE+CVVHRDIK SNVMLD S
Sbjct: 443 GNGTFLTWPMRVKIVLGLGSALFYLHEEWEQCVVHRDIKPSNVMLDES 490
>UniRef100_Q6ZFS6 Putative receptor-type protein kinase LRK1 [Oryza sativa]
Length = 681
Score = 321 bits (822), Expect = 3e-86
Identities = 202/497 (40%), Positives = 283/497 (56%), Gaps = 48/497 (9%)
Query: 40 PASSIHFKYPSFDPSD-ANIVYQGSAAPRDGEVNFNINE--NYSCQVGRVFYSEKVLLWD 96
P +++ F YP+F SD NI QG A+ G V+ + N GRV Y+ V LWD
Sbjct: 6 PVAALSFNYPTFASSDNQNIDIQGQASVSVGYVDISANSVSGMGNSAGRVVYAPPVQLWD 65
Query: 97 SNTGKLTDFTTHYTF-VINTQGRSPSLYGHGLAFFLVPYGFEIPLNSDGGF-MGLFNTTT 154
+ TG++ FTT ++F +I RS G G+AFFL Y +P+ +GG +GL N T
Sbjct: 66 AATGEVASFTTRFSFNIIAPSDRSKK--GDGMAFFLTSYPSRLPVGHEGGENLGLTNQTV 123
Query: 155 --MVSSSNQIVHVEFDSFANR-EFREARGHVGININSIISSATTPW-NASKHSGDTAEVW 210
+ + N+ V VEFD+F N + H+GI++NS++S N S TA V
Sbjct: 124 GNVSTGQNRFVAVEFDTFVNPFDPNTTNDHIGIDVNSVVSVTNESLPNFSLIGNMTATV- 182
Query: 211 IRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEWITVGFSAATSYVQELNYLL 270
YN+ ++ L++ T+ P +LS +DL + +PE +TVGFSA+T E + L
Sbjct: 183 -DYNNNSRILSIKLWINETTTPY---TLSSMVDLKRALPENVTVGFSASTGSAFEQHQLT 238
Query: 271 SWEFNSTLA-------------------------TSGDSKTKETRLVIILTVSCGVIVIG 305
SW F S+ + T S ++ +V T+ + VI
Sbjct: 239 SWYFKSSSSFEQKLAAKVASPPPPSSPSPPPPSLTPITSHSRRGGVVAGATLGAVMFVIL 298
Query: 306 VGALVAYALFWRKRKRSNKQKEEAMHLTSMNDD------LERGAGPRRFTYKELDLATNN 359
+ A+VA + R+ K+ + ++ H + +DD +E G GPRRF Y EL AT +
Sbjct: 299 LFAMVAVLVRRRQSKKRREAEDGGWHGSDDDDDGEPIVEIEMGMGPRRFPYHELVDATKS 358
Query: 360 FSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGS-RQGKKEYVTEVKVISQLRHRNLV 418
F+ + KLGQGGFGAVY+GY +L L VA+K+ ++ S +QG+KEY +E+KVIS+LRHRNLV
Sbjct: 359 FAPEEKLGQGGFGAVYRGYLRELGLAVAIKRFAKNSSKQGRKEYKSEIKVISRLRHRNLV 418
Query: 419 KLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEEWER 478
+L+GWCH + E LLVYE PN SLD HL G T L+W +R I GL S LLYLHEEW++
Sbjct: 419 QLIGWCHGRTELLLVYELFPNRSLDVHLHGNGTFLTWPMRINIVHGLGSALLYLHEEWDQ 478
Query: 479 CVVHRDIKSSNVMLDSS 495
CVVHRDIK SNVMLD S
Sbjct: 479 CVVHRDIKPSNVMLDES 495
>UniRef100_Q6ZFB7 Putative receptor-type protein kinase LRK1 [Oryza sativa]
Length = 676
Score = 316 bits (810), Expect = 8e-85
Identities = 195/488 (39%), Positives = 284/488 (57%), Gaps = 23/488 (4%)
Query: 21 ASSLYILTLFYMSLIFFNIPASSIHFKYPSFDPSDANIVYQGSAAPRDGE---VNFNINE 77
A++ +++L M ++ A+++ F Y SF + NI QGSAA E +
Sbjct: 14 AAAAALVSLLVMLRCLPSVVATTVSFNYSSFSNASKNITLQGSAALAGAEWIELTKGKGN 73
Query: 78 NYSC--QVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYG 135
N S +GR+ Y+ V LWD+ TG++ FTT ++F I + +S G G+ FFLV Y
Sbjct: 74 NLSSGGTMGRMVYTPPVQLWDAATGEVASFTTRFSFNITPKNKSNK--GDGMTFFLVSYP 131
Query: 136 FEIPLNSDGGFMGLFNTT-TMVSSSNQIVHVEFDSFANREFREARG---HVGININSIIS 191
+P GG +GL + T ++ ++ V VEFD++ N F + H+GI++N++ S
Sbjct: 132 SRMPYMGYGGALGLTSQTFDNATAGDRFVAVEFDTY-NNSFLDPDATYDHIGIDVNALRS 190
Query: 192 SATTPWNASKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEW 251
T + G+ + + YNS + ++V ++ P +LS +DL +PE
Sbjct: 191 VKTESLPSYILIGNMTAI-VDYNSNSSIMSVKLWANGSTTPY---NLSSKVDLKSALPEK 246
Query: 252 ITVGFSAATSYVQELNYLLSWEFNSTLATSGDSKTKETRLVIILTVSCGVIVIGVGALVA 311
