

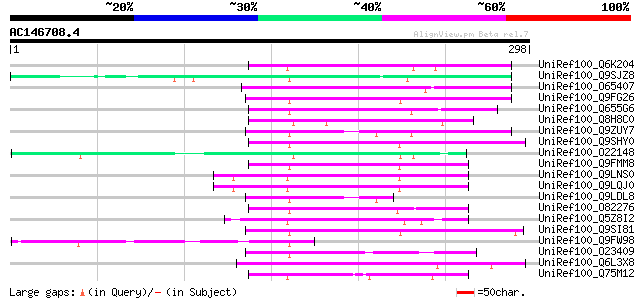
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146708.4 + phase: 0
(298 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6K204 Hypothetical protein B1469H02.35 [Oryza sativa] 70 9e-11
UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcrip... 67 4e-10
UniRef100_O65407 Hypothetical protein F18E5.40 [Arabidopsis thal... 65 2e-09
UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like... 64 6e-09
UniRef100_Q655G6 Hypothetical protein P0009H10.11 [Oryza sativa] 63 1e-08
UniRef100_Q8H8C0 Hypothetical protein OJ1134F05.9 [Oryza sativa] 62 2e-08
UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis tha... 60 9e-08
UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana] 59 2e-07
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 58 3e-07
UniRef100_Q9FMM8 Non-LTR retroelement reverse transcriptase-like... 57 8e-07
UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana] 56 1e-06
UniRef100_Q9LQJ0 F28G4.15 protein [Arabidopsis thaliana] 56 1e-06
UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis tha... 54 5e-06
UniRef100_O82276 Putative non-LTR retroelement reverse transcrip... 54 7e-06
UniRef100_Q5Z8I2 Hypothetical protein P0622F03.7 [Oryza sativa] 53 9e-06
UniRef100_Q9SI81 F23N19.5 [Arabidopsis thaliana] 53 1e-05
UniRef100_Q9FW98 Putative non-LTR retroelement reverse transcrip... 50 9e-05
UniRef100_O23409 SIMILARITY to probable splicing factor CEPRP21 ... 47 5e-04
UniRef100_Q6L3X8 Hypothetical protein [Solanum demissum] 47 5e-04
UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa] 47 6e-04
>UniRef100_Q6K204 Hypothetical protein B1469H02.35 [Oryza sativa]
Length = 212
Score = 69.7 bits (169), Expect = 9e-11
Identities = 45/158 (28%), Positives = 75/158 (46%), Gaps = 7/158 (4%)
Query: 138 WKPPSAPWVKVNTDGSVLNNS--GACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGFIL 195
W+ P A W+K+N DGS + + + GG++RDH G F+ + +GR + E++
Sbjct: 51 WRKPEAGWIKLNFDGSSKHATKIASIGGVYRDHEGAFVLGYAERIGRATSSVAELAALRR 110
Query: 196 VMEHAALHGWYNIWLESDASSALMVFKNPSLVPIL--LRNRWHNARTLNV--QVISSHIF 251
+E +GW +W E D+ + + V + + V LR A L + + SH++
Sbjct: 111 GLELVVRNGWRRVWAEGDSKTVVDVVCDRANVRSEEDLRQCREIAALLPLIDDMAVSHVY 170
Query: 252 REGNVCVDRLANLGHSVV-GEVWLSTLPSDFHQVFYED 288
R GN A LGH V VW + P + + +D
Sbjct: 171 RSGNKVAHGFARLGHKAVRPRVWRAAPPEEVLRFLQQD 208
>UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 321
Score = 67.4 bits (163), Expect = 4e-10
