

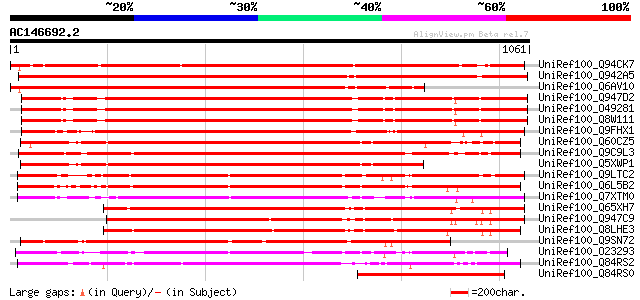
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146692.2 - phase: 0
(1061 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q94CK7 Hypothetical protein At5g12350 [Arabidopsis tha... 1624 0.0
UniRef100_Q942A5 P0431G06.4 protein [Oryza sativa] 1563 0.0
UniRef100_Q6AV10 Hypothetical protein OJ1354_D07.10 [Oryza sativa] 1317 0.0
UniRef100_Q947D2 Zinc finger protein [Arabidopsis thaliana] 1036 0.0
UniRef100_O49281 F22K20.5 protein [Arabidopsis thaliana] 1036 0.0
UniRef100_Q8W111 At1g76950/F22K20_5 [Arabidopsis thaliana] 1035 0.0
UniRef100_Q9FHX1 TMV resistance protein-like [Arabidopsis thaliana] 1020 0.0
UniRef100_Q60CZ5 Putative zinc finger protein [Solanum demissum] 997 0.0
UniRef100_Q9C9L3 Putative regulator of chromosome condensation; ... 947 0.0
UniRef100_Q5XWP1 P0431G06.4-like [Solanum tuberosum] 907 0.0
UniRef100_Q9LTC2 Chromosome condensation regulator-like protein ... 889 0.0
UniRef100_Q6L5B2 Putative regulator of chromosome condensation p... 881 0.0
UniRef100_Q7XTM0 OSJNBa0070M12.5 protein [Oryza sativa] 880 0.0
UniRef100_Q65XH7 Ptative chromosome condensation factor [Oryza s... 871 0.0
UniRef100_Q947C9 Putative chromosome condensation factor [Tritic... 865 0.0
UniRef100_Q8LHE3 Putative chromosome condensation factor [Oryza ... 859 0.0
UniRef100_Q9SN72 Hypothetical protein F1P2.210 [Arabidopsis thal... 769 0.0
UniRef100_O23293 Disease resistance N like protein [Arabidopsis ... 750 0.0
UniRef100_Q84RS2 ZR1 protein [Medicago sativa] 703 0.0
UniRef100_Q84RS0 ZR4 protein [Medicago sativa] 558 e-157
>UniRef100_Q94CK7 Hypothetical protein At5g12350 [Arabidopsis thaliana]
Length = 1062
Score = 1624 bits (4206), Expect = 0.0
Identities = 832/1091 (76%), Positives = 912/1091 (83%), Gaps = 74/1091 (6%)
Query: 1 MNSDLSRTGAVERDIEQT-----------------------------------FEWKEEK 25
M SDLSR G VERDIEQ F EEK
Sbjct: 1 MASDLSRAGPVERDIEQAIIALKKGAYLLKYGRRGKPKFCPFRLSNDETVLIWFSGNEEK 60
Query: 26 HLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDRSLDLICKDKDEAEVWFSGLKA 85
HLKLSHVSRIISGQRT RYPRPEKEYQSFSLIY++RSLD+ DKDEAEVWF+GLKA
Sbjct: 61 HLKLSHVSRIISGQRT----RYPRPEKEYQSFSLIYSERSLDV---DKDEAEVWFTGLKA 113
Query: 86 LISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDHLRLHSPY 145
LIS H R RTESRSDG PSEANSPRTYTRRSSPLHSPF SN+S QKD +HLR+HSP+
Sbjct: 114 LISHCHQRNRRTESRSDGTPSEANSPRTYTRRSSPLHSPFSSNDSLQKDGSNHLRIHSPF 173
Query: 146 ESPPKNGLDKAL-DVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTMGMDAFRV 204
ESPPKNGLDKA D+ LYAVP K F+P DSA+ SVHS GGSDSMHGHM+ MGMDAFRV
Sbjct: 174 ESPPKNGLDKAFSDMALYAVPPKGFYPSDSATISVHS---GGSDSMHGHMRGMGMDAFRV 230
Query: 205 SLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALE 264
S+SSAVSSSS GSGHDDGDALGDVFIWGEG G+GV+GGGN RVGS +K+DSL PKALE
Sbjct: 231 SMSSAVSSSSHGSGHDDGDALGDVFIWGEGIGEGVLGGGNRRVGSSFDIKMDSLLPKALE 290
Query: 265 SAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIE 324
S +VLDVQNIACGG+HA LVTKQGE FSWGEES GRLGHGVDS++ PKLIDAL+ TNIE
Sbjct: 291 STIVLDVQNIACGGQHAVLVTKQGESFSWGEESEGRLGHGVDSNIQQPKLIDALNTTNIE 350
Query: 325 LVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCG 384
LVACGE+H+CAVTLSGDLYTWG G ++G+LGHGN+VSHWVPKRVN LEGIHVS I+CG
Sbjct: 351 LVACGEFHSCAVTLSGDLYTWGKG--DFGVLGHGNEVSHWVPKRVNFLLEGIHVSSIACG 408
Query: 385 PWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVV 444
P+HTAVVTSAGQLFTFGDGTFG LGHGD+KSV +PREV+SLKGLRT+RA+CGVWHTAAVV
Sbjct: 409 PYHTAVVTSAGQLFTFGDGTFGVLGHGDKKSVFIPREVDSLKGLRTVRAACGVWHTAAVV 468
Query: 445 EVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVA-LVEHNFCQVACGHSLT 503
EVMVG+SSSSNCSSGKLFTWGDGDKGRLGHG+KE KLVPTCVA LVE NFCQVACGHSLT
Sbjct: 469 EVMVGSSSSSNCSSGKLFTWGDGDKGRLGHGNKEPKLVPTCVAALVEPNFCQVACGHSLT 528
Query: 504 VALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNE 563
VALTTSGHVY MGSPVYGQLGN ADGK P RVEGKL KSFVEEIACGAYHVAVLT R E
Sbjct: 529 VALTTSGHVYTMGSPVYGQLGNSHADGKTPNRVEGKLHKSFVEEIACGAYHVAVLTSRTE 588
Query: 564 VYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWVSGVDQSM 623
VYTWGKG+NGRLGHGD DDRN+PTLV++LKDK VKSIACGTNFTAA+C+H+W SG+DQSM
Sbjct: 589 VYTWGKGSNGRLGHGDVDDRNSPTLVESLKDKQVKSIACGTNFTAAVCIHRWASGMDQSM 648
Query: 624 CSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCFNKLRKTL 683
CSGCR PF+FKRKRHNCYNCGLVFCHSC+SKKSLKA MAPNPNKPYRVCD CFNKL+KT+
Sbjct: 649 CSGCRQPFSFKRKRHNCYNCGLVFCHSCTSKKSLKACMAPNPNKPYRVCDKCFNKLKKTM 708
Query: 684 ETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMESFKQVESRSSKKNKK 743
ETD SSHSS+SRRGSINQGS + IDKDDK D+RS QLARFS MES +QV+SR KKNKK
Sbjct: 709 ETDPSSHSSLSRRGSINQGS-DPIDKDDKFDSRSDGQLARFSLMESMRQVDSR-HKKNKK 766
Query: 744 LEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRATSPISRRPS 803
EFNSSRVSP+P+G SQRGALNI+KSFNPVFG+SKKFFSASVPGSRIVSRATSPISRRPS
Sbjct: 767 YEFNSSRVSPIPSGSSQRGALNIAKSFNPVFGASKKFFSASVPGSRIVSRATSPISRRPS 826
Query: 804 PPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQLQEIELERTS 863
PPRSTTPTPTL GL TPK VVDD K+TND+LSQEV+KLRSQVESLTRKAQLQE+ELERT+
Sbjct: 827 PPRSTTPTPTLSGLATPKFVVDDTKRTNDNLSQEVVKLRSQVESLTRKAQLQEVELERTT 886
Query: 864 KQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGTAKSVKS-PSIASFGSNEL 922
KQLK+A+AI EET +CKAAKEVIKSLTAQLKDMAERLPVG+A++VKS PS+ SFGS
Sbjct: 887 KQLKEALAITNEETTRCKAAKEVIKSLTAQLKDMAERLPVGSARTVKSPPSLNSFGS--- 943
Query: 923 SFAAIDRLNI--QATSPEADLTGSNTQLLSNGSSTVSNRSTGQNKQSQSDSTNRNGSRTK 980
S ID NI QA S E++ G T + SNG+ T NG T
Sbjct: 944 SPGRIDPFNILNQANSQESEPNGITTPMFSNGTMT---------------PAFGNGEAT- 987
Query: 981 DSESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVYE 1040
+E+R+E EWVEQDEPGVYITLT+L GG DLKRVRFSRKRFSE QAE WWA+NR RVYE
Sbjct: 988 -NEARNEKEWVEQDEPGVYITLTALAGGARDLKRVRFSRKRFSEIQAEQWWADNRGRVYE 1046
Query: 1041 QYNVRMVDKSS 1051
QYNVRMVDK+S
Sbjct: 1047 QYNVRMVDKAS 1057
>UniRef100_Q942A5 P0431G06.4 protein [Oryza sativa]
Length = 1086
Score = 1563 bits (4048), Expect = 0.0
Identities = 777/1042 (74%), Positives = 872/1042 (83%), Gaps = 15/1042 (1%)
Query: 19 FEWKEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDRSLDLICKDKDEAEV 78
F KEEKHL+LS VSRI+ GQRT IFQRYPRPEKE QSFSLI +DRSLD+ICKDKDEAEV
Sbjct: 53 FSGKEEKHLRLSQVSRIVPGQRTAIFQRYPRPEKECQSFSLISHDRSLDIICKDKDEAEV 112
Query: 79 WFSGLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDH 138
WF+GLK LIS SH RKWRTESRSD I S A SPRTYTRRSSPL SPF SN+S KD DH
Sbjct: 113 WFAGLKTLISHSHQRKWRTESRSDIISSGATSPRTYTRRSSPLSSPFSSNDSVHKDGSDH 172
Query: 139 LRLHSPYESPPKNGLDKAL-DVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTM 197
RL SP+ SPPKN LDKA DVVLYAVP K FFP DS + SVHS+SSG SD+ +GH + +
Sbjct: 173 YRLRSPFGSPPKNALDKAFSDVVLYAVPPKGFFPSDSNAGSVHSMSSGHSDNTNGHPRGI 232
Query: 198 GMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDS 257
MDAFRVS SSA+SSSS GSGHDDGDALGDVFIWGEGTG+G++GGG+ RVGS K+D
Sbjct: 233 PMDAFRVSYSSAISSSSHGSGHDDGDALGDVFIWGEGTGEGILGGGSSRVGSSSSAKMDY 292
Query: 258 LFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDA 317
L PK LE AV LDVQNI+CGGRHAALVTKQGEI+SWGEESGGRLGHGVD DV PKLID+
Sbjct: 293 LVPKPLEFAVRLDVQNISCGGRHAALVTKQGEIYSWGEESGGRLGHGVDCDVAQPKLIDS 352
Query: 318 LSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIH 377
L++ NIELVACGEYHTCAVTLSGDLYTWG+G + +GLLGHGN VSHWVPKRV+GPLEGIH
Sbjct: 353 LAHMNIELVACGEYHTCAVTLSGDLYTWGDGTFKFGLLGHGNDVSHWVPKRVDGPLEGIH 412
Query: 378 VSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGV 437
VS ISCGPWHTA+VTSAGQLFTFGDG+FG LGHGDR S+S+PREVESLKGLRT+RA+CGV
Sbjct: 413 VSSISCGPWHTALVTSAGQLFTFGDGSFGVLGHGDRASLSVPREVESLKGLRTVRAACGV 472