+ VGFSAAT E + L SW FN TL + + +R ++ + G I+ V
Sbjct: 247 VAVGFSAATGSSFEQHQLRSWYFNLTLEQKQPTG-QHSRGGVVAGATVGAILFIVLLFTM 305
Query: 312 YALFWRKRKRSNKQKEEAMHLTSMND---DLERGAGPRRFTYKELDLATNNFSKDRKLGQ 368
A+ R+R+R ++EE S D ++E G GPRRF Y L AT +F+ + KLGQ
Sbjct: 306 VAILVRRRQRKKMREEEEDD--SEGDPIVEIEMGTGPRRFPYHILVNATKSFAAEEKLGQ 363
Query: 369 GGFGAVYKGYFADLDLQVAVKKISR-GSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDK 427
GGFGAVY+G +L L VA+K+ ++ S+QG+KEY +E+KVIS+LRHRNLV+L+GWCH +
Sbjct: 364 GGFGAVYRGNLRELGLDVAIKRFAKDSSKQGRKEYKSEIKVISRLRHRNLVQLIGWCHGR 423
Query: 428 GEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKS 487
E LLVYE +PN SLD HL G T L+W +R I LGL + LLYLHEEWE+CVVHRDIK
Sbjct: 424 DELLLVYELVPNRSLDVHLHGNGTFLTWPMRINIVLGLGNALLYLHEEWEQCVVHRDIKP 483
Query: 488 SNVMLDSS 495
SN+MLD S
Sbjct: 484 SNIMLDES 491
>UniRef100_Q6ZFS5 Putative receptor-type protein kinase LRK1 [Oryza sativa]
Length = 683
Score = 315 bits (807), Expect = 2e-84
Identities = 205/514 (39%), Positives = 294/514 (56%), Gaps = 60/514 (11%)
Query: 34 LIFFNI-------PASSIHFKYPSFDPS-DANIVYQGSAAPRDGEVNFNINENYSCQVGR 85
+IFF++ P +++ F YP+F S + I +G+A+ G ++ + N + VGR
Sbjct: 14 VIFFSVCYMQPPAPVAALSFNYPTFASSHNQYIEIEGNASVSVGYIDISAN-SVGNNVGR 72
Query: 86 VFYSEKVLLWDSNTGKLTDFTTHYTF-VINTQGRSPSLYGHGLAFFLVPYGFEIPLNSDG 144
VFY V LWD+ TG++ FTT ++F +I RS G G+AFFL Y +P+ +G
Sbjct: 73 VFYKPPVQLWDAATGEVASFTTRFSFNIIAPSDRSKK--GDGMAFFLTSYPSRLPVGHEG 130
Query: 145 GF-MGLFNTTT--MVSSSNQIVHVEFDSFANR-EFREARGHVGININSIISSATTPW-NA 199
G +GL N T + + N+ V VEFD+F N + H+GI++NS++S N
Sbjct: 131 GENLGLTNQTVGNVSTGQNRFVAVEFDTFVNPFDPNTTNDHIGIDVNSVVSVTNESLPNF 190
Query: 200 SKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEWITVGFSAA 259
S TA V YN+ ++ L+V ++ P +LS +DL + +PE ITVGFSA+
Sbjct: 191 SLIGNMTATV--DYNNNSRILSVKLWINGSTTPY---TLSSMVDLKRALPENITVGFSAS 245
Query: 260 TSYVQELNYLLSWEFNSTLA-------------------------TSGDSKTKETRLVII 294
E + L SW F S+ + T S ++ +V
Sbjct: 246 IGSAYEQHQLTSWYFKSSSSFEQKLAAKVASPPPPSSPSPPPPSLTPITSHSRRGGVVAG 305
Query: 295 LTVSCGVIVIGVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDD------LERGAGPRRF 348
TV + VI + A+VA + R+ K+ + ++ H + +DD +E G GPRRF
Sbjct: 306 ATVGAVMFVILLFAMVAVLVRRRQSKKRREAEDGGWHGSDDDDDGEPIVEIEMGMGPRRF 365
Query: 349 TYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISR-GSRQGKKEYVTEVK 407
Y EL AT +F+ + KLGQGGFGAVY+GY +L L VA+K+ ++ S+QG+KEY +E+K
Sbjct: 366 PYHELVDATKSFATEEKLGQGGFGAVYRGYLRELGLAVAIKRFAKDSSKQGRKEYKSEIK 425
Query: 408 VISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVR----HKIAL 463
VIS+LRHRNLV+L+GWCH + E LLVYE +PN SLD HL G T L+W +R H+I +
Sbjct: 426 VISRLRHRNLVQLIGWCHGRTELLLVYELVPNRSLDVHLHGNGTFLTWPMRLGCCHRINI 485
Query: 464 --GLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
GL S LLYLHEEW++CVVHRDIK SNVMLD S
Sbjct: 486 VHGLGSALLYLHEEWDQCVVHRDIKPSNVMLDES 519
>UniRef100_Q6ZFT2 Putative receptor-type protein kinase LRK1 [Oryza sativa]
Length = 688
Score = 313 bits (803), Expect = 6e-84
Identities = 198/498 (39%), Positives = 285/498 (56%), Gaps = 39/498 (7%)