Identities = 80/305 (26%), Positives = 121/305 (39%), Gaps = 46/305 (15%)
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDFSSIKALMSSLPRHCSSQVRDL 60
+C C ET H+ C + +WS L +PR Q+R
Sbjct: 34 VCQICQGGEETILHVLRDCPAMAGIWSRL-------------------VPR---DQIRQF 71
Query: 61 YVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQV------RVHAFIGLSGA----VSTGKC 110
+ A+++ W N+R + + + V R G +G V K
Sbjct: 72 FTASLLE------WIYKNLRERGSWPTVFVMAVWWGWKWRCGNIFGGNGKCRDRVKFIKD 125
Query: 111 IAADAAILDAFRIPPHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDH 168
+A + AI +AF R R V W P WVK+NTDG+ N G GG+ RDH
Sbjct: 126 LAEEVAIANAFVKGNEVRVSRVERLVSWVSPEDGWVKLNTDGASRGNPGFATAGGVLRDH 185
Query: 169 LGTFLGAFVGNLGRCSVFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKNPSLV- 227
G ++G F N+G CS E+ G + A G + LE D S ++ F +
Sbjct: 186 NGAWIGGFAVNIGVCSAPLAELWGVYYGLFIAWGRGARRVELEVD-SKMVVGFLTTGIAD 244
Query: 228 --PI-LLRNRWHNARTLNVQVISSHIFREGNVCVDRLANLGHSV-VGEVWLSTLPSDFHQ 283
P+ L ++ + V SH++RE N D LAN S+ +G L + P
Sbjct: 245 SHPLSFLLRLCYDFLSKGWIVRISHVYREANRLADGLANYAFSLSLGLHLLESRPDVVSS 304
Query: 284 VFYED 288
+ +D
Sbjct: 305 ILLDD 309
>UniRef100_O65407 Hypothetical protein F18E5.40 [Arabidopsis thaliana]
Length = 229
Score = 65.1 bits (157), Expect = 2e-09
Identities = 47/162 (29%), Positives = 73/162 (45%), Gaps = 8/162 (4%)
Query: 134 VSVCWKPPSAPWVKVNTDGSV-LNNSGACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSG 192
V V WK P +K+N DGS + GG+FR+H FL + ++G + E +
Sbjct: 59 VKVVWKKPENGRIKLNFDGSRGREGQASIGGVFRNHKAEFLLGYSESIGEATSTMAEFAA 118
Query: 193 FILVMEHAALHGWYNIWLESDASSALMVFKNPSLVPILLRNRWHN-----ARTLNVQVIS 247
+E A +G ++WLE DA + + + N+ N LN +
Sbjct: 119 LKRGLELALENGLTDLWLEGDAKIIMDIISRRGRLRCEKTNKHVNYIKVVMPELN-NCVL 177
Query: 248 SHIFREGNVCVDRLANLGHSVVG-EVWLSTLPSDFHQVFYED 288
SH++REGN D+LA LGH +VW P + ++D
Sbjct: 178 SHVYREGNRVADKLAKLGHQFQDPKVWRVQPPEIVLPIMHDD 219
>UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like [Arabidopsis
thaliana]
Length = 676
Score = 63.5 bits (153), Expect = 6e-09
Identities = 46/159 (28%), Positives = 73/159 (44%), Gaps = 6/159 (3%)
Query: 136 VCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGF 193
+ W+ P+ WV +NTDG+ N G GG+ RD G++L F N+G CS E+ G
Sbjct: 506 IAWRKPAEGWVTMNTDGASHGNPGQATAGGVIRDEHGSWLVGFALNIGVCSAPLAELWGV 565
Query: 194 ILVMEHAALHGWYNIWLESDASSALMVFKN---PSLVPILLRNRWHNARTLNVQVISSHI 250
+ A GW + LE D++ + ++ S L H + + V +H+
Sbjct: 566 YYGLVVAWERGWRRVRLEVDSALVVGFLQSGIGDSHPLAFLVRLCHGFISKDWIVRITHV 625