Query: 438 WHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQV 496
WHTAAVVEVMVGNSSSSNCSSGK+FTWGDGDKGRLGHGDK+++ VPTCV ALVE NFCQV
Sbjct: 473 WHTAAVVEVMVGNSSSSNCSSGKIFTWGDGDKGRLGHGDKDSRFVPTCVAALVEPNFCQV 532
Query: 497 ACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVA 556
ACGH LTVALTTSGHVY MGS VYGQLGNPQADG LP RVEGKL K+FVEEI+CGAYHVA
Sbjct: 533 ACGHCLTVALTTSGHVYTMGSAVYGQLGNPQADGLLPVRVEGKLHKNFVEEISCGAYHVA 592
Query: 557 VLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWV 616
VLT R EVYTWGKGANGRLGHG TDD+N PTLV+ALKDK V+S+ CG NFTAAIC+HKWV
Sbjct: 593 VLTSRTEVYTWGKGANGRLGHGGTDDKNTPTLVEALKDKQVRSVVCGINFTAAICIHKWV 652
Query: 617 SGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCF 676
SG DQSMCSGCR PFN +RKRHNCYNC LVFCHSCSSKKSLKAS+APNPNKPYRVCD C+
Sbjct: 653 SGSDQSMCSGCRQPFNLRRKRHNCYNCALVFCHSCSSKKSLKASLAPNPNKPYRVCDSCY 712
Query: 677 NKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMESFKQVESR 736
+KL K LETD++S ++RG++ QG E +++L+TRS QL+R SSMESFK ++SR
Sbjct: 713 SKLTKGLETDTNSS---TKRGTVVQGFSE--TNEEELETRSNTQLSRLSSMESFKNMDSR 767
Query: 737 SSKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRATS 796
SKKNKK EFNS+RVSPVPNG S LNIS+SFNPVFGSSKKFFSASVPGSRIVSRATS
Sbjct: 768 YSKKNKKFEFNSTRVSPVPNGSSHWSGLNISRSFNPVFGSSKKFFSASVPGSRIVSRATS 827
Query: 797 PISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQLQE 856
P+SRR SPPRSTTPTPTLGGLT+P+++ +DAK TNDSLS EV+ LRSQVE+LTRK+ L E
Sbjct: 828 PVSRRTSPPRSTTPTPTLGGLTSPRVIANDAKPTNDSLSHEVLNLRSQVENLTRKSHLLE 887
Query: 857 IELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGTAKSVKSPSIAS 916
+ELERT+KQLK+AI IAGEETAKCKAAKEVIKSLTAQLK MAERLP G K+ K P ++
Sbjct: 888 VELERTTKQLKEAIVIAGEETAKCKAAKEVIKSLTAQLKGMAERLPGGVTKNSKLPPLSG 947
Query: 917 FG-SNELSFAAIDRLNIQATSPEADLTGSNTQLLSNGSSTVSNRSTGQNKQSQSDSTNRN 975
F +ELS A + L ++ E G N L SNG S+V ++ +N
Sbjct: 948 FPMPSELSSMATESLGSPSSVGEQISNGPNGLLASNGPSSV-------RIKAGHPEVGKN 1000
Query: 976 GSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENR 1035
GSR ++ES E EWVEQDEPGVYITLT+LPGG DLKRVRFSRKRFSE QAE WW ENR
Sbjct: 1001 GSRLPEAESCHEAEWVEQDEPGVYITLTALPGGARDLKRVRFSRKRFSETQAEQWWQENR 1060
Query: 1036 VRVYEQYNVRMVDKSSVGVGSE 1057
RVY+ YNVRMV+KS+ + +E
Sbjct: 1061 TRVYQHYNVRMVEKSASSIDNE 1082
>UniRef100_Q6AV10 Hypothetical protein OJ1354_D07.10 [Oryza sativa]
Length = 901
Score = 1317 bits (3408), Expect = 0.0
Identities = 658/886 (74%), Positives = 734/886 (82%), Gaps = 50/886 (5%)
Query: 2 NSDL--SRTGAVERDIEQT-----------------------------------FEWKEE 24
+SDL +R GAVERDIEQ F KEE
Sbjct: 5 SSDLGGARPGAVERDIEQAITALKKGAYLLKYGRRGKPKFCPFRLSNDESILIWFSGKEE 64
Query: 25 KHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDRSLDLICKDKDEAEVWFSGLK 84
K L+LSHVSRII GQRT IFQRYPRPEKE QSFSLI +DRSLD+ICKDKDEAEVWF+GLK
Sbjct: 65 KQLRLSHVSRIIPGQRTAIFQRYPRPEKECQSFSLISHDRSLDIICKDKDEAEVWFAGLK 124
Query: 85 ALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDHLRLHSP 144
LI+RSH RKWRTESRSD + S SPRTYTRRSSPL SPF SN+S KD ++ RL SP
Sbjct: 125 TLITRSHQRKWRTESRSDMLSSGTTSPRTYTRRSSPLSSPFSSNDSVHKDGVENYRLRSP 184
Query: 145 YESPPKNGLDKAL-DVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTMGMDAFR 203
+ SPPK GL+KA D+V YA P K FFP DS + SVHS+SSG SD+ + H + + MDAFR
Sbjct: 185 FGSPPKVGLEKAFSDIVSYAAPPKPFFPSDSNAGSVHSVSSGQSDNTNLHSRGIPMDAFR 244
Query: 204 VSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKAL 263
VSLSSAVSSSS GSGHDDGDALGDVFIWGEGTG+G++GGGN RVG+ GV +D L PK L
Sbjct: 245 VSLSSAVSSSSHGSGHDDGDALGDVFIWGEGTGEGILGGGNSRVGNSSGVNMDCLIPKPL 304
Query: 264 ESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNI 323
E AV LDVQNI+CGGRHA LVTKQGEI+SWGEESGGRLGHGVD DV PKLIDAL+N NI
Sbjct: 305 EFAVKLDVQNISCGGRHATLVTKQGEIYSWGEESGGRLGHGVDCDVPQPKLIDALANMNI 364
Query: 324 ELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISC 383
ELVACGEYHTCAVTLSGDLYTWGNG N GLLGHGN+VSHWVPKRVNGPLEGIHVS ISC
Sbjct: 365 ELVACGEYHTCAVTLSGDLYTWGNGTSNSGLLGHGNEVSHWVPKRVNGPLEGIHVSAISC 424
Query: 384 GPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAV 443
GPWHTA+VTSAGQLFTFGDG+FG LGHGDR+S+S+PREVESLKGLRT+RA+CGVWHTAAV
Sbjct: 425 GPWHTAIVTSAGQLFTFGDGSFGVLGHGDRQSLSVPREVESLKGLRTVRAACGVWHTAAV 484
Query: 444 VEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQVACGHSL 502
VEVMVGNSSSSNCSSGK+FTWGDGDKGRLGHGDK+ +LVPTCV ALVE NFCQ+ACGH +
Sbjct: 485 VEVMVGNSSSSNCSSGKIFTWGDGDKGRLGHGDKDTRLVPTCVAALVEPNFCQIACGHCM 544
Query: 503 TVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRN 562
TVALTTSGHVY MGSPVYGQLGNPQADG LP RVEGKL K+FVEEI+CGAYHVAVLT R
Sbjct: 545 TVALTTSGHVYTMGSPVYGQLGNPQADGMLPVRVEGKLHKNFVEEISCGAYHVAVLTSRT 604
Query: 563 EVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWVSGVDQS 622
EVYTWGKGANGRLGHGDTDDR++PTLV+ALKDK V+++ CG NFTAAIC+HKWVSGVDQS
Sbjct: 605 EVYTWGKGANGRLGHGDTDDRSSPTLVEALKDKQVRTVVCGINFTAAICIHKWVSGVDQS 664
Query: 623 MCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCFNKLRKT 682
MCSGCRLPFN +RKRHNCYNC LVFCH+CSSKKSLKAS+APNPNKPYRVCD C++KL K
Sbjct: 665 MCSGCRLPFNLRRKRHNCYNCALVFCHACSSKKSLKASLAPNPNKPYRVCDSCYSKLNKG 724
Query: 683 LETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMESFKQVESRSSKKNK 742
ETD S ++RGS+ QG + + D++L+T+S QL+R SS+ESFK ++SR+SKKNK
Sbjct: 725 PETD---RYSSAKRGSVIQGFNDSV--DEELETKSNAQLSRLSSLESFKHMDSRTSKKNK 779
Query: 743 KLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRATSPISRRP 802
K EFNSSRVSP+PNG S LNISKS FGSSKKFFSASVPGSRIVSRATSP+SRR
Sbjct: 780 KFEFNSSRVSPIPNGSSHWSGLNISKS----FGSSKKFFSASVPGSRIVSRATSPVSRRA 835
Query: 803 SPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESL 848
SPPRSTTPTPTLGGLT+P++V D K NDS+SQEV+ LRSQV L
Sbjct: 836 SPPRSTTPTPTLGGLTSPRVV--DGVKPNDSISQEVLSLRSQVHIL 879
>UniRef100_Q947D2 Zinc finger protein [Arabidopsis thaliana]
Length = 1103
Score = 1036 bits (2679), Expect = 0.0
Identities = 564/1079 (52%), Positives = 725/1079 (66%), Gaps = 87/1079 (8%)
Query: 24 EKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDR--SLDLICKDKDEAEVWFS 81
EK LKL+ VS+I+ GQRT +FQRY RPEK+Y SFSL+YN + SLDLICKDK EAE+W
Sbjct: 58 EKRLKLASVSKIVPGQRTAVFQRYLRPEKDYLSFSLLYNGKKKSLDLICKDKVEAEIWIG 117
Query: 82 GLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDHLRL 141
GLK LIS + + + S G S ++ R T SSP + SS S H
Sbjct: 118 GLKTLISTGQGGRSKIDGWSGGGLS-VDASRELTS-SSP-------SSSSASASRGHSSP 168
Query: 142 HSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTMGMDA 201
+P+ P A V +KS LD+ + K G D
Sbjct: 169 GTPFNIDPITSPKSAEPEVPPTDSEKSHVALDNKNMQT---------------KVSGSDG 213
Query: 202 FRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPK 261
FRVS+SSA SSSS GS DD DALGDV+IWGE D VV G + S L + D L PK
Sbjct: 214 FRVSVSSAQSSSSHGSAADDSDALGDVYIWGEVICDNVVKVGIDKNASYLTTRTDVLVPK 273
Query: 262 ALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNT 321
LES +VLDV IACG RHAA VT+QGEIF+WGEESGGRLGHG+ DV HP+L+++L+ T
Sbjct: 274 PLESNIVLDVHQIACGVRHAAFVTRQGEIFTWGEESGGRLGHGIGKDVFHPRLVESLTAT 333
Query: 322 N-IELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSY 380
+ ++ VACGE+HTCAVTL+G+LYTWG+G +N GLLGHG+ +SHW+PKR+ G LEG+HV+
Sbjct: 334 SSVDFVACGEFHTCAVTLAGELYTWGDGTHNVGLLGHGSDISHWIPKRIAGSLEGLHVAS 393
Query: 381 ISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHT 440
+SCGPWHTA++TS G+LFTFGDGTFG LGHGD+++V PREVESL GLRT+ SCGVWHT
Sbjct: 394 VSCGPWHTALITSYGRLFTFGDGTFGVLGHGDKETVQYPREVESLSGLRTIAVSCGVWHT 453
Query: 441 AAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQVACG 499
AAVVE++V S+SS+ SSGKLFTWGDGDK RLGHGDK+ +L PTCV AL+++NF ++ACG
Sbjct: 454 AAVVEIIVTQSNSSSVSSGKLFTWGDGDKNRLGHGDKDPRLKPTCVPALIDYNFHKIACG 513
Query: 500 HSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLT 559