Query: 29 LFYMSLIFFNIPA--SSIHFKYPSFDPS-DANIVYQGSAAPRDGEVNFNINENYSCQVGR 85
+ + S+ + PA +++ F YP+F S + I +G+A+ G ++ + N + VGR
Sbjct: 14 VIFFSVCYIQPPAHFAALSFNYPTFASSHNQYIEIEGNASVSVGYIDISAN-SVGNNVGR 72
Query: 86 VFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGFEIPLNSDGG 145
VF+ V LWD+ TG++ FTT ++F I RS G G+ FFL Y +P GG
Sbjct: 73 VFHKPPVQLWDAATGEVASFTTRFSFNIIPGNRSKK--GDGMTFFLTSYPSRLPEGDAGG 130
Query: 146 F-MGLFNTTTMVSSS-NQIVHVEFDSFANR-EFREARGHVGININSIISSATTPW-NASK 201
+GL N T VS+ N+ V VEFD+F N + H+GI++NS++S N S
Sbjct: 131 QNLGLTNQTVGVSTGENRFVAVEFDTFVNPFDPNATNDHIGIDVNSVVSVTNESLPNFSL 190
Query: 202 HSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEWITVGFSAATS 261
TA V YN+ ++ L+V ++ P +LS +DL + +PE +T+GFSA+
Sbjct: 191 IGNMTATV--DYNNNSRILSVKLWINGSTTPY---TLSSMVDLKRALPENVTIGFSASIG 245
Query: 262 YVQELNYLLSWEFNSTLA-----------------TSGDSKTKETRLVIILTVSCGVIVI 304
E + L SW F ST + S ++ +V TV + VI
Sbjct: 246 SAYEQHQLTSWYFKSTSSFEQKLAAEVASPPPPSSPSPPPTSRRGGVVAGATVGVVMFVI 305
Query: 305 GVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDD------LERGAGPRRFTYKELDLATN 358
+ +V + R+ K+ + ++ + H + +DD +E G GPRRF Y +L AT
Sbjct: 306 LLFTMVQVLVRRRQSKKRREAQDGSWHGSDDDDDGELIMEIEMGTGPRRFPYHKLVDATK 365
Query: 359 NFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGS-RQGKKEYVTEVKVISQLRHRNL 417
+F+ + KLGQGGFGAVY+GY +L L VA+K+ ++ S +QG+KEY +E+KVIS+LRHRNL
Sbjct: 366 SFAPEEKLGQGGFGAVYRGYLRELGLAVAIKRFAKNSSKQGRKEYKSEIKVISRLRHRNL 425
Query: 418 VKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEEWE 477
V+L+GWCH + E LLVYE PN SLD HL G T L+W +R I GL S LLYLHEEW+
Sbjct: 426 VQLIGWCHGRTELLLVYELFPNRSLDVHLHGNGTFLTWPMRINIVHGLGSALLYLHEEWD 485
Query: 478 RCVVHRDIKSSNVMLDSS 495
+CVVHRDIK SNVMLD S
Sbjct: 486 QCVVHRDIKPSNVMLDES 503
>UniRef100_Q7XIH4 Putative lectin-like protein kinase [Oryza sativa]
Length = 588
Score = 299 bits (766), Expect = 1e-79
Identities = 162/365 (44%), Positives = 240/365 (65%), Gaps = 15/365 (4%)
Query: 144 GGFMGLFNTTTMV--SSSNQIVHVEFDSFANREFREARGHVGININSIISSATTPWNASK 201
GG +G+F +T + S++ IV VEFD+F N E+ ++ H+GI++NSI S+A +
Sbjct: 7 GGLLGVFTNSTGMNPSAAAPIVAVEFDTFQN-EWDQSSDHIGIDVNSINSTAVKLLSDRS 65
Query: 202 HSGDTAEVW--IRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEWITVGFSAA 259
S T + + YN++T+ L V + + + L+ ++DL ++P + +GFSAA
Sbjct: 66 LSNVTEPMVASVSYNNSTRMLAVMLQ-MAPQDGGKRYELNSTVDLKSLLPAQVAIGFSAA 124
Query: 260 TSYVQELNYLLSWEFNSTLATSGDSKTKETRLVIILTVSCGVIV---IGVGALVAYALFW 316
+ + +E + +L+W FNSTL S + + TR V GV+V + VGA + +
Sbjct: 125 SGWSEERHQVLTWSFNSTLVASEERRENATRGRPAAAVLAGVVVASVVVVGASICLFVMI 184
Query: 317 RKRKRSNKQKEEAMHLTS-----MNDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGGF 371
R+R+ S ++ E + MND+ E+G GPRRF Y +L ATN+FS+D KLG+GGF
Sbjct: 185 RRRRISRRRTREEYEMGGSDDFDMNDEFEQGTGPRRFLYSQLATATNDFSEDGKLGEGGF 244
Query: 372 GAVYKGYFAD-LDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEF 430
G+VY+G ++ + VAVK+IS+ S+QG+KEY +EV +IS+LRHRNLV+L+GWCH +G+F
Sbjct: 245 GSVYRGVLSEPAGVHVAVKRISKTSKQGRKEYASEVSIISRLRHRNLVQLVGWCHGRGDF 304
Query: 431 LLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNV 490