Query: 251 FREGNVCVDRLANLGHSV-VGEVWLSTLPSDFHQVFYED 288
+RE N D LAN ++ G + L + P + ED
Sbjct: 626 YREANRLADGLANYAFTLPFGFLLLDSCPEHVSSILLED 664
>UniRef100_Q655G6 Hypothetical protein P0009H10.11 [Oryza sativa]
Length = 240
Score = 62.8 bits (151), Expect = 1e-08
Identities = 43/151 (28%), Positives = 70/151 (45%), Gaps = 9/151 (5%)
Query: 138 WKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGFIL 195
W+ P W+K+N DGS +++G + GG++RDH G FL + +G + E++
Sbjct: 79 WRKPEEGWMKLNFDGSSKHSTGIASIGGVYRDHDGAFLLGYAERIGTATSSVAELAALRR 138
Query: 196 VMEHAALHGWYNIWLESDASSALMVFKNPSLVPI-----LLRNRWHNARTLNVQVISSHI 250
+E A +GW +W E D+ + + V + + V L R L+ + SH+
Sbjct: 139 GLELAVRNGWRRVWAEGDSKAVVDVVCDRADVQSEEDLRLCREIAALLPQLDDMAV-SHV 197
Query: 251 FREGNVCVDRLANLGHSVV-GEVWLSTLPSD 280
R GN A LGH VW + P +
Sbjct: 198 RRGGNKVAHGFAELGHRAARPRVWRAAPPDE 228
>UniRef100_Q8H8C0 Hypothetical protein OJ1134F05.9 [Oryza sativa]
Length = 210
Score = 62.0 bits (149), Expect = 2e-08
Identities = 41/137 (29%), Positives = 62/137 (44%), Gaps = 8/137 (5%)
Query: 138 WKPPSAPWVKVNTDGSVLNNSGAC--GGLFRDHLGTFLGAFVGNL--GRCSVFDTEVSGF 193
W+ P W+K+N DGS + +GA GG FRDH G F+ + + G S F E++
Sbjct: 47 WRKPEVGWMKLNFDGSRNDATGAASIGGAFRDHEGAFVAGYAERMIVGGASSFTAELAAL 106
Query: 194 ILVMEHAALHGWYNIWLESDASSALMVFKNPSLVPILLRNRWHNARTLNVQVIS----SH 249
+E AA +GW +W E D+ + + V + V R + + SH
Sbjct: 107 RRGLELAARYGWRRVWAEGDSRAVVDVVHGRAGVRSEEDRRQCGEIAALLPALDGVAVSH 166
Query: 250 IFREGNVCVDRLANLGH 266
+ R+ N A LGH
Sbjct: 167 LCRDENKVAHGFAKLGH 183
>UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis thaliana]
Length = 314
Score = 59.7 bits (143), Expect = 9e-08
Identities = 49/167 (29%), Positives = 74/167 (43%), Gaps = 22/167 (13%)
Query: 136 VCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGF 193
+ W P W K+NTDG+ N G + GG+ RD G + G F N+G CS E+ G
Sbjct: 144 IAWSKPEEGWWKLNTDGASRGNPGLASAGGVLRDEEGAWRGGFALNIGVCSAPLAELWG- 202
Query: 194 ILVMEHAALHGWYNIW------LESDASSALMV-FKNPSLVPI----LLRNRWHNARTLN 242
+G Y W LE + S ++V F + + L H+ + +
Sbjct: 203 -------VYYGLYIAWERRVTRLEIEVDSEIVVGFLKIGINEVHPLSFLVRLCHDFISRD 255
Query: 243 VQVISSHIFREGNVCVDRLANLGHSV-VGEVWLSTLPSDFHQVFYED 288
+V SH++RE N D LAN S+ +G LS +P + +D
Sbjct: 256 WRVRISHVYREANRLADGLANYAFSLPLGFHSLSLVPDSLRFILLDD 302
>UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana]
Length = 1055
Score = 58.5 bits (140), Expect = 2e-07
Identities = 50/165 (30%), Positives = 68/165 (40%), Gaps = 6/165 (3%)
Query: 138 WKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGFIL 195