HSLTV LTTSG V+ MGS VYGQLGN Q DGKLP VE KL FVEEI+CGAYHVA LT
Sbjct: 514 HSLTVGLTTSGQVFTMGSTVYGQLGNLQTDGKLPCLVEDKLASEFVEEISCGAYHVAALT 573
Query: 560 LRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWVSGV 619
RNEVYTWGKGANGRLGHGD +DR PT+V+ALKD+HVK IACG+N+TAAICLHKWVSG
Sbjct: 574 SRNEVYTWGKGANGRLGHGDLEDRKVPTIVEALKDRHVKYIACGSNYTAAICLHKWVSGA 633
Query: 620 DQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCFNKL 679
+QS CS CRL F F RKRHNCYNCGLV CHSCSSKK+ +A++AP+ + YRVCD C+ KL
Sbjct: 634 EQSQCSTCRLAFGFTRKRHNCYNCGLVHCHSCSSKKAFRAALAPSAGRLYRVCDSCYVKL 693
Query: 680 RKTLETDSSS--HSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARF--SSMESFKQVES 735
K E + ++ +S+V R N+ D+LD +S +LA+F S+M+ KQ++S
Sbjct: 694 SKVSEINDTNRRNSAVPRLSGENR---------DRLD-KSEIRLAKFGTSNMDLIKQLDS 743
Query: 736 RSSKKNKKLE-FNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRA 794
+++K+ KK + F+ R S +P+ + A + + + ++ K A S I SR+
Sbjct: 744 KAAKQGKKTDTFSLGRNSQLPSLLQLKDA--VQSNIGDMRRATPKLAQAP---SGISSRS 798
Query: 795 TSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQL 854
SP SRR SPPRS TP P+ GL P + D+ KKTN+ L+QE++KLR+QV+SLT+K +
Sbjct: 799 VSPFSRRSSPPRSATPMPSTSGLYFPVGIADNMKKTNEILNQEIVKLRTQVDSLTQKCEF 858
Query: 855 QEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGTAKSVK---- 910
QE+EL+ + K+ ++A+A+A EE+AK +AAKE IKSL AQLKD+AE+LP G +SVK
Sbjct: 859 QEVELQNSVKKTQEALALAEEESAKSRAAKEAIKSLIAQLKDVAEKLPPG--ESVKLACL 916
Query: 911 -------------------------SPSIASFGSNELSFAAIDRLNIQA--TSPEADLTG 943
+ SI+S + +FA N+Q+ +P A
Sbjct: 917 QNGLDQNGFHFPEENGFHPSRSESMTSSISSVAPFDFAFANASWSNLQSPKQTPRASERN 976
Query: 944 SNT----QLLSNGSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVY 999
SN LS+ S +S R + Q Q++S N + ++ ++ E EW+EQ EPGVY
Sbjct: 977 SNAYPADPRLSSSGSVISERI--EPFQFQNNSDNGSSQTGVNNTNQVEAEWIEQYEPGVY 1034
Query: 1000 ITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVYEQYNVRMVDKSSVGVGSED 1058
ITL +L G DL+RVRFSR+RF E QAE WW+ENR +VYE+YNVR+ +KS+ D
Sbjct: 1035 ITLVALHDGTRDLRRVRFSRRRFGEHQAETWWSENREKVYEKYNVRVSEKSTASQTHRD 1093
>UniRef100_O49281 F22K20.5 protein [Arabidopsis thaliana]
Length = 1108
Score = 1036 bits (2679), Expect = 0.0
Identities = 564/1079 (52%), Positives = 725/1079 (66%), Gaps = 87/1079 (8%)
Query: 24 EKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDR--SLDLICKDKDEAEVWFS 81
EK LKL+ VS+I+ GQRT +FQRY RPEK+Y SFSL+YN + SLDLICKDK EAE+W
Sbjct: 63 EKRLKLASVSKIVPGQRTAVFQRYLRPEKDYLSFSLLYNGKKKSLDLICKDKVEAEIWIG 122
Query: 82 GLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDHLRL 141
GLK LIS + + + S G S ++ R T SSP + SS S H
Sbjct: 123 GLKTLISTGQGGRSKIDGWSGGGLS-VDASRELTS-SSP-------SSSSASASRGHSSP 173
Query: 142 HSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTMGMDA 201
+P+ P A V +KS LD+ + K G D
Sbjct: 174 GTPFNIDPITSPKSAEPEVPPTDSEKSHVALDNKNMQT---------------KVSGSDG 218
Query: 202 FRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPK 261
FRVS+SSA SSSS GS DD DALGDV+IWGE D VV G + S L + D L PK
Sbjct: 219 FRVSVSSAQSSSSHGSAADDSDALGDVYIWGEVICDNVVKVGIDKNASYLTTRTDVLVPK 278
Query: 262 ALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNT 321
LES +VLDV IACG RHAA VT+QGEIF+WGEESGGRLGHG+ DV HP+L+++L+ T
Sbjct: 279 PLESNIVLDVHQIACGVRHAAFVTRQGEIFTWGEESGGRLGHGIGKDVFHPRLVESLTAT 338
Query: 322 N-IELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSY 380
+ ++ VACGE+HTCAVTL+G+LYTWG+G +N GLLGHG+ +SHW+PKR+ G LEG+HV+
Sbjct: 339 SSVDFVACGEFHTCAVTLAGELYTWGDGTHNVGLLGHGSDISHWIPKRIAGSLEGLHVAS 398
Query: 381 ISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHT 440
+SCGPWHTA++TS G+LFTFGDGTFG LGHGD+++V PREVESL GLRT+ SCGVWHT
Sbjct: 399 VSCGPWHTALITSYGRLFTFGDGTFGVLGHGDKETVQYPREVESLSGLRTIAVSCGVWHT 458
Query: 441 AAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQVACG 499
AAVVE++V S+SS+ SSGKLFTWGDGDK RLGHGDK+ +L PTCV AL+++NF ++ACG
Sbjct: 459 AAVVEIIVTQSNSSSVSSGKLFTWGDGDKNRLGHGDKDPRLKPTCVPALIDYNFHKIACG 518
Query: 500 HSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLT 559
HSLTV LTTSG V+ MGS VYGQLGN Q DGKLP VE KL FVEEI+CGAYHVA LT
Sbjct: 519 HSLTVGLTTSGQVFTMGSTVYGQLGNLQTDGKLPCLVEDKLASEFVEEISCGAYHVAALT 578
Query: 560 LRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWVSGV 619
RNEVYTWGKGANGRLGHGD +DR PT+V+ALKD+HVK IACG+N+TAAICLHKWVSG
Sbjct: 579 SRNEVYTWGKGANGRLGHGDLEDRKVPTIVEALKDRHVKYIACGSNYTAAICLHKWVSGA 638
Query: 620 DQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCFNKL 679
+QS CS CRL F F RKRHNCYNCGLV CHSCSSKK+ +A++AP+ + YRVCD C+ KL
Sbjct: 639 EQSQCSTCRLAFGFTRKRHNCYNCGLVHCHSCSSKKAFRAALAPSAGRLYRVCDSCYVKL 698
Query: 680 RKTLETDSSS--HSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARF--SSMESFKQVES 735
K E + ++ +S+V R N+ D+LD +S +LA+F S+M+ KQ++S
Sbjct: 699 SKVSEINDTNRRNSAVPRLSGENR---------DRLD-KSEIRLAKFGTSNMDLIKQLDS 748
Query: 736 RSSKKNKKLE-FNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRA 794
+++K+ KK + F+ R S +P+ + A + + + ++ K A S I SR+
Sbjct: 749 KAAKQGKKTDTFSLGRNSQLPSLLQLKDA--VQSNIGDMRRATPKLAQAP---SGISSRS 803
Query: 795 TSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQL 854
SP SRR SPPRS TP P+ GL P + D+ KKTN+ L+QE++KLR+QV+SLT+K +
Sbjct: 804 VSPFSRRSSPPRSATPMPSTSGLYFPVGIADNMKKTNEILNQEIVKLRTQVDSLTQKCEF 863
Query: 855 QEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGTAKSVK---- 910
QE+EL+ + K+ ++A+A+A EE+AK +AAKE IKSL AQLKD+AE+LP G +SVK
Sbjct: 864 QEVELQNSVKKTQEALALAEEESAKSRAAKEAIKSLIAQLKDVAEKLPPG--ESVKLACL 921
Query: 911 -------------------------SPSIASFGSNELSFAAIDRLNIQA--TSPEADLTG 943
+ SI+S + +FA N+Q+ +P A
Sbjct: 922 QNGLDQNGFHFPEENGFHPSRSESMTSSISSVAPFDFAFANASWSNLQSPKQTPRASERN 981
Query: 944 SNT----QLLSNGSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVY 999
SN LS+ S +S R + Q Q++S N + ++ ++ E EW+EQ EPGVY
Sbjct: 982 SNAYPADPRLSSSGSVISERI--EPFQFQNNSDNGSSQTGVNNTNQVEAEWIEQYEPGVY 1039
Query: 1000 ITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVYEQYNVRMVDKSSVGVGSED 1058
ITL +L G DL+RVRFSR+RF E QAE WW+ENR +VYE+YNVR+ +KS+ D
Sbjct: 1040 ITLVALHDGTRDLRRVRFSRRRFGEHQAETWWSENREKVYEKYNVRVSEKSTASQTHRD 1098
>UniRef100_Q8W111 At1g76950/F22K20_5 [Arabidopsis thaliana]
Length = 1103
Score = 1035 bits (2677), Expect = 0.0
Identities = 564/1079 (52%), Positives = 724/1079 (66%), Gaps = 87/1079 (8%)
Query: 24 EKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDR--SLDLICKDKDEAEVWFS 81
EK LKL+ VS+I+ GQRT +FQRY RPEK+Y SFSL+YN + SLDLICKDK EAE+W
Sbjct: 58 EKRLKLASVSKIVPGQRTAVFQRYLRPEKDYLSFSLLYNGKKKSLDLICKDKVEAEIWIG 117
Query: 82 GLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDHLRL 141
GLK LIS + + S G S ++ R T SSP + SS S H
Sbjct: 118 GLKTLISTGQGGRSKINGWSGGGLS-VDASRELTS-SSP-------SSSSASASRGHSSP 168
Query: 142 HSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTMGMDA 201
+P+ P A V +KS LD+ + K G D
Sbjct: 169 GTPFNIDPITSPKSAEPEVPPTDSEKSHVALDNKNMQT---------------KVSGSDG 213
Query: 202 FRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPK 261
FRVS+SSA SSSS GS DD DALGDV+IWGE D VV G + S L + D L PK
Sbjct: 214 FRVSVSSAQSSSSHGSAADDSDALGDVYIWGEVICDNVVKVGIDKNASYLTTRTDVLVPK 273
Query: 262 ALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNT 321
LES +VLDV IACG RHAA VT+QGEIF+WGEESGGRLGHG+ DV HP+L+++L+ T
Sbjct: 274 PLESNIVLDVHQIACGVRHAAFVTRQGEIFTWGEESGGRLGHGIGKDVFHPRLVESLTAT 333
Query: 322 N-IELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSY 380
+ ++ VACGE+HTCAVTL+G+LYTWG+G +N GLLGHG+ +SHW+PKR+ G LEG+HV+
Sbjct: 334 SSVDFVACGEFHTCAVTLAGELYTWGDGTHNVGLLGHGSDISHWIPKRIAGSLEGLHVAS 393
Query: 381 ISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHT 440