LLVYE +PNGSLD+HL+G L W R++IALGL S LLYLH +E+CVVHRDIK SN+
Sbjct: 305 LLVYELVPNGSLDAHLYGGGATLPWPTRYEIALGLGSALLYLHSGYEKCVVHRDIKPSNI 364
Query: 491 MLDSS 495
MLDS+
Sbjct: 365 MLDSA 369
>UniRef100_Q6YX21 Putative receptor kinase Lecrk [Oryza sativa]
Length = 750
Score = 297 bits (761), Expect = 4e-79
Identities = 197/500 (39%), Positives = 277/500 (55%), Gaps = 48/500 (9%)
Query: 41 ASSIHFKYPSFDPSD-ANIVYQGSAA---------PRDGEVNFNINENYSCQV--GRVFY 88
A ++ F + +F P + NI +G AA G V+ + N + S + GRV Y
Sbjct: 39 ALALTFNHTNFGPDEQTNIRLEGDAAFSADFSFSGDGGGWVDISANRHGSIEDSRGRVSY 98
Query: 89 SEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSL--YGHGLAFFLVPYGFEIPLNSDGGF 146
+ V LWD+ TG++ FTT ++FVIN + + G G+AFFL + +P S
Sbjct: 99 ALPVPLWDAATGEVASFTTGFSFVINPPKQDGGIDNKGAGMAFFLAGFPSRLP-GSYPYN 157
Query: 147 MGLFNTTT--MVSSSNQIVHVEFDSFANREFREARG---HVGININSIISSATTPWNASK 201
+GL N T + + ++ V VEFD+F + + H+G+++NS++S T +
Sbjct: 158 LGLTNQTADQVAAGDDRFVAVEFDTFNDTIVHDPDATYDHLGVDVNSVVSKTTLTLPSFT 217
Query: 202 HSGDTAEVWIRYNSTTKNLTVSWKY-QTTSNPQENT--SLSISIDLMKVMPEWITVGFSA 258
G+ V + Y++ + L + S P+ +LS +DL V+PE + VGFSA
Sbjct: 218 LVGNMTAV-VEYDNVSSILAMRLHLGYGLSGPRHRPDYNLSYKVDLKSVLPEQVAVGFSA 276
Query: 259 ATSYVQELNYLLSWEFNSTLATSG--------------DSKTKETRLVIILTVSCGVIVI 304
ATS EL+ L SW F+S+L S + +V TV + V+
Sbjct: 277 ATSTSVELHQLRSWYFSSSLEPKAAPPPVAPPSPSPPPTSGSGSGGVVAGATVGAALFVV 336
Query: 305 GVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDD--------LERGAGPRRFTYKELDLA 356
+ A+VA + R+R + K +E A +DD +E G GPRRF Y L A
Sbjct: 337 LLLAMVAVVVLVRRRHQRKKMRE-AEEANDDDDDTEGDPIMEIENGTGPRRFAYHVLVNA 395
Query: 357 TNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKK-ISRGSRQGKKEYVTEVKVISQLRHR 415
T +F+ + KLGQGGFGAVY+GY + L VA+K+ I S QG++EY +E+KVIS+LRHR
Sbjct: 396 TKSFAAEEKLGQGGFGAVYRGYLREQGLAVAIKRFIKDSSNQGRREYKSEIKVISRLRHR 455
Query: 416 NLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEE 475
NLV+L+GWCH E LLVYE +PN SLD HL G T L+W +R KI LGL S L YLHEE
Sbjct: 456 NLVQLIGWCHGHDELLLVYELVPNRSLDIHLHGNGTFLTWPMRVKIILGLGSALFYLHEE 515
Query: 476 WERCVVHRDIKSSNVMLDSS 495
WE+CVVHRDIK SNVMLD S
Sbjct: 516 WEQCVVHRDIKPSNVMLDES 535
>UniRef100_Q6ZFA9 Putative receptor-type protein kinase LRK1 [Oryza sativa]
Length = 541
Score = 286 bits (731), Expect = 1e-75
Identities = 167/380 (43%), Positives = 232/380 (60%), Gaps = 19/380 (5%)
Query: 127 LAFFLVPYGFEIPLNSDGGFMGLFNT--TTMVSSSNQIVHVEFDSFANREF--REARGHV 182
+AFFL Y +P N+ +GL N T + + ++ V VEFD++ NR+F H+
Sbjct: 1 MAFFLGSYPSRLPKNASSSGLGLTNKSYTNVSTGEDRFVAVEFDTYLNRDFDPNATYDHI 60
Query: 183 GININSIISSATTPWNASKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISI 242
GI++NSI+S +G + YNS++ L+V T+ P N LS +
Sbjct: 61 GIDVNSIVSVTNESLPDFSLNGSMTAT-VDYNSSSSILSVKLWINDTTKPPYN--LSDKV 117
Query: 243 DLMKVMPEWITVGFSAATSYVQELNYLLSWEFNSTLATSGDSKTKETRLVIILTVSCGVI 302
DL +PE +T+GFSAAT EL+ L SW FNS+ S + K + TV +
Sbjct: 118 DLKSALPEKVTIGFSAATGASVELHQLTSWYFNSS--PSFEHKHGRAGVEAGATVGATLF 175
Query: 303 VIGVGALVAYALFWRKRKRSNKQKEEAMHLTS--MNDD------LERGAGPRRFTYKELD 354
V+ + VA L R+R ++ K+ E+ ++S ++DD +E G GPRRF Y EL