W P WVKVNTDG+ N G + GG+ RD G + G F N+GRCS E+ G
Sbjct: 561 WVSPCVGWVKVNTDGASRGNPGLASAGGVLRDCTGAWCGGFSLNIGRCSAPQAELWGVYY 620
Query: 196 VMEHAALHGWYNIWLESDASSALMVFK---NPSLVPILLRNRWHNARTLNVQVISSHIFR 252
+ A + LE D+ + K + S L H + V H++R
Sbjct: 621 GLYFAWEKKVPRVELEVDSEVIVGFLKTGISDSHPLSFLVRLCHGFLQKDWLVRIVHVYR 680
Query: 253 EGNVCVDRLANLGHSV-VGEVWLSTLPSDFHQVFYEDRCSMPRIR 296
E N D LAN S+ +G +P + ED R R
Sbjct: 681 EANRLADGLANYAFSLSLGFHSFDLVPDAMSSLLREDTLGSTRPR 725
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 58.2 bits (139), Expect = 3e-07
Identities = 67/273 (24%), Positives = 108/273 (39%), Gaps = 32/273 (11%)
Query: 2 CSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDF--SSIKALMSSLPRHCSSQVRD 59
C C ET +HL +C F W+ G ++ S + + L H S
Sbjct: 1092 CVRCPSHGETVNHLLFKCPFARLTWAISPLPAPPGGEWAESLFRNMHHVLSVHKSQPEES 1151
Query: 60 LYVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCIAADAAILD 119
+ A + ++ +W RN++ F + ++ V ++ D +
Sbjct: 1152 DHHALIPWILWRLWKNRNDLVFKGREFTAPQVILKA----------------TEDMDAWN 1195
Query: 120 AFRIP-PHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNSGAC--GGLFRDHLGTFLGAF 176
+ P P S V W+PPS WVK NTDG+ + G C G + R+H G L
Sbjct: 1196 NRKEPQPQVTSSTRDRCVKWQPPSHGWVKCNTDGAWSKDLGNCGVGWVLRNHTGRLLWLG 1255
Query: 177 VGNL-GRCSVFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKN----PSLVPIL- 230
+ L + SV +TEV + + + + ESD+ + + +N PSL P +
Sbjct: 1256 LRALPSQQSVLETEVEALRWAVLSLSRFNYRRVIFESDSQYLVSLIQNEMDIPSLAPRIQ 1315
Query: 231 -LRNRWHNARTLNVQVISSHIFREGNVCVDRLA 262
+RN + + Q REGN DR A
Sbjct: 1316 DIRNLLRHFEEVKFQFTR----REGNNVADRTA 1344
>UniRef100_Q9FMM8 Non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 308
Score = 56.6 bits (135), Expect = 8e-07
Identities = 43/131 (32%), Positives = 59/131 (44%), Gaps = 5/131 (3%)
Query: 138 WKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGFIL 195
W P WVKVNTDG+ N G + GG+ RD G + G F N+GRCS E+ G
Sbjct: 140 WVLPCVGWVKVNTDGASRGNPGLASAGGVLRDCEGAWCGGFSLNIGRCSAQHAELWGVYY 199
Query: 196 VMEHAALHGWYNIWLESDASSALMVFK---NPSLVPILLRNRWHNARTLNVQVISSHIFR 252
+ A + LE D+ + + K + S L HN + V +++R
Sbjct: 200 GLYFAWEKKVPRVELEVDSEAIVGFLKTGISDSHPLSFLVRLCHNFLQKDWLVRIVYVYR 259
Query: 253 EGNVCVDRLAN 263
E N D LAN
Sbjct: 260 EANCLADGLAN 270
>UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana]
Length = 253
Score = 55.8 bits (133), Expect = 1e-06
Identities = 45/156 (28%), Positives = 67/156 (42%), Gaps = 10/156 (6%)
Query: 118 LDAFRIPPHR-----RSMREIVSVCWKPPSAPWVKVNTDGSVLNNS--GACGGLFRDHLG 170