+SCGPWHTA++TS G+LFTFGDGTFG LGHGD+++V PREVESL GLRT+ SCGVWHT
Sbjct: 394 VSCGPWHTALITSYGRLFTFGDGTFGVLGHGDKETVQYPREVESLSGLRTIAVSCGVWHT 453
Query: 441 AAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQVACG 499
AAVVE++V S+SS+ SSGKLFTWGDGDK RLGHGDK+ +L PTCV AL+++NF ++ACG
Sbjct: 454 AAVVEIIVTQSNSSSVSSGKLFTWGDGDKNRLGHGDKDPRLKPTCVPALIDYNFHKIACG 513
Query: 500 HSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLT 559
HSLTV LTTSG V+ MGS VYGQLGN Q DGKLP VE KL FVEEI+CGAYHVA LT
Sbjct: 514 HSLTVGLTTSGQVFTMGSTVYGQLGNLQTDGKLPCLVEDKLASEFVEEISCGAYHVAALT 573
Query: 560 LRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWVSGV 619
RNEVYTWGKGANGRLGHGD +DR PT+V+ALKD+HVK IACG+N+TAAICLHKWVSG
Sbjct: 574 SRNEVYTWGKGANGRLGHGDLEDRKVPTIVEALKDRHVKYIACGSNYTAAICLHKWVSGA 633
Query: 620 DQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCFNKL 679
+QS CS CRL F F RKRHNCYNCGLV CHSCSSKK+ +A++AP+ + YRVCD C+ KL
Sbjct: 634 EQSQCSTCRLAFGFTRKRHNCYNCGLVHCHSCSSKKAFRAALAPSAGRLYRVCDSCYVKL 693
Query: 680 RKTLETDSSS--HSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARF--SSMESFKQVES 735
K E + ++ +S+V R N+ D+LD +S +LA+F S+M+ KQ++S
Sbjct: 694 SKVSEINDTNRRNSAVPRLSGENR---------DRLD-KSEIRLAKFGTSNMDLIKQLDS 743
Query: 736 RSSKKNKKLE-FNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRA 794
+++K+ KK + F+ R S +P+ + A + + + ++ K A S I SR+
Sbjct: 744 KAAKQGKKTDTFSLGRNSQLPSLLQLKDA--VQSNIGDMRRATPKLAQAP---SGISSRS 798
Query: 795 TSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQL 854
SP SRR SPPRS TP P+ GL P + D+ KKTN+ L+QE++KLR+QV+SLT+K +
Sbjct: 799 VSPFSRRSSPPRSATPMPSTSGLYFPVGIADNMKKTNEILNQEIVKLRTQVDSLTQKCEF 858
Query: 855 QEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGTAKSVK---- 910
QE+EL+ + K+ ++A+A+A EE+AK +AAKE IKSL AQLKD+AE+LP G +SVK
Sbjct: 859 QEVELQNSVKKTQEALALAEEESAKSRAAKEAIKSLIAQLKDVAEKLPPG--ESVKLACL 916
Query: 911 -------------------------SPSIASFGSNELSFAAIDRLNIQA--TSPEADLTG 943
+ SI+S + +FA N+Q+ +P A
Sbjct: 917 QNGLDQNGFHFPEENGFHPSRSESMTSSISSVAPFDFAFANASWSNLQSPKQTPRASERN 976
Query: 944 SNT----QLLSNGSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVY 999
SN LS+ S +S R + Q Q++S N + ++ ++ E EW+EQ EPGVY
Sbjct: 977 SNAYPADPRLSSSGSVISERI--EPFQFQNNSDNGSSQTGVNNTNQVEAEWIEQYEPGVY 1034
Query: 1000 ITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVYEQYNVRMVDKSSVGVGSED 1058
ITL +L G DL+RVRFSR+RF E QAE WW+ENR +VYE+YNVR+ +KS+ D
Sbjct: 1035 ITLVALHDGTRDLRRVRFSRRRFGEHQAETWWSENREKVYEKYNVRVSEKSTASQTHRD 1093
>UniRef100_Q9FHX1 TMV resistance protein-like [Arabidopsis thaliana]
Length = 1073
Score = 1020 bits (2638), Expect = 0.0
Identities = 565/1062 (53%), Positives = 719/1062 (67%), Gaps = 84/1062 (7%)
Query: 24 EKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDR--SLDLICKDKDEAEVWFS 81
EK LKL+ VS+I+ GQRT +FQRY RP+K+Y SFSLIY++R +LDLICKDK EAEVW +
Sbjct: 53 EKRLKLATVSKIVPGQRTAVFQRYLRPDKDYLSFSLIYSNRKRTLDLICKDKVEAEVWIA 112
Query: 82 GLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDHLRL 141
GLKALIS R + + SDG S A+S R L SP S+ + +D
Sbjct: 113 GLKALISGQAGRS-KIDGWSDGGLSIADS------RDLTLSSPTNSSVCASRDFNI---A 162
Query: 142 HSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSG----GSDSMHGHMKTM 197
SPY S + FP S + +S+SS SDS + ++
Sbjct: 163 DSPYNS--------------------TNFP--RTSRTENSVSSERSHVASDSPNMLVRGT 200
Query: 198 GMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDS 257
G DAFRVS+SS SSSS GS DD DALGDV+IWGE + V G + LG + D
Sbjct: 201 GSDAFRVSVSSVQSSSSHGSAPDDCDALGDVYIWGEVLCENVTKFGADKNIGYLGSRSDV 260
Query: 258 LFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDA 317
L PK LES VVLDV +IACG +HAALV++QGE+F+WGE SGGRLGHG+ DV P+LI++
Sbjct: 261 LIPKPLESNVVLDVHHIACGVKHAALVSRQGEVFTWGEASGGRLGHGMGKDVTGPQLIES 320
Query: 318 LSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIH 377
L+ T+I+ VACGE+HTCAVT++G++YTWG+G +N GLLGHG VSHW+PKR++GPLEG+
Sbjct: 321 LAATSIDFVACGEFHTCAVTMTGEIYTWGDGTHNAGLLGHGTDVSHWIPKRISGPLEGLQ 380
Query: 378 VSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGV 437
++ +SCGPWHTA++TS GQLFTFGDGTFG LGHGD+++V PREVESL GLRT+ +CGV
Sbjct: 381 IASVSCGPWHTALITSTGQLFTFGDGTFGVLGHGDKETVFYPREVESLSGLRTIAVACGV 440
Query: 438 WHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQV 496
WH AA+VEV+V +SSSS SSGKLFTWGDGDK RLGHGDKE +L PTCV AL++H F +V
Sbjct: 441 WHAAAIVEVIVTHSSSS-VSSGKLFTWGDGDKSRLGHGDKEPRLKPTCVSALIDHTFHRV 499
Query: 497 ACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVA 556
ACGHSLTV LTTSG VY MGS VYGQLGNP ADGKLP VE KL K VEEIACGAYHVA
Sbjct: 500 ACGHSLTVGLTTSGKVYTMGSTVYGQLGNPNADGKLPCLVEDKLTKDCVEEIACGAYHVA 559
Query: 557 VLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWV 616
VLT RNEV+TWGKGANGRLGHGD +DR PTLVDALK++HVK+IACG+NFTAAICLHKWV
Sbjct: 560 VLTSRNEVFTWGKGANGRLGHGDVEDRKAPTLVDALKERHVKNIACGSNFTAAICLHKWV 619
Query: 617 SGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCF 676
SG +QS CS CR F F RKRHNCYNCGLV CHSCSSKKSLKA++APNP KPYRVCD C
Sbjct: 620 SGTEQSQCSACRQAFGFTRKRHNCYNCGLVHCHSCSSKKSLKAALAPNPGKPYRVCDSCH 679
Query: 677 NKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARF---SSMESFKQV 733
+KL K E + S +V R S + D+LD ++ +LA+ S+++ KQ+
Sbjct: 680 SKLSKVSEANIDSRKNVMPRLS--------GENKDRLD-KTEIRLAKSGIPSNIDLIKQL 730
Query: 734 ESRSSKKNKKLE-FNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVS 792
++R++++ KK + F+ R S P + N++ G K A P S S
Sbjct: 731 DNRAARQGKKADTFSLVRTSQTPLTQLKDALTNVADLRR---GPPK---PAVTPSS---S 781
Query: 793 RATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKA 852
R SP SRR SPPRS TP P GL + + KKTN+ L+QEV++LR+Q ESL +
Sbjct: 782 RPVSPFSRRSSPPRSVTPIPLNVGLGFSTSIAESLKKTNELLNQEVVRLRAQAESLRHRC 841
Query: 853 QLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGT--AKSVK 910
++QE E++++ K++++A+++A EE+AK +AAKEVIKSLTAQ+KD+A LP G A++ +
Sbjct: 842 EVQEFEVQKSVKKVQEAMSLAAEESAKSEAAKEVIKSLTAQVKDIAALLPPGAYEAETTR 901
Query: 911 SPSIAS-FGSNELSFA---------AIDRLNIQATSPEADLTGSNTQLLSNGSS-TVSNR 959
+ ++ + F N F + + SP A S L N S ++
Sbjct: 902 TANLLNGFEQNGFHFTNANGQRQSRSDSMSDTSLASPLAMPARSMNGLWRNSQSPRNTDA 961
Query: 960 STGQ---------NKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVI 1010
S G+ N S+ +R+ + + + S+ E EW+EQ EPGVYITL +L G
Sbjct: 962 SMGELLSEGVRISNGFSEDGRNSRSSAASASNASQVEAEWIEQYEPGVYITLLALGDGTR 1021
Query: 1011 DLKRVRFSRKRFSEKQAENWWAENRVRVYEQYNVRMVDKSSV 1052
DLKRVRFSR+RF E+QAE WW+ENR RVYE+YN+R D+SSV
Sbjct: 1022 DLKRVRFSRRRFREQQAETWWSENRERVYEKYNIRGTDRSSV 1063
>UniRef100_Q60CZ5 Putative zinc finger protein [Solanum demissum]
Length = 1082
Score = 997 bits (2577), Expect = 0.0
Identities = 542/1055 (51%), Positives = 699/1055 (65%), Gaps = 87/1055 (8%)
Query: 22 KEEKHLKLSHVSRIISGQRT---------------PIFQRYPRPEKEYQSFSLIYNDRSL 66
KEEK L+L HVSRII GQRT IFQ+YPRPEKEYQSFSLI NDRSL
Sbjct: 78 KEEKQLELCHVSRIIPGQRTVSLSIPTTFFPYKGKAIFQQYPRPEKEYQSFSLICNDRSL 137
Query: 67 DLICKDKDEAEVWFSGLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFG 126
DLICKDKDEAEVW +GLKA+I+R RK + ++RS+ + S++ + T +S +
Sbjct: 138 DLICKDKDEAEVWITGLKAIITRGRSRKGKYDARSETVFSDSPHGQRVTTSTSSI----- 192
Query: 127 SNESSQKDSGDHLRLHSPYESPPKNGLDKAL-DVVLYAVPQKSFFPLDSASASVHSISSG 185
D GD+ R ES P++ L KA D++ Y KS +++ S ++ S+S+G
Sbjct: 193 -------DQGDNQRT----ESLPQSRLGKAYADIIQYTAAGKSPTLVETGSFNLSSLSAG 241
Query: 186 GSDSMHGHMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNH 245
D+ + T D FRVSLSSA+SSSSQGS +D D LGDVFIWGEGTG+G++GGG H
Sbjct: 242 AVDNSNARSSTA--DTFRVSLSSALSSSSQGSCLEDFDNLGDVFIWGEGTGNGLLGGGKH 299
Query: 246 RVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGV 305