Sbjct: 176 VVLLFT-VAAILIRRRRIKNRKEAEDEQDISSDSLDDDGEPIVEIEMGTGPRRFPYYELV 234
Query: 355 LATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISR-GSRQGKKEYVTEVKVISQLR 413
AT +F+ + KLGQGGFGAVY+GY + L VA+K+ ++ S+QG+KEY +E+KVIS+LR
Sbjct: 235 EATKSFAAEEKLGQGGFGAVYRGYLREQGLAVAIKRFAKDSSKQGRKEYKSEIKVISRLR 294
Query: 414 HRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLH 473
HRNLV+L+GWCH + E LL+YE +PN SLD HL G T L+W +R KI LG+ S L YLH
Sbjct: 295 HRNLVQLIGWCHGRDELLLIYELVPNRSLDIHLHGNGTFLTWPMRVKIVLGIGSALFYLH 354
Query: 474 EEWERCVVHRDIKSSNVMLD 493
EEW +CVVHRDIK SNVMLD
Sbjct: 355 EEWGQCVVHRDIKPSNVMLD 374
>UniRef100_Q7X756 OSJNBb0096E05.1 protein [Oryza sativa]
Length = 746
Score = 274 bits (700), Expect = 5e-72
Identities = 176/495 (35%), Positives = 267/495 (53%), Gaps = 30/495 (6%)
Query: 27 LTLFYMSLIFFNIPASSIHFKYPSFDPSDANIVYQGSAAPRDGEVNFNINENYSCQV-GR 85
L L + L+ +PA+S+ F Y + + + A G + + E ++ + GR
Sbjct: 25 LLLLLLHLLLVAVPATSLTFSYDADSFVSEDFRQEDDAMVTAGRIEL-LGEEFAARARGR 83
Query: 86 VFYSEKVLLWDSNTGKLTDFTTHYTFVINT-QGRSPSLYGHGLAFFLVPYGFEIPLNSDG 144
Y V LWD TG+ F + F I + GR +L GHG+ FFL P+ ++P
Sbjct: 84 ALYKRPVQLWDGATGEEASFAASFNFTIRSVAGRGNALAGHGMTFFLAPFMPDMPQECYE 143
Query: 145 GFMGLFN----------TTTMVSSSNQIVHVEFDSFANREFREARGHVGININSIISS-- 192
G +GLF+ T S + V VEFD+ + R HVG++IN++ S
Sbjct: 144 GCLGLFDQSLTRNTASATMGNASGAASFVAVEFDTHMDGWDPSGR-HVGVDINNVDSRRG 202
Query: 193 --ATTPWNASKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPE 250
P ++ +G + Y+S + L V+ + +LS ++ L V+PE
Sbjct: 203 NYVVLPEDSLVDAG-VMSATVSYDSGARRLDVALAIGGGA-ATATYNLSAAVHLRSVLPE 260
Query: 251 WITVGFSAATSYVQELNY-LLSWEFNSTLAT-SGDSKTKETRLVIILTVSCGVIVIGVGA 308
+ VGFSAAT N+ +LS+ F+STL T + + T +S V G+
Sbjct: 261 QVAVGFSAATGDQFASNHTVLSFTFSSTLPTRTTNPPPPSTSSAKTAHLSAAVAAAGIAL 320
Query: 309 LV---AYALFWRKRKRSNKQKEEAMHLTSMNDD----LERGAGPRRFTYKELDLATNNFS 361
L+ A + R+ ++ ++ + + S++DD +E G GPRR Y +L AT F+
Sbjct: 321 LLLVLAITILIRRARKRRRRDDGVSYDDSLDDDDEEDMESGTGPRRIPYAQLAAATGGFA 380
Query: 362 KDRKLGQGGFGAVYKGYFADLDLQVAVKKISRG-SRQGKKEYVTEVKVISQLRHRNLVKL 420
+ KLG+GG G+VY G+ +L VA+K +RG S +G+KEY +EV VIS+LRHRNLV+L
Sbjct: 381 EIGKLGEGGSGSVYGGHVRELGRDVAIKVFTRGASMEGRKEYRSEVTVISRLRHRNLVQL 440
Query: 421 LGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEEWERCV 480
+GWCH + LLVYE + NGSLD HL+ + L+W +R++I GLAS +LYLH+EW++CV
Sbjct: 441 MGWCHGRRRLLLVYELVRNGSLDGHLYSNKETLTWPLRYQIINGLASAVLYLHQEWDQCV 500
Query: 481 VHRDIKSSNVMLDSS 495
VH DIK SN+MLD S
Sbjct: 501 VHGDIKPSNIMLDES 515
>UniRef100_Q7XL08 OSJNBa0065J03.1 protein [Oryza sativa]
Length = 746
Score = 271 bits (694), Expect = 2e-71
Identities = 175/495 (35%), Positives = 266/495 (53%), Gaps = 30/495 (6%)
Query: 27 LTLFYMSLIFFNIPASSIHFKYPSFDPSDANIVYQGSAAPRDGEVNFNINENYSCQV-GR 85
L L + L+ +PA+S+ F Y + + + A G + + E ++ + GR
Sbjct: 25 LLLLLLHLLLVAVPATSLTFSYDADSFVSEDFRQEDDAMVTAGRIEL-LGEEFAARARGR 83
Query: 86 VFYSEKVLLWDSNTGKLTDFTTHYTFVINT-QGRSPSLYGHGLAFFLVPYGFEIPLNSDG 144
Y V LWD TG+ F + F I + GR +L GHG+ FFL P+ ++P
Sbjct: 84 ALYKRPVQLWDGATGEEASFAASFNFTIRSVAGRGNALAGHGMTFFLAPFMPDMPQECYE 143
Query: 145 GFMGLFN----------TTTMVSSSNQIVHVEFDSFANREFREARGHVGININSIISS-- 192