LD ++ H+ R+ RE + W PP W K+NTDG+ N GG+ RD G
Sbjct: 60 LDVWKAHVHKMGVTTRTAREERLIAWSPPRVGWFKLNTDGASRGNPRLATAGGVVRDGDG 119
Query: 171 TFLGAFVGNLGRCSVFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFK---NPSLV 227
+ F N+G CS E+ G + A G + +E D+ + + + S
Sbjct: 120 NWCYGFSLNIGICSAPLAELWGAYYGLNIAWERGVTQLEMEIDSEMVVGFLRTGIDDSHP 179
Query: 228 PILLRNRWHNARTLNVQVISSHIFREGNVCVDRLAN 263
L H + + V SH++RE N D LAN
Sbjct: 180 LSFLVRLCHGLLSKDWSVRISHVYREANRLADGLAN 215
>UniRef100_Q9LQJ0 F28G4.15 protein [Arabidopsis thaliana]
Length = 272
Score = 55.8 bits (133), Expect = 1e-06
Identities = 45/156 (28%), Positives = 67/156 (42%), Gaps = 10/156 (6%)
Query: 118 LDAFRIPPHR-----RSMREIVSVCWKPPSAPWVKVNTDGSVLNNS--GACGGLFRDHLG 170
LD ++ H+ R+ RE + W PP W K+NTDG+ N GG+ RD G
Sbjct: 79 LDVWKAHVHKMGVTTRTAREERLIAWSPPRVGWFKLNTDGASRGNPRLATAGGVVRDGDG 138
Query: 171 TFLGAFVGNLGRCSVFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFK---NPSLV 227
+ F N+G CS E+ G + A G + +E D+ + + + S
Sbjct: 139 NWCYGFSLNIGICSAPLAELWGAYYGLNIAWERGVTQLEMEIDSEMVVGFLRTGIDDSHP 198
Query: 228 PILLRNRWHNARTLNVQVISSHIFREGNVCVDRLAN 263
L H + + V SH++RE N D LAN
Sbjct: 199 LSFLVRLCHGLLSKDWSVRISHVYREANRLADGLAN 234
>UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis thaliana]
Length = 947
Score = 53.9 bits (128), Expect = 5e-06
Identities = 33/93 (35%), Positives = 45/93 (47%), Gaps = 16/93 (17%)
Query: 136 VCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGF 193
V WKPP WVK+NTDG+ N G GG+ RD +G + G F ++G CS E+ G
Sbjct: 839 VAWKPPDGEWVKLNTDGASRGNLGLATTGGVLRDGIGHWCGGFALDIGVCSAPLAELWG- 897
Query: 194 ILVMEHAALHGWYNIW------LESDASSALMV 220
+G Y W +E + S L+V
Sbjct: 898 -------VYYGLYMAWERRFTRVELEVDSELVV 923
>UniRef100_O82276 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 53.5 bits (127), Expect = 7e-06
Identities = 42/132 (31%), Positives = 60/132 (44%), Gaps = 7/132 (5%)
Query: 138 WKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGFIL 195
W+ PS WVK+ TDG+ N G A GG R+ G +LG F N+G C+ E+ G
Sbjct: 1063 WQVPSDGWVKITTDGASRGNHGLAAAGGAIRNGQGEWLGGFALNIGSCAAPLAELWGAYY 1122
Query: 196 VMEHAALHGWYNIWLESDASSALMVF----KNPSLVPILLRNRWHNARTLNVQVISSHIF 251
+ A G+ + L+ D + N + L+R T + V SH++
Sbjct: 1123 GLLIAWDKGFRRVELDLDCKLVVGFLSTGVSNAHPLSFLVR-LCQGFFTRDWLVRVSHVY 1181
Query: 252 REGNVCVDRLAN 263
RE N D LAN
Sbjct: 1182 REANRLADGLAN 1193
>UniRef100_Q5Z8I2 Hypothetical protein P0622F03.7 [Oryza sativa]
Length = 217
Score = 53.1 bits (126), Expect = 9e-06