R+G G +ID+ PK+LES+VVLDVQNI+CG RHA LVTKQGE FSWGEE+GGRLGHG
Sbjct: 300 RIGKSSGTRIDANTPKSLESSVVLDVQNISCGNRHAMLVTKQGEAFSWGEEAGGRLGHGA 359
Query: 306 DSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWV 365
++DV HPKLI N+EL+ACGEYH+CAVT SGDLYTWG+GA + GLLGH ++ SHW+
Sbjct: 360 ETDVSHPKLIKNFKGMNVELIACGEYHSCAVTSSGDLYTWGDGAKSSGLLGHRSEASHWI 419
Query: 366 PKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESL 425
PK+V G +EG+ VS++SCGPWHTA++TSAG+LFTFGDGTFGALGHGDR PREVE+
Sbjct: 420 PKKVCGLMEGLRVSHVSCGPWHTALITSAGRLFTFGDGTFGALGHGDRSGCITPREVETF 479
Query: 426 KGLRTMRASCGVWHTAAVVEVMVG-NSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPT 484
GL+T++ +CGVWHTAAVVE+M G +S S+ SG LFTWGDGDKG+LGHGD + +L P
Sbjct: 480 NGLKTLKVACGVWHTAAVVELMSGLDSRPSDAPSGTLFTWGDGDKGKLGHGDNKPRLAPQ 539
Query: 485 CV-ALVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKS 543
C+ ALV+ +F +VAC +++TVALTT+G VY MGS VYGQLG P A+G P VE L+ S
Sbjct: 540 CITALVDKSFSEVACSYAMTVALTTTGRVYTMGSNVYGQLGCPLANGMSPICVEDYLVDS 599
Query: 544 FVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACG 603
VEEI+CG++HVAVLT R EVYTWGKG NG+LGHGD +++ PTLVD L+DK VK I CG
Sbjct: 600 TVEEISCGSHHVAVLTSRTEVYTWGKGENGQLGHGDCENKCTPTLVDILRDKQVKRIVCG 659
Query: 604 TNFTAAICLHKWVSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAP 663
+NF+AAIC+H W D S+C GCR+PFNF+RKRHNCYNCG VFC +CSSKKSLKAS+AP
Sbjct: 660 SNFSAAICVHNWALSADNSICFGCRIPFNFRRKRHNCYNCGFVFCKACSSKKSLKASLAP 719
Query: 664 NPNKPYRVCDGCFNKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLAR 723
+ +KPYRVCD C++KL+K +E++ S + G+ + E DK+ +R
Sbjct: 720 STSKPYRVCDDCYDKLQKAIESEPFSRVPKVKAGNALYKASEQTDKESGFPLLV-GHTSR 778
Query: 724 FSSMESFKQVESRSSKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSA 783
SS +SF + + R S +++ +R S N R + ++SKS F SK FSA
Sbjct: 779 LSSSDSFNRAQGRIS----RVDQYENRASSFQNENPPRESFSLSKSPISAFRVSKSLFSA 834
Query: 784 SVPGSRIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRS 843
S+P +R+V ++TSP+ + S S P P + T ++VVD+ K NDSLSQEV +L++
Sbjct: 835 SLPSTRVVPQSTSPLLGKASALWSAIPAP-YPPVRTAEVVVDNLKPINDSLSQEVKQLKA 893
Query: 844 QVES--------------LTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKS 889
Q+E+ L K+QL E EL R +KQL DA A A E K +AAK VIKS
Sbjct: 894 QLEAMLLGLFKNANGLEELASKSQLLEAELGRKTKQLMDATAKAAVEAEKRRAAKHVIKS 953
Query: 890 LTAQLKDMAERLPVGTAKSVKSPSIASFGSNELSFAAIDRLNIQATSPEADLTGSNTQLL 949
LTAQLK++ ERLP ++S + +D N++ TS N
Sbjct: 954 LTAQLKEVTERLP----------------EEQISTSNLD-FNVEQTS-------FNRTRP 989
Query: 950 SNGSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGGV 1009
SNG + T S S+N S K + E + Q EPGVY+ L SLP G
Sbjct: 990 SNGKCVTTTTLT-----ECSGSSNTVVSAKKSRGQKPER--MLQVEPGVYLYLISLPDGG 1042
Query: 1010 IDLKRVRFSRKRFSEKQAENWWAENRVRVYEQYNV 1044
+LKRV FSRK FSE +AE WW EN ++ E+YN+
Sbjct: 1043 NELKRVHFSRKCFSEDEAEKWWNENGQKICEKYNI 1077
>UniRef100_Q9C9L3 Putative regulator of chromosome condensation; 48393-44372
[Arabidopsis thaliana]
Length = 1028
Score = 947 bits (2447), Expect = 0.0
Identities = 503/1029 (48%), Positives = 668/1029 (64%), Gaps = 59/1029 (5%)
Query: 19 FEWKEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDRSLDLICKDKDEAEV 78
+ KEEK +KLS V RI+ GQRTP F+RYPRPEKEYQSFSLI DRSLDLICKDKDEAEV
Sbjct: 53 YSGKEEKQIKLSQVLRIVPGQRTPTFKRYPRPEKEYQSFSLICPDRSLDLICKDKDEAEV 112
Query: 79 WFSGLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDH 138
W GLK+LI+R KW+T + + +E +P + RR SP + ++
Sbjct: 113 WVVGLKSLITRVKVSKWKTTIKPEITSAECPTP--HARRVSPFVTILDQVIQPSNETSTQ 170
Query: 139 LRLHSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTMG 198
RL + D+V P + +++ S ++ + +++
Sbjct: 171 TRLGKVFS-----------DIVAITAPPSNNNQTEASGNLFCPFSPTPANVENSNLRFST 219
Query: 199 MDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSL 258
D FR+SLSSAVS+SS GS H+D DALGDVF+WGE DGV+ G + + S D+L
Sbjct: 220 NDPFRLSLSSAVSTSSHGSYHEDFDALGDVFVWGESISDGVLSGTGNSLNS---TTEDAL 276
Query: 259 FPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDAL 318
PKALES +VLD QNIACG HA LVTKQGEIFSWGE GG+LGHG+++D PK I ++
Sbjct: 277 LPKALESTIVLDAQNIACGKCHAVLVTKQGEIFSWGEGKGGKLGHGLETDAQKPKFISSV 336
Query: 319 SNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHV 378
+ +ACG++HTCA+T SGDLY+WG+G +N LLGHGN+ S W+PKRV G L+G++V
Sbjct: 337 RGLGFKSLACGDFHTCAITQSGDLYSWGDGTHNVDLLGHGNESSCWIPKRVTGDLQGLYV 396
Query: 379 SYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVW 438
S ++CGPWHTAVV S+GQLFTFGDGTFGALGHGDR+S S+PREVESL GL + +CGVW
Sbjct: 397 SDVACGPWHTAVVASSGQLFTFGDGTFGALGHGDRRSTSVPREVESLIGLIVTKVACGVW 456
Query: 439 HTAAVVEVMVGNSSSS-NCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQV 496
HTAAVVEV S + + S G++FTWGDG+KG+LGHGD + KL+P CV +L N CQV
Sbjct: 457 HTAAVVEVTNEASEAEVDSSRGQVFTWGDGEKGQLGHGDNDTKLLPECVISLTNENICQV 516
Query: 497 ACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVA 556
ACGHSLTV+ T+ GHVY MGS YGQLGNP A G P RVEG ++++ VEEIACG+YHVA
Sbjct: 517 ACGHSLTVSRTSRGHVYTMGSTAYGQLGNPTAKGNFPERVEGDIVEASVEEIACGSYHVA 576
Query: 557 VLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWV 616
VLT ++E+YTWGKG NG+LGHG+ +++ P +V L++K VK+I CG+NFTA IC+HKWV
Sbjct: 577 VLTSKSEIYTWGKGLNGQLGHGNVENKREPAVVGFLREKQVKAITCGSNFTAVICVHKWV 636
Query: 617 SGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCF 676
G + S+C+GCR PFNF+RKRHNCYNCGLVFC CSS+KSL+A++AP+ NKPYRVC GCF
Sbjct: 637 PGSEHSLCAGCRNPFNFRRKRHNCYNCGLVFCKVCSSRKSLRAALAPDMNKPYRVCYGCF 696
Query: 677 NKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSME-SFKQVES 735
KL+K+ E+ S+ +S +R+ +N + + D L + + AR SS + S E
Sbjct: 697 TKLKKSRESSPSTPTSRTRK-LLNMRKSTDVSERDSLTQKFLSVNARLSSADSSLHYSER 755
Query: 736 RSSKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRAT 795
R +++ K E N+S V P NG Q SK + K +PGS + SR T
Sbjct: 756 RHHRRDLKPEVNNSNVFPSMNGSLQPVGSPFSKG-STALPKIPKNMMVKIPGSGMSSRTT 814
Query: 796 SPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQLQ 855
SP+S + + PR + ++ ++K+ DS +Q++ L+ QVE L KA
Sbjct: 815 SPVSVKSTSPRRSY-----------EVAAAESKQLKDSFNQDMAGLKEQVEQLASKAHQL 863
Query: 856 EIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGTAKSVKSPSIA 915
E ELE+T +QLK A+A +E + ++AKEVI+SLT QLK+MAE K + SI+
Sbjct: 864 EEELEKTKRQLKVVTAMAADEAEENRSAKEVIRSLTTQLKEMAE-------KQSQKDSIS 916
Query: 916 SFGSNELSFAAIDRLNIQATSPEADLTGSNTQLLSNGSSTVSNRSTGQNKQSQSDSTNRN 975
+ N + T E T + T ++ S VS S QN+ + + + N
Sbjct: 917 T--------------NSKHTDKEKSETVTQTSNQTHIRSMVSQDS--QNENNLTSKSFAN 960
Query: 976 GSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENR 1035
G R + + E V QDEPGVY+TL SLPGG +LKRVRFSRK+F+E+QAE WW EN
Sbjct: 961 GHR----KQNDKPEKVVQDEPGVYLTLLSLPGGGTELKRVRFSRKQFTEEQAEKWWGENG 1016
Query: 1036 VRVYEQYNV 1044
+V E++N+
Sbjct: 1017 AKVCERHNI 1025
>UniRef100_Q5XWP1 P0431G06.4-like [Solanum tuberosum]
Length = 1979
Score = 907 bits (2345), Expect = 0.0
Identities = 461/827 (55%), Positives = 594/827 (71%), Gaps = 27/827 (3%)
Query: 22 KEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDRSLDLICKDKDEAEVWFS 81
KEEK L+L HVSRII GQRT IFQ+YPRPEKEYQSFSLI NDRSLDLICKDKDEAEVW +
Sbjct: 1153 KEEKQLELCHVSRIIPGQRTAIFQQYPRPEKEYQSFSLICNDRSLDLICKDKDEAEVWIT 1212
Query: 82 GLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDHLRL 141
GLKA+I+R RK + ++RS+ + S++ + T +S + D GD+ R
Sbjct: 1213 GLKAIITRGRSRKGKYDARSETVFSDSPHGQRVTTSTSSI------------DQGDNQRT 1260
Query: 142 HSPYESPPKNGLDKAL-DVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTMGMD 200
ES P++ L KA D++ Y KS +++ S ++ S+S+G D+ + T D
Sbjct: 1261 ----ESLPQSRLGKAYADIIQYTAAGKSPTLVETGSFNLSSLSAGAVDNSNARSSTA--D 1314
Query: 201 AFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFP 260