G +GLF+ T S + V VEFD+ + R HVG++IN++ S
Sbjct: 144 GCLGLFDQSLTRNTASATMGNASGAASFVAVEFDTHMDGWDPSGR-HVGVDINNVDSRRG 202
Query: 193 --ATTPWNASKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPE 250
P ++ +G + Y+S + L V+ + +LS ++ L V+PE
Sbjct: 203 NYVVLPEDSLVDAG-VMSATVSYDSGARRLDVALAIGGGA-ATATYNLSAAVHLRSVLPE 260
Query: 251 WITVGFSAATSYVQELNY-LLSWEFNSTLATSGDSKTK-ETRLVIILTVSCGVIVIGVGA 308
+ VGFSAAT N+ +LS+ F+STL T + + T +S V G+
Sbjct: 261 QVAVGFSAATGDQFASNHTVLSFTFSSTLPTRTTNPPQPSTSSAKTAHLSAAVAAAGIAL 320
Query: 309 ---LVAYALFWRKRKRSNKQKEEAMHLTSMNDD----LERGAGPRRFTYKELDLATNNFS 361
++A + R+ ++ ++ + + S++DD +E G GPRR Y L AT F+
Sbjct: 321 QLLVLAITILIRRARKRRRRDDGVSYDDSIDDDDEEDMESGTGPRRIPYAHLAAATGGFA 380
Query: 362 KDRKLGQGGFGAVYKGYFADLDLQVAVKKISRG-SRQGKKEYVTEVKVISQLRHRNLVKL 420
+ KLG+GG G+VY G+ +L VA+K +RG S +G+KEY +EV VIS+LRHRNLV+L
Sbjct: 381 EIGKLGEGGSGSVYGGHVRELGRNVAIKVFTRGASMEGRKEYRSEVTVISRLRHRNLVQL 440
Query: 421 LGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEEWERCV 480
+GWCH + LLVYE + NGSLD HL+ + L+W +R++I GLAS +LYLH+EW++CV
Sbjct: 441 MGWCHGRRRLLLVYELVRNGSLDGHLYSNKETLTWPLRYQIINGLASAVLYLHQEWDQCV 500
Query: 481 VHRDIKSSNVMLDSS 495
VH DIK SN+MLD S
Sbjct: 501 VHGDIKPSNIMLDES 515
>UniRef100_Q6UY59 Lectin-like receptor kinase 7;2 [Medicago truncatula]
Length = 669
Score = 268 bits (685), Expect = 3e-70
Identities = 164/460 (35%), Positives = 249/460 (53%), Gaps = 38/460 (8%)
Query: 60 YQGSAAPRDGEVNFNIN------ENYSCQVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVI 113
+Q S DG N N + + FY ++ +++ G ++ F+T + F I
Sbjct: 28 FQSSHLYLDGIANLTSNGLLRLTNDTKQEKAHAFYPNPIVFKNTSNGSVSSFSTTFVFAI 87
Query: 114 NTQGRSPSLYGHGLAFFLVPYGFEIPLNSDGGFMGLFNTTTMVSSSNQIVHVEFDSFANR 173
Q P+L GHG+ F + P +P + ++GLFN + +SSN + VE D+ +
Sbjct: 88 RPQ--YPTLSGHGIVFVVSPTK-GLPNSLQSQYLGLFNKSNNGNSSNHVFGVELDTIISS 144
Query: 174 EFREAR-GHVGININSIISSATTPW----------NASKHSGDTAEVWIRYNSTTKNLTV 222
EF + HVGI+IN + S+ +TP N + SG+ +VWI Y+ K + V
Sbjct: 145 EFNDINDNHVGIDINDLKSAKSTPAGYYDVNGQLKNLTLFSGNPMQVWIEYDGEKKKIDV 204
Query: 223 SWKYQTTSNPQENTSLSISIDLMKVMPEWITVGFSAATSYVQELNYLLSWEFN-----ST 277
+ P++ LS++ DL ++ + VGFS+AT V +Y+L W F
Sbjct: 205 TLAPINVVKPKQPL-LSLTRDLSPILNNSMYVGFSSATGSVFTSHYILGWSFKVNGQAEN 263
Query: 278 LATSGDSKTK---ETRLVIILTVSCGVIVIGVGALVAYALFWRKRKRSNKQKEEAMHLTS 334
L S K E + + LTV ++++ + ++ + + ++R
Sbjct: 264 LVISELPKLPRFGEKKESMFLTVGLPLVLLSLVFMITLGVIYYIKRRKK--------FAE 315
Query: 335 MNDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRG 394
+ +D E GP RF +K+L AT F + LG GGFG VYKG L+VAVK++S
Sbjct: 316 LLEDWEHEYGPHRFKFKDLYFATKGFKEKGLLGVGGFGRVYKGVMPGSKLEVAVKRVSHE 375
Query: 395 SRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGK-RTPL 453
SRQG +E+V+E+ I +LRHRNLV LLG+C KGE LLVY++MPNGSLD++L+ + R L
Sbjct: 376 SRQGMREFVSEIVSIGRLRHRNLVPLLGYCRRKGELLLVYDYMPNGSLDNYLYNQPRVTL 435
Query: 454 SWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLD 493
+W R +I G+A GL YLHEEWE+ V+HRDIK+SNV+LD
Sbjct: 436 NWSQRFRIIKGVALGLFYLHEEWEQVVIHRDIKASNVLLD 475
>UniRef100_Q9M2S4 Probable serine/threonine-specific protein kinase [Arabidopsis