Identities = 47/151 (31%), Positives = 68/151 (44%), Gaps = 20/151 (13%)
Query: 124 PPHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNS---GACGGLFRDHLGTFLGAFVGNL 180
PP R I V W PP WVK+N DGSV N+ + GG+ RD G L AF
Sbjct: 35 PPTR-----ITLVRWTPPRHGWVKLNFDGSVHNDGSGRASIGGVIRDDHGRVLLAFAERT 89
Query: 181 GRCSVFDTEVSGFILVMEHAALHGWYN-IWLESDASSALMVFKNPS----LVPILLRN-R 234
++ E I ++ A HGW + + +E D + + + + S + P +L +
Sbjct: 90 PHATIGVVEARALIRGLQLALDHGWNDRLLVEGDDLTLVRLLRCESTHTRIPPAMLDDIL 149
Query: 235 W--HNARTLNVQVISSHIFREGNVCVDRLAN 263
W + R VQ H +REGN D L +
Sbjct: 150 WLLDSFRVCEVQ----HAYREGNQVADALCH 176
>UniRef100_Q9SI81 F23N19.5 [Arabidopsis thaliana]
Length = 233
Score = 52.8 bits (125), Expect = 1e-05
Identities = 47/168 (27%), Positives = 72/168 (41%), Gaps = 8/168 (4%)
Query: 136 VCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGF 193
V W PS W K+NTDG+ N G GG R+ G + F N+GRC E+ G
Sbjct: 63 VRWSKPSLGWCKLNTDGASHGNPGLATAGGALRNEYGEWCFGFALNIGRCLAPLAELWGV 122
Query: 194 ILVMEHAALHGWYNIWLESDASSALMVFKN--PSLVPI-LLRNRWHNARTLNVQVISSHI 250
+ A G + LE D+ + + S P+ L H + + V H+
Sbjct: 123 YYGLFMAWDRGITRLELEVDSEMVVGFLRTGIGSSHPLSFLVRMCHGFLSRDWIVRIGHV 182
Query: 251 FREGNVCVDRLANLGHSV-VGEVWLSTLPSDFHQVFYEDR--CSMPRI 295
+RE N D LAN + +G ++ P+ + +D S+PR+
Sbjct: 183 YREANRLADGLANYAFDLPLGYHAFASPPNSLDSILRDDELGISVPRL 230
>UniRef100_Q9FW98 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1382
Score = 49.7 bits (117), Expect = 9e-05
Identities = 43/179 (24%), Positives = 77/179 (42%), Gaps = 24/179 (13%)
Query: 2 CSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLD---FSSIKALMSSLPRHCSSQVR 58
C FC + +T +H+FL C F +W + + V L FS+++ + + SS
Sbjct: 1104 CVFC-NRDDTVEHVFLFCPFAAQIWEEIKGKCAVKLGRNGFSTMRQWIFDFLKRGSSHAN 1162
Query: 59 DLYVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCIAADAAIL 118
L H IW ARNN + ++ V V +++ +++ + + + +
Sbjct: 1163 TLLAVTFWH----IWEARNNTKNNNGTVHPQRVVIKILSYVDM--------ILKHNTKTV 1210
Query: 119 DAFRIPPHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGA 175
D R + R W+PP A +N+D ++ ++S G L RD+ G L A
Sbjct: 1211 DGQRGGNTQAIPR------WQPPPASVWMINSDAAIFSSSRTMGVGALIRDNTGKCLVA 1263
>UniRef100_O23409 SIMILARITY to probable splicing factor CEPRP21 [Arabidopsis
thaliana]
Length = 559
Score = 47.4 bits (111), Expect = 5e-04
Identities = 42/136 (30%), Positives = 59/136 (42%), Gaps = 15/136 (11%)
Query: 136 VCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGF 193
+ W P WVKV+TDG+ N G A GG+ RD G ++G F L + G