FRVSLSSA+SSSSQGS +D D LGDVFIWGEGTG+G++GGG HR+G G +ID+ P
Sbjct: 1315 TFRVSLSSALSSSSQGSCLEDFDNLGDVFIWGEGTGNGLLGGGKHRIGKSSGTRIDANTP 1374
Query: 261 KALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSN 320
K+LES+VVLDVQNI+CG RHA LVTKQGE FSWGEE+GGRLGHG ++DV HPKLI
Sbjct: 1375 KSLESSVVLDVQNISCGNRHAMLVTKQGEAFSWGEEAGGRLGHGAETDVSHPKLIKNFKG 1434
Query: 321 TNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSY 380
N+EL+ACGEYH+CAVT SGDLYTWG+GA + GLLGH ++ SHW+PK+V G +EG+ VS+
Sbjct: 1435 MNVELIACGEYHSCAVTSSGDLYTWGDGAKSSGLLGHRSEASHWIPKKVCGLMEGLRVSH 1494
Query: 381 ISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHT 440
+SCGPWHTA++TSAG+LFTFGDGTFGALGHGDR PREVE+ GL+T++ +CGVWHT
Sbjct: 1495 VSCGPWHTALITSAGRLFTFGDGTFGALGHGDRSGCITPREVETFNGLKTLKVACGVWHT 1554
Query: 441 AAVVEVMVG-NSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQVAC 498
AAVVE+M G +S S+ SG LFTWGDGDKG+LGHGD + +L P C+ ALV+ +F +VAC
Sbjct: 1555 AAVVELMSGLDSRPSDAPSGTLFTWGDGDKGKLGHGDNKPRLAPQCITALVDKSFSEVAC 1614
Query: 499 GHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVL 558
+++TVALTT+G VY MGS VYGQLG P A+G P VE L+ S VEEI+CG++HVAVL
Sbjct: 1615 SYAMTVALTTTGRVYTMGSNVYGQLGCPLANGMSPICVEDYLVDSTVEEISCGSHHVAVL 1674
Query: 559 TLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWVSG 618
T R EVYTWGKG NG+LGHGD +++ PTLVD L+DK VK I CG+NF+AAIC+H W
Sbjct: 1675 TSRTEVYTWGKGENGQLGHGDCENKCTPTLVDILRDKQVKRIVCGSNFSAAICVHNWALS 1734
Query: 619 VDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCFNK 678
D S+C GCR+PFNF+RKRHNCYNCG VFC +CSSKKSLKAS+AP+ +KPYRVCD C++K
Sbjct: 1735 ADNSICFGCRIPFNFRRKRHNCYNCGFVFCKACSSKKSLKASLAPSTSKPYRVCDDCYDK 1794
Query: 679 LRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMESFKQVESRSS 738
L+K +E++ S + G+ + E DK+ +R SS +SF + + R S
Sbjct: 1795 LQKAIESEPFSRVPKVKAGNALYKASEQTDKESGFPLLV-GHTSRLSSSDSFNRAQGRIS 1853
Query: 739 KKNKKLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRATSPI 798
+++ +R S N R + ++SKS F SK FSAS+P +R+V ++TSP+
Sbjct: 1854 ----RVDQYENRASSFQNENPPRESFSLSKSPISAFRVSKSLFSASLPSTRVVPQSTSPL 1909
Query: 799 SRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQV 845
+ S S P P + T ++VVD+ K NDSLSQEV +L++QV
Sbjct: 1910 LGKASALWSAIPAP-YPPVRTAEVVVDNLKPINDSLSQEVKQLKAQV 1955
>UniRef100_Q9LTC2 Chromosome condensation regulator-like protein protein [Arabidopsis
thaliana]
Length = 1067
Score = 889 bits (2296), Expect = 0.0
Identities = 490/1055 (46%), Positives = 665/1055 (62%), Gaps = 101/1055 (9%)
Query: 16 EQTFEW---KEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYN--DRSLDLIC 70
E+T W EEK LKL VSRI+ GQRT +F+R+ RPEK++ SFSL+YN +RSLDLIC
Sbjct: 53 EKTLIWFSRGEEKGLKLFEVSRIVPGQRTAVFKRFLRPEKDHLSFSLLYNNRERSLDLIC 112
Query: 71 KDKDEAEVWFSGLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNES 130
KDK E EVWF+ LK LI +S +R+ R+E IP +S R S
Sbjct: 113 KDKAETEVWFAALKFLIEKSRNRRARSE-----IPEIHDSDTFSVGRQS----------- 156
Query: 131 SQKDSGDHLRLHSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSDSM 190
+D V +P+ S + S G +
Sbjct: 157 --------------------------IDFVPSNIPRGR----TSIDLGYQNNSDVGYE-- 184
Query: 191 HGHMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSG 250
G+M D FR+S+SS S SS GSG DD ++LGDV++WGE +G++ G S
Sbjct: 185 RGNMLRPSTDGFRISVSSTPSCSSGGSGPDDIESLGDVYVWGEVWTEGILPDGT---ASN 241
Query: 251 LGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVL 310
VK D L P+ LES VVLDV I CG RH ALVT+QGE+F+WGEE GGRLGHG+ D+
Sbjct: 242 ETVKTDVLTPRPLESNVVLDVHQIVCGVRHVALVTRQGEVFTWGEEVGGRLGHGIQVDIS 301
Query: 311 HPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVN 370
PKL++ L+ TNI+ VACGEYHTC V+ SGDL++WG+G +N GLLGHG+ +SHW+PKRV+
Sbjct: 302 RPKLVEFLALTNIDFVACGEYHTCVVSTSGDLFSWGDGIHNVGLLGHGSDISHWIPKRVS 361
Query: 371 GPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRT 430
GPLEG+ V ++CG WH+A+ T+ G+LFTFGDG FG LGHG+R+SVS P+EV+SL GL+T
Sbjct: 362 GPLEGLQVLSVACGTWHSALATANGKLFTFGDGAFGVLGHGNRESVSYPKEVQSLNGLKT 421
Query: 431 MRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVA-LV 489
++ +C +WHTAA+VEVM ++++ SS KLFTWGDGDK RLGHG+KE L+PTCV+ L+
Sbjct: 422 VKVACSIWHTAAIVEVM--GQTATSMSSRKLFTWGDGDKNRLGHGNKETYLLPTCVSSLI 479
Query: 490 EHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIA 549
++NF ++ACGH+ TVALTTSGHV+ MG +GQLGN +DGKLP V+ +L+ FVEEIA
Sbjct: 480 DYNFHKIACGHTFTVALTTSGHVFTMGGTAHGQLGNSISDGKLPCLVQDRLVGEFVEEIA 539
Query: 550 CGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAA 609
CGA+HVAVLT R+EV+TWGKGANGRLGHGDT+D+ PTLV+AL+D+HVKS++CG+NFT++
Sbjct: 540 CGAHHVAVLTSRSEVFTWGKGANGRLGHGDTEDKRTPTLVEALRDRHVKSLSCGSNFTSS 599
Query: 610 ICLHKWVSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPY 669
IC+HKWVSG DQS+CSGCR F F RKRHNCYNCGLV CH+CSSKK+LKA++AP P KP+
Sbjct: 600 ICIHKWVSGADQSICSGCRQAFGFTRKRHNCYNCGLVHCHACSSKKALKAALAPTPGKPH 659
Query: 670 RVCDGCFNKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMES 729
RVCD C++KL+ +S S+V+R + S++ + D+ TRS L + +
Sbjct: 660 RVCDACYSKLK---AAESGYSSNVNRNVATPGRSIDGSVRTDRETTRSSKVL-----LSA 711
Query: 730 FKQVESRSSKKNKKLEFNSSRVSPVPNGGSQR------GALNISKSFNPVFGSSKK---- 779
K SS+ E +++R S VP+ + I +F PV +
Sbjct: 712 NKNSVMSSSRPGFTPESSNARASQVPSLQQLKDIAFPSSLSAIQNAFKPVVAPTTTPPRT 771
Query: 780 -FFSASVPGSRIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEV 838
S P R++SP +RRPSPPR++ G + + V+D +KTN+ ++QE+
Sbjct: 772 LVIGPSSPSPPPPPRSSSPYARRPSPPRTS-------GFS--RSVIDSLRKTNEVMNQEM 822
Query: 839 IKLRSQVESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMA 898
KL SQV++L ++ Q E+ER K KDA +A +++K KAA E +KS+ QLK++
Sbjct: 823 TKLHSQVKNLKQRCNNQGTEIERFQKAAKDASELAARQSSKHKAATEALKSVAEQLKELK 882
Query: 899 ERLPVGTAKSVKSPSIASFGSNELSFAAIDRLNIQATSPEADLTGSNT--QLLSNGSSTV 956
E+LP ++S SI S L+ + + TS + T T Q+ SN S T
Sbjct: 883 EKLPPEVSESEAFESINSQAEAYLNANKVSETSPLTTSGQEQETYQKTEEQVPSNSSITE 942
Query: 957 SNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVR 1016
++ S S++ ST + SR ES+ EQ EPGVY+T G +RVR
Sbjct: 943 TSSS------SRAPSTEASSSRISGKESK------EQFEPGVYVTYEVDMNGNKIFRRVR 990
Query: 1017 FSRKRFSEKQAENWWAENRVRVYEQYNVRMVDKSS 1051
FS+KRF E QAE+WW +N+ R+ + Y+ SS
Sbjct: 991 FSKKRFDEHQAEDWWTKNKDRLLKCYSSNSSSSSS 1025
>UniRef100_Q6L5B2 Putative regulator of chromosome condensation protein [Oryza sativa]
Length = 1064
Score = 881 bits (2276), Expect = 0.0
Identities = 494/1059 (46%), Positives = 669/1059 (62%), Gaps = 85/1059 (8%)
Query: 16 EQTFEW---KEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYND--RSLDLIC 70
E T W +EK L+LS VS++I GQRT +F+R+ RPEK+Y SFSLIY + RSLDL+C
Sbjct: 56 ENTLVWYSHNKEKCLRLSSVSKVIPGQRTAVFRRFLRPEKDYLSFSLIYKNGQRSLDLVC 115
Query: 71 KDKDEAEVWFSGLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNES 130
KD+ E EVWFS L++LI+ R +DG T R S F + +
Sbjct: 116 KDQAEVEVWFSTLESLITSC-----RLNFLNDG----------QTDRVS-----FSEDVT 155
Query: 131 SQKDSGDHLRLHSPYESPPK--NGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSD 188
+DS + Y++ + + ++ + Y+ P L+S A V GSD
Sbjct: 156 IYQDS-------TSYDTTLDIASSITRSFNSAGYSTPNS----LNSIRADV------GSD 198
Query: 189 SMHGHMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVG 248
++ + G D RVS+SSA SSSSQ SG DD ++LGDV++WGE + + G+
Sbjct: 199 RVNMLRASTG-DNSRVSISSAPSSSSQSSGLDDIESLGDVYVWGEVWTEVLPSEGSSNY- 256
Query: 249 SGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSD 308
L K D L PK LES VVLDVQ IACG RH L T+QGE+F+WGEE GGRLGHG D+D
Sbjct: 257 --LCSKTDFLIPKPLESDVVLDVQQIACGSRHIGLTTRQGEVFTWGEELGGRLGHGTDTD 314
Query: 309 VLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKR 368
+ PKL+++L+ +N+E +ACGE+HTC VT SGDLY WG+G+YN GLLGHG VSHW+PKR