thaliana]
Length = 684
Score = 264 bits (675), Expect = 4e-69
Identities = 186/492 (37%), Positives = 258/492 (51%), Gaps = 52/492 (10%)
Query: 34 LIFFNIPASSI--HFKYPSFDPSDANIVYQGSA--APRDGEVNFNINENYSCQVGRVFYS 89
LIF SS+ F + F + N+ G A AP + +G FYS
Sbjct: 12 LIFLTHLVSSLIQDFSFIGFKKASPNLTLNGVAEIAPTGA---IRLTTETQRVIGHAFYS 68
Query: 90 EKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGFEIPLNSDGGFMGL 149
+ + F+T +F I +L GHGLAF + P ++ + ++GL
Sbjct: 69 LPIRFKPIGVNRALSFST--SFAIAMVPEFVTLGGHGLAFAITPTP-DLRGSLPSQYLGL 125
Query: 150 FNTTTMVSSSNQIVHVEFDSFANREFREAR-GHVGININSIISSATTPW-----NASKHS 203
N++ V+ S+ VEFD+ + EF + HVGI+INS+ SS +TP N++K
Sbjct: 126 LNSSR-VNFSSHFFAVEFDTVRDLEFEDINDNHVGIDINSMESSISTPAGYFLANSTKKE 184
Query: 204 -----GDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEWITVGFSA 258
G + WI Y+S K L V K S + + LS +DL V+ + + VGFSA
Sbjct: 185 LFLDGGRVIQAWIDYDSNKKRLDV--KLSPFSEKPKLSLLSYDVDLSSVLGDEMYVGFSA 242
Query: 259 ATSYVQELNYLLSWEFNST------------LATSGDSKTKETRLVIILTVS--CGVIVI 304
+T + +Y+L W FN + S K K+ R +IL VS C +++
Sbjct: 243 STGLLASSHYILGWNFNMSGEAFSLSLPSLPRIPSSIKKRKKKRQSLILGVSLLCSLLIF 302
Query: 305 GVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNNFSKDR 364
V LVA +LF ++ + + EE E GP RF+Y+EL ATN F
Sbjct: 303 AV--LVAASLFVVRKVKDEDRVEE----------WELDFGPHRFSYRELKKATNGFGDKE 350
Query: 365 KLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWC 424
LG GGFG VYKG D VAVK+IS SRQG +E+++EV I LRHRNLV+LLGWC
Sbjct: 351 LLGSGGFGKVYKGKLPGSDEFVAVKRISHESRQGVREFMSEVSSIGHLRHRNLVQLLGWC 410
Query: 425 HDKGEFLLVYEFMPNGSLDSHLFGKRTP--LSWGVRHKIALGLASGLLYLHEEWERCVVH 482
+ + LLVY+FMPNGSLD +LF + L+W R KI G+ASGLLYLHE WE+ V+H
Sbjct: 411 RRRDDLLLVYDFMPNGSLDMYLFDENPEVILTWKQRFKIIKGVASGLLYLHEGWEQTVIH 470
Query: 483 RDIKSSNVMLDS 494
RDIK++NV+LDS
Sbjct: 471 RDIKAANVLLDS 482
>UniRef100_Q6UY58 Lectin-like receptor kinase 7;3 [Medicago truncatula]
Length = 682
Score = 264 bits (674), Expect = 5e-69
Identities = 180/485 (37%), Positives = 259/485 (53%), Gaps = 45/485 (9%)
Query: 34 LIFFNIPASSI--HFKYPSF-DPSDANIVYQGSAAPRDGEVNFNINENYSCQVGRVFYSE 90
L+FF +P S Y F D N+ G A + NE S +G FY +
Sbjct: 9 LLFFFVPVLSQVDQLLYTGFKDVGPKNLTLNGIAEIEKNGIIRLTNET-SRLLGHAFYPQ 67
Query: 91 KVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGFEIPLNSDGGFMGLF 150
+ + TGK+ F++ +F + P L GHG+AF +VP L S ++GL
Sbjct: 68 PFQIKNKTTGKVFSFSS--SFALACVPEYPKLGGHGMAFTIVPSKDLKALPSQ--YLGLL 123
Query: 151 NTTTMVSSSNQIVHVEFDSFANREFREAR-GHVGININSIISSATTP----------WNA 199
N++ + + SN + VEFD+ + EF + HVGI+INS+ S+AT N
Sbjct: 124 NSSDVGNFSNHLFAVEFDTVQDFEFGDINYNHVGIDINSMRSNATITAGYYSDDDMVHNI 183
Query: 200 SKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEWITVGFSAA 259
S G VW+ Y+S+ + ++V+ TSN + L+ +DL + + + VGFSA+
Sbjct: 184 SIKGGKPILVWVDYDSSLELISVT--LSPTSNKPKKPILTFHMDLSPLFLDTMYVGFSAS 241
Query: 260 TSYVQELNYLLSWEFN----------STLATSGDSKTKETRLVIILTVSCGVIVIGVGAL 309
T + +Y+L W F S L K K T L++ L++ +IV+
Sbjct: 242 TGLLASSHYVLGWSFKINGPAPFLDLSKLPKLPHPKKKHTSLILGLSLGSALIVL---CS 298
Query: 310 VAYALFWRKRKRSNKQKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNNFSKDRKLGQG 369