Sbjct: 26 IAWTKPPEGWVKVSTDGASRGNPGPAAAGGVIRDEDGLWVGGFALQLAFVRLRWQSCGGR 85
Query: 194 I-LVMEHAALHGWYNIWLESDASSALMVFKNPSLVPILLRNRWHNARTLNVQVISSHIFR 252
+ L ++ + G +L+S S A + L LL W V SH++R
Sbjct: 86 VELEVDSELVVG----FLQSGISEAHPLAFLVRLCHGLLSKDW--------LVRVSHVYR 133
Query: 253 EGNVCVDRLANLGHSV 268
E N D LAN S+
Sbjct: 134 EANRLADGLANYAFSL 149
>UniRef100_Q6L3X8 Hypothetical protein [Solanum demissum]
Length = 1155
Score = 47.4 bits (111), Expect = 5e-04
Identities = 45/171 (26%), Positives = 72/171 (41%), Gaps = 5/171 (2%)
Query: 131 REIVSVCWKPPSAPWVKVNTDGSVLNNSGACGGLFRDHLGTFLGAFVGNLGRCSVFDTEV 190
+ ++ V W P + K+NTDGS +N GG+ R+ LG + AF LG + E
Sbjct: 985 KNVIRVNWIKPPTMFAKLNTDGSCVNGRCGGGGILRNALGQVIMAFTIKLGEGTSSWAEA 1044
Query: 191 SGFILVMEHAALHGWYNIWLESDASSALMVFKNPSLVPILLRNRWHNARTLNVQ--VISS 248
+ M+ G I E+D+ +P + + + + I +
Sbjct: 1045 MSMLHGMQLCIQRGVNMIIGETDSILLAKAITENWSIPWRMYIPVKKIQKMVEEHGFIIN 1104
Query: 249 HIFREGNVCVDRLANLGHSV-VGEVWLS--TLPSDFHQVFYEDRCSMPRIR 296
H RE N D+LA++ S V V+ S LPS + DR ++P R
Sbjct: 1105 HCLREANQPADKLASISLSTDVNHVFKSYANLPSLVKGLVNLDRMNLPTFR 1155
>UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa]
Length = 1936
Score = 47.0 bits (110), Expect = 6e-04
Identities = 42/134 (31%), Positives = 65/134 (48%), Gaps = 10/134 (7%)
Query: 138 WKPPSAPWVKVNTDGSVLNNS--GACGGLFRDHLGTFLGAFVGNLGRCS-VFDTEVSGFI 194
W+ P W+K+N DGS NS G G + R+ LG + + +L CS + E+ +
Sbjct: 1777 WERPRNGWMKLNVDGSFDINSEKGGIGMILRNCLGNVIFSSCRSLDSCSGPLEAELHACV 1836
Query: 195 LVMEHAALHGW--YNIWLESDASSALMVFKNPSLVPILLRNRWHNARTL---NVQVISSH 249
+ H ALH W I +E+D SS + + +P +L N A++L + Q+ S
Sbjct: 1837 EGL-HLALH-WTLLPIQVETDCSSVIQLLNHPDKDRSVLANIAQEAKSLMAGDRQIAISK 1894
Query: 250 IFREGNVCVDRLAN 263
+ R NV LAN
Sbjct: 1895 VQRSQNVISHFLAN 1908
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.137 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 490,762,874
Number of Sequences: 2790947
Number of extensions: 18806538
Number of successful extensions: 49385
Number of sequences better than 10.0: 124
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 89
Number of HSP's that attempted gapping in prelim test: 49253
Number of HSP's gapped (non-prelim): 152
length of query: 298
length of database: 848,049,833
effective HSP length: 126
effective length of query: 172
effective length of database: 496,390,511
effective search space: 85379167892
effective search space used: 85379167892
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146708.4