Sbjct: 315 ICRPKLVESLAVSNVEYIACGEFHTCVVTASGDLYDWGDGSYNAGLLGHGTGVSHWLPKR 374
Query: 369 VNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGL 428
V+GPLEG+ V ++CG WH+A+ S+G+LFTFGDGTFG+LGHGDR+SV+ P+EVE+L G
Sbjct: 375 VSGPLEGLQVLSVACGSWHSALTMSSGKLFTFGDGTFGSLGHGDRESVAYPKEVEALSGF 434
Query: 429 RTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-A 487
R M+ +CGVWH+AA+VE+ +S+N S KLFTWGDGDK RLGHGDKEAKLVPTCV A
Sbjct: 435 RAMKVACGVWHSAAIVEI--SGQASTNAMSRKLFTWGDGDKNRLGHGDKEAKLVPTCVQA 492
Query: 488 LVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEE 547
LV+HNF QVACGHS+TVAL TSGHV+ MGS GQLGNP+ADGK P V+ KL VEE
Sbjct: 493 LVDHNFHQVACGHSMTVALATSGHVFTMGSSNNGQLGNPKADGKQPCMVQDKLGNELVEE 552
Query: 548 IACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFT 607
I+CG+ HVA LT R+EVYTWG GANGRLGHG +D+ PTLVDALKD+HVKSI+CG+NFT
Sbjct: 553 ISCGSNHVAALTSRSEVYTWGMGANGRLGHGSVEDKKKPTLVDALKDRHVKSISCGSNFT 612
Query: 608 AAICLHKWVSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNK 667
IC+HKWVSG DQS+CSGCR PF F RKRHNCYNCGLV CH+CSS+K LKA++AP P K
Sbjct: 613 TCICIHKWVSGADQSVCSGCRQPFGFTRKRHNCYNCGLVHCHACSSRKVLKAALAPTPGK 672
Query: 668 PYRVCDGCFNKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSM 727
P+RVCD CF KL+ D+ SS ++R I + S+++ DK ++ + R ++LA S
Sbjct: 673 PHRVCDSCFMKLK---AADTGVISSYNKRNVITRRSIDIKDKLERPEIRP-SRLATTSPA 728
Query: 728 ESFKQVESRSSKKNKKL--EFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASV 785
E K E+++ + K + + S VP Q + + +F V + K
Sbjct: 729 EPVKYQETKNVRNETKPADPMSMMKASQVP-AMLQFKDMAFAGTFGTVPTTVKSMTMGGQ 787
Query: 786 PGSRIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQV 845
+ + SP S++PSPP +T +P +G K+ D KKTN+ L+Q++ KL+SQV
Sbjct: 788 MQMGMPMFSPSPPSKKPSPPPATA-SPLIG-----KVDNDGLKKTNELLNQDISKLQSQV 841
Query: 846 ESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTA------------Q 893
L +K + Q+ +L++ ++ K A ++A EE+A+ + +K L + Q
Sbjct: 842 NKLKQKCETQDEQLQKAERKAKQAASMASEESARRNTVLDFVKHLDSEDILMISQPSILQ 901
Query: 894 LKDMAERLPVGTAKSVKSPS------IASFGSNELSFAAIDRLNI--QATSPEADLTGSN 945
LK +A+R+P A ++K+ +A SN + + +I +S L S
Sbjct: 902 LKVIADRVPGDVADNLKTLQSQSERFLAGQSSNLVEITGLTGHDIGHHRSSSTGSLPVSQ 961
Query: 946 TQLLSNGS-STVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTS 1004
N S S+++ S + ++ G + E + +EQ EPGVY+TL
Sbjct: 962 DGSSGNASGSSIAMASDSPCHRIMENNLKAPGDFAPKYGTHGEVQLIEQFEPGVYVTLIQ 1021
Query: 1005 LPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVYEQYN 1043
L G KRVRFS++RF+E+QAE WW EN+ RV+++YN
Sbjct: 1022 LRDGTKVFKRVRFSKRRFAEQQAEEWWRENQERVFKKYN 1060
>UniRef100_Q7XTM0 OSJNBa0070M12.5 protein [Oryza sativa]
Length = 1057
Score = 880 bits (2274), Expect = 0.0
Identities = 495/1090 (45%), Positives = 654/1090 (59%), Gaps = 131/1090 (12%)
Query: 16 EQTFEW---KEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYND--RSLDLIC 70
E T W +EK LKLS VSR++SGQRT +FQR+ PEK++ SFSLIYND RSLDLIC
Sbjct: 40 ESTLIWVSNNKEKSLKLSSVSRVLSGQRTLVFQRFLLPEKDHLSFSLIYNDGKRSLDLIC 99
Query: 71 KDKDEAEVWFSGLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNES 130
KDK EAEVWF+GL LIS H DGI R+ L G + S
Sbjct: 100 KDKVEAEVWFAGLNVLISPGQHGS--QHQHIDGI------------RNGALSFECGRDSS 145
Query: 131 SQKDSGDHLRLHSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSDSM 190
S S + D++ Y+ + S + SD
Sbjct: 146 LSSSSAYTTDSFENKLSSANSAKDRSSGEFTYS---------ERTDVSDMQVKGASSD-- 194
Query: 191 HGHMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSG 250
R+S+SSA+S+SS GSG DD ++ GDV++WGE D G+
Sbjct: 195 -----------IRISVSSALSTSSHGSG-DDSESFGDVYVWGEVMCDTTCRQGSDSNAYS 242
Query: 251 LGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVL 310
D L PK LES V+LDV +ACG +HAALVT+Q E+F+WGEE GRLGHG + +
Sbjct: 243 ATAATDILVPKPLESNVMLDVSYVACGVKHAALVTRQAEVFTWGEECSGRLGHGAGTSIF 302
Query: 311 HPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVN 370
P+L+++LS N+E +ACGE+HTCA+T +GDLYTWG+G +N GLLGHG+ VSHW+PKRV+
Sbjct: 303 QPRLVESLSICNVETIACGEFHTCAITATGDLYTWGDGTHNAGLLGHGSNVSHWIPKRVS 362
Query: 371 GPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRT 430
GPLEG+ VS +SCG WHTA++TS+G+L+TFGDGTFG LGHG+R+++S P+EVESLKGLRT
Sbjct: 363 GPLEGLQVSAVSCGTWHTALITSSGKLYTFGDGTFGVLGHGNRETISYPKEVESLKGLRT 422
Query: 431 MRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALV 489
+ SCGVWHTAAVVEV++ + SN SSGKLFTWGDGDK RLGHGD+ +KL PTCV +L+
Sbjct: 423 ISVSCGVWHTAAVVEVIM---AQSNTSSGKLFTWGDGDKYRLGHGDRSSKLKPTCVPSLI 479
Query: 490 EHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIA 549
++NF + CGH+LT+ LTTSGH++ GS VYGQLGNP DG+ P VE KL V E+A
Sbjct: 480 DYNFHKAVCGHTLTIGLTTSGHIFTAGSSVYGQLGNPNNDGRYPRLVEEKLGGGGVVEVA 539
Query: 550 CGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAA 609
CGAYHVAVLT EVYTWGKGANGRLGHGD DR PT V+AL+D+ VK IACG+ FTAA
Sbjct: 540 CGAYHVAVLTQSGEVYTWGKGANGRLGHGDIADRKTPTFVEALRDRSVKRIACGSGFTAA 599
Query: 610 ICLHKWVSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPY 669
IC HK VSG++QS CS CR PF F RKRHNCYNCGLV CHSCSSKK+L+A+++PNP KPY
Sbjct: 600 ICQHKSVSGMEQSQCSSCRQPFGFTRKRHNCYNCGLVHCHSCSSKKALRAALSPNPGKPY 659
Query: 670 RVCDGCFNKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMES 729
RVCD C+ KL K L++ + + N + K DK+D++ N++A +S +
Sbjct: 660 RVCDSCYLKLSKVLDSGIGHNKN-------NTPRIPGDSKADKMDSKG-NRVASANSSDM 711
Query: 730 FKQVESRSSKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSR 789
K ++ +++K+ KK ++ F + F +
Sbjct: 712 IKNLDVKAAKQTKKYDY--------------------PPQFPAILQLKDIPFIGAADQQP 751
Query: 790 IVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLT 849
S +SP+ R P+ S++ L++ D + N+ L QEV KL+ +V SL
Sbjct: 752 NDSTYSSPLLRLPNLNSSSS-------LSSESF--DILRDANELLKQEVQKLKEEVNSLR 802
Query: 850 RKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGT--AK 907
++ + Q+ +L+++ + +A+ +A EE +K KAAK+VIKSLTAQLK+MAERLP + K
Sbjct: 803 QQREQQDADLQKSEAKAHEAMTLASEEASKSKAAKDVIKSLTAQLKEMAERLPPASCDTK 862
Query: 908 SVKSP-------------------------------SIASFGSNELSFAA--IDRLNIQA 934
+ P S+AS N A I N
Sbjct: 863 QTRQPYLPGGAVSPDTGRENQKRYEPGSFQYPQTPTSVASARFNGFLAQAHQISEPNGNT 922
Query: 935 TSPEADLTGSN--------TQLLSNGSSTVSNRSTGQNKQSQSD--STNRNGSRTKDSES 984
P +N Q ++NG T T + + +++ N +G + S S
Sbjct: 923 MVPHDSRHENNGNTKEFPVAQQMTNGGMTGYRPRTEDHDRRETERFQINLHGFNMRGSSS 982
Query: 985 RS---ETEWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVYEQ 1041
S E EW+EQ EPGVY+TL SL G +LKRVRFSR+RF E QAE+WW +NR +VY++
Sbjct: 983 PSNQVEAEWIEQYEPGVYLTLVSLRDGTKELKRVRFSRRRFGEHQAESWWNDNREKVYDK 1042
Query: 1042 YNVRMVDKSS 1051
YNVR D+ S
Sbjct: 1043 YNVRGTDRIS 1052
>UniRef100_Q65XH7 Ptative chromosome condensation factor [Oryza sativa]
Length = 917
Score = 871 bits (2251), Expect = 0.0
Identities = 458/927 (49%), Positives = 609/927 (65%), Gaps = 90/927 (9%)
Query: 193 HMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLG 252
H + D R S+SSA S+SS GS DD D+LGDV++WGE + V RVGS
Sbjct: 2 HTRGASSDVLRASISSAPSTSSHGSAQDDCDSLGDVYVWGEVFCENSV-----RVGSDTI 56
Query: 253 V----KIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSD 308
+ K D L PK LES +VLDV ++ CG RHAALVT+ G++F+WGE+SGGRLGHG D
Sbjct: 57 IRSTEKTDFLLPKPLESRLVLDVYHVDCGVRHAALVTRNGDVFTWGEDSGGRLGHGTRED 116
Query: 309 VLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKR 368
+HP+L+++L+ N++ VACGE+HTCAVT +G+LYTWG+G +N GLLGHG HW+PKR
Sbjct: 117 SVHPRLVESLAACNVDFVACGEFHTCAVTTTGELYTWGDGTHNVGLLGHGTDAGHWIPKR 176
Query: 369 VNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGL 428
++G L+G+ V+Y+SCG WHTA++TS GQLFTFGDG+FG LGHG+ S+S P+EVESL GL
Sbjct: 177 ISGALDGLPVAYVSCGTWHTALITSMGQLFTFGDGSFGVLGHGNLTSISCPKEVESLSGL 236