+A+ F+ RK N EA L GP R+TY+EL AT F + + LGQG
Sbjct: 299 MAFG-FYIYRKIKNADVIEAWELE---------VGPHRYTYQELKKATKGFKEKQLLGQG 348
Query: 370 GFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGE 429
GFG VY G +QVAVK++S S+QG +E+V+E+ I +LRHRNLV LLGWC +G+
Sbjct: 349 GFGKVYNGILPKSKIQVAVKRVSHESKQGLREFVSEIASIGRLRHRNLVMLLGWCRRRGD 408
Query: 430 FLLVYEFMPNGSLDSHLF-GKRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSS 488
LLVY++M NGSLD +LF LSW R KI G+ASGLLYLHE +E+ V+HRD+K+S
Sbjct: 409 LLLVYDYMANGSLDKYLFEDSEYVLSWEQRFKIIKGVASGLLYLHEGYEQVVIHRDVKAS 468
Query: 489 NVMLD 493
NV+LD
Sbjct: 469 NVLLD 473
>UniRef100_Q9SZD5 Serine/threonine-specific kinase like protein [Arabidopsis
thaliana]
Length = 669
Score = 260 bits (665), Expect = 6e-68
Identities = 159/436 (36%), Positives = 236/436 (53%), Gaps = 39/436 (8%)
Query: 84 GRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGFEIPLNSD 143
G VFY+ V +S G ++ F+T + F I + + L GHGLAF + P +P +S
Sbjct: 60 GHVFYNSPVRFKNSPNGTVSSFSTTFVFAIVSNVNA--LDGHGLAFVISPTK-GLPYSSS 116
Query: 144 GGFMGLFNTTTMVSSSNQIVHVEFDSFANREFREA-RGHVGININSIISSATTPWNASKH 202
++GLFN T SN IV VEFD+F N+EF + HVGI+INS+ S + +
Sbjct: 117 SQYLGLFNLTNNGDPSNHIVAVEFDTFQNQEFDDMDNNHVGIDINSLSSEKASTAGYYED 176
Query: 203 SGDT-----------AEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEW 251
T + WI Y+S+ + L V+ P+ LS++ DL + +
Sbjct: 177 DDGTFKNIRLINQKPIQAWIEYDSSRRQLNVTIHPIHLPKPKIPL-LSLTKDLSPYLFDS 235
Query: 252 ITVGFSAATSYVQELNYLLSWEFNSTLATSG-----------DSKTKETRLVIILTVSCG 300
+ VGF++AT ++ +Y+L W F S DS++ + ++ +++S
Sbjct: 236 MYVGFTSATGRLRSSHYILGWTFKLNGTASNIDISRLPKLPRDSRSTSVKKILAISLSLT 295
Query: 301 VIVIGVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNNF 360
+ I V ++Y LF +++K L + +D E GP RF YK+L +AT F
Sbjct: 296 SLAILVFLTISYMLFLKRKK-----------LMEVLEDWEVQFGPHRFAYKDLYIATKGF 344
Query: 361 SKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKL 420
LG+GGFG VYKG + ++ +AVKK+S SRQG +E+V E+ I +LRH NLV+L
Sbjct: 345 RNSELLGKGGFGKVYKGTLSTSNMDIAVKKVSHDSRQGMREFVAEIATIGRLRHPNLVRL 404
Query: 421 LGWCHDKGEFLLVYEFMPNGSLDSHLFGK-RTPLSWGVRHKIALGLASGLLYLHEEWERC 479
LG+C KGE LVY+ MP GSLD L+ + L W R KI +ASGL YLH +W +
Sbjct: 405 LGYCRRKGELYLVYDCMPKGSLDKFLYHQPEQSLDWSQRFKIIKDVASGLCYLHHQWVQV 464
Query: 480 VVHRDIKSSNVMLDSS 495
++HRDIK +NV+LD S
Sbjct: 465 IIHRDIKPANVLLDDS 480
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 835,432,600
Number of Sequences: 2790947
Number of extensions: 36797327
Number of successful extensions: 123108
Number of sequences better than 10.0: 14533
Number of HSP's better than 10.0 without gapping: 4466
Number of HSP's successfully gapped in prelim test: 10067
Number of HSP's that attempted gapping in prelim test: 101332
Number of HSP's gapped (non-prelim): 15751
length of query: 495
length of database: 848,049,833
effective HSP length: 132
effective length of query: 363
effective length of database: 479,644,829
effective search space: 174111072927
effective search space used: 174111072927
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146709.1