Query: 429 RTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVA- 487
+T+ +CGVWHTAA+VEV+V +SSSS S+GKLFTWGDGDK RLGHGDKE++L PTCVA
Sbjct: 237 KTIAVACGVWHTAAIVEVIVTHSSSS-VSAGKLFTWGDGDKHRLGHGDKESRLKPTCVAS 295
Query: 488 LVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEE 547
L++++F +VACGHSLTV LTTSG V +MG+ VYGQLGNP +DG+LP VE ++ V +
Sbjct: 296 LIDYDFYRVACGHSLTVCLTTSGKVLSMGNSVYGQLGNPNSDGRLPCLVEDRIAGEHVLQ 355
Query: 548 IACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFT 607
+ACG+YHVAVLT R+EV+TWGKGANGRLGHGD +DR PT V+ALKD+ V+ IACG NFT
Sbjct: 356 VACGSYHVAVLTGRSEVFTWGKGANGRLGHGDIEDRKVPTQVEALKDRAVRHIACGANFT 415
Query: 608 AAICLHKWVSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNK 667
AAICLHKWVSG DQS CS C+ PF F RKRHNCYNCGLV C++C+S+K+L+A++APNP K
Sbjct: 416 AAICLHKWVSGADQSQCSSCQQPFGFTRKRHNCYNCGLVHCNACTSRKALRAALAPNPGK 475
Query: 668 PYRVCDGCFNKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSM 727
PYRVCD CF KL+ L++D S ++R I + + + DT++ + S+M
Sbjct: 476 PYRVCDSCFLKLKNALDSD-----SFNKRKDI----VSHLAGESNGDTKASKTILS-SNM 525
Query: 728 ESFKQVESRSSKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPG 787
+ + ++S+++++ KK + S +P + Q + +S S + + S + S
Sbjct: 526 DIIRSLDSKAARQGKKTDALSFLRTPQVSSLLQLRDIALSGSAD-MNRSVPRAVRTSAVR 584
Query: 788 SRIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVES 847
S SRA SP SR+ SPPRSTTP PT GL+ K D+ KTN+ L+QE+ +L +QV++
Sbjct: 585 SVTTSRAVSPFSRKSSPPRSTTPVPTTHGLSFSKSATDNLAKTNELLNQEIDRLHAQVDN 644
Query: 848 LTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLP----- 902
L + + QE+EL +++K++++A+ + EE+AK KAAKEVIKSLTAQLKDMAER+P
Sbjct: 645 LRHRCEHQEVELHKSAKKVQEAMTLVAEESAKSKAAKEVIKSLTAQLKDMAERIPPEQGT 704
Query: 903 --VGTAKSVKSPSIASFGSNELSFAAIDRLNIQATSPEADLTGSNTQLLSNGSSTVSNRS 960
V AK V P+ + A +N+ L SN Q L++G S N
Sbjct: 705 YDVSEAKPVHVPN-----GIDSHIAIYSSINVAHQPQNELLNASNAQSLNSGRSLHPNGI 759
Query: 961 TGQNK-------------------------------QSQSDSTNRNGSRTKD-------- 981
+ Q++ S SD R D
Sbjct: 760 SSQHRLLGNATEASEGSAQSHRITSPCKLDVPHRRAHSNSDDMLTASHRGDDNVSIDAMS 819
Query: 982 -----------------SESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSE 1024
S S+ + EW+EQ EPGVYITLT+L G DLKRVRFSR+RF E
Sbjct: 820 LQNGEDGYKPRGTVSSISSSQVQAEWIEQYEPGVYITLTTLLDGTRDLKRVRFSRRRFGE 879
Query: 1025 KQAENWWAENRVRVYEQYNVRMVDKSS 1051
QAE WW ENR +VYE+YNVR ++ S
Sbjct: 880 HQAEKWWNENREKVYERYNVRSSERVS 906
>UniRef100_Q947C9 Putative chromosome condensation factor [Triticum monococcum]
Length = 907
Score = 865 bits (2234), Expect = 0.0
Identities = 465/912 (50%), Positives = 609/912 (65%), Gaps = 80/912 (8%)
Query: 199 MDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSL 258
+D R S+SSA S+SS GS DD D+LGDV++WGE D + V G K D L
Sbjct: 8 LDVLRASISSAPSTSSHGSAQDDCDSLGDVYVWGEVVCDNSARTSSDTVIRSTG-KTDFL 66
Query: 259 FPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDAL 318
PK LES +VLDV ++ CG RHA+LVT+ GE+F+WGE+SGGRLGHG D +HP+L+++L
Sbjct: 67 LPKPLESNLVLDVYHVDCGVRHASLVTRNGEVFTWGEDSGGRLGHGTREDSVHPRLVESL 126
Query: 319 SNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHV 378
+ NI+ VACGE+HTCAVT +G+LYTWG+G +N GLLGHG V HW+P+R++GPLEG+ +
Sbjct: 127 AACNIDFVACGEFHTCAVTTTGELYTWGDGTHNVGLLGHGTDVGHWIPRRISGPLEGLQI 186
Query: 379 SYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVW 438
+Y+SCG WHTA++TS GQLFTFGDG+FG LGHGD KS+S PREVESL GL+T+ +CGVW
Sbjct: 187 AYVSCGTWHTALITSMGQLFTFGDGSFGVLGHGDLKSISCPREVESLSGLKTIAVACGVW 246
Query: 439 HTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVA-LVEHNFCQVA 497
HTAA+VEV+V SS+ S+GKLFTWGDGDK RLGHGDKEA+L PTCVA L++++F +VA
Sbjct: 247 HTAAIVEVIV-TQSSATVSAGKLFTWGDGDKHRLGHGDKEARLKPTCVASLIDYDFYRVA 305
Query: 498 CGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAV 557
CGHSLTV LTTSG V+ MG+ VYGQLGN +DG+ P VE K+ V +IACG+YHVAV
Sbjct: 306 CGHSLTVGLTTSGKVWTMGNSVYGQLGNLNSDGR-PCLVEDKIASEHVLQIACGSYHVAV 364
Query: 558 LTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWVS 617
LT R+EV+TWGKGANGRLGHGD +DR PT V+AL+D+ V+ IACG NFTAAICLHKWVS
Sbjct: 365 LTSRSEVFTWGKGANGRLGHGDIEDRKVPTTVEALRDRAVRHIACGANFTAAICLHKWVS 424
Query: 618 GVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCFN 677
G DQS CS CR PF F RKRHNCYNCGLV C++C+S+K+L+A++AP+P K +RVCD C++
Sbjct: 425 GADQSQCSSCRQPFGFTRKRHNCYNCGLVHCNACTSRKALRAALAPSPGKLHRVCDSCYS 484
Query: 678 KLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMESFKQVESRS 737
KL+ ++ SS +++ +L + + R + S+M+ + ++S++
Sbjct: 485 KLK-------NASSSANKK--------DLAPGESNGEARVGKSILS-SNMDMIRSLDSKA 528
Query: 738 SKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIV--SRAT 795
+K+ KK + S +P + Q + S +P + + +A P +R V SRA
Sbjct: 529 AKQGKKTDALSFLRNPQVSSLLQLRDIAFSGGADPNRPAVPR--AARTPAARSVTSSRAV 586
Query: 796 SPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQLQ 855
SP SRR SPPRSTTP PT GL+ K D+ K N+ LSQEV +LR+QV+SL +
Sbjct: 587 SPFSRRSSPPRSTTPVPTTHGLSLSKSATDNLVKANELLSQEVERLRAQVDSLRNRCDHH 646
Query: 856 EIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLP-------VGTAKS 908
E+EL +++K++++A+ + EE+AK KAAKEVIKSLTAQLKDMAERLP AK
Sbjct: 647 ELELHKSAKKVQEAMTLVAEESAKSKAAKEVIKSLTAQLKDMAERLPPEHGAYDFNEAKQ 706
Query: 909 VK-----SPSIASFGS--NELSFAAIDRLNI----QATSPEADLTGSNT-QLLSNGS--- 953
V P +A++ S ++ A + LN SP ++ SN +LL N S
Sbjct: 707 VHVPNGVEPHVATYSSMNGKVHQARNELLNAPSPNSGRSPHSNGGISNQHKLLGNISENS 766
Query: 954 --STVSNRSTG--------QNKQSQSDSTNRNGSRTKD---------------------- 981
ST S R T + S SD SR D
Sbjct: 767 EGSTHSLRITSPHGSDRPHRRAHSNSDEMLSASSRGDDNVSIDARSLQNGDDSYKPRGTI 826
Query: 982 --SESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVY 1039
S ++ + EW+EQ EPGVYITLT+L G DLKRVRFSR+RF E QAENWW ENR +VY
Sbjct: 827 SISSNQVQAEWIEQYEPGVYITLTTLRDGTRDLKRVRFSRRRFGEHQAENWWNENREKVY 886
Query: 1040 EQYNVRMVDKSS 1051
E+YNVR ++ S
Sbjct: 887 EKYNVRSSERVS 898
>UniRef100_Q8LHE3 Putative chromosome condensation factor [Oryza sativa]
Length = 927
Score = 859 bits (2219), Expect = 0.0
Identities = 455/920 (49%), Positives = 610/920 (65%), Gaps = 88/920 (9%)
Query: 193 HMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLG 252
H K D RVS SSA S+SS GS DD D+ GDV++WGE D V G+ V
Sbjct: 2 HTKGASSDVIRVSTSSAPSTSSHGSAQDDCDSSGDVYVWGEVICDNSVRTGSDTVVRST- 60
Query: 253 VKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHP 312
V+ D L PK LES +VLD ++ CG +H+ALVTK GE+F+WGEESGGRLGHG D +HP
Sbjct: 61 VRTDVLRPKPLESNLVLDAYHVDCGVKHSALVTKNGEVFTWGEESGGRLGHGSREDSIHP 120
Query: 313 KLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGP 372
+LI++L+ N+++VACGE+HTCAVT +G+LYTWG+G +N GLLGHG VSHW+PKR+ G
Sbjct: 121 RLIESLAVCNVDIVACGEFHTCAVTTAGELYTWGDGTHNVGLLGHGKDVSHWIPKRIAGA 180
Query: 373 LEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMR 432
LEG+ V+Y+SCG WHTA++T+ GQLFTFGDGTFG LGHG+R+S+S P+EVESL GL+T+
Sbjct: 181 LEGLAVAYVSCGTWHTALITTMGQLFTFGDGTFGVLGHGNRESISCPKEVESLSGLKTIS 240
Query: 433 ASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVA-LVEH 491
+CGVWHTAA+VEV+V SSSS SSGKLFTWGDGDK RLGHGDKE +L PTCVA L+++
Sbjct: 241 VACGVWHTAAIVEVIVTQSSSS-ISSGKLFTWGDGDKHRLGHGDKEPRLKPTCVASLIDY 299
Query: 492 NFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACG 551
+F ++ACGHSLTV LTTSG V +MG+ VYGQLGNP++DGK+P VE +++ V ++ACG
Sbjct: 300 DFHRIACGHSLTVGLTTSGKVLSMGNTVYGQLGNPRSDGKIPCLVE-EIVGENVVQVACG 358
Query: 552 AYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAIC 611
+YHVAVLT+++EV+TWGKGANGRLGHGD +DR PTLV+AL+D+ V+ IACG NFTAAIC
Sbjct: 359 SYHVAVLTIKSEVFTWGKGANGRLGHGDIEDRKIPTLVEALRDRSVRHIACGANFTAAIC 418
Query: 612 LHKWVSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRV 671
HKWVSG +QS C+ CR PF F RKRHNC+NCGLV C++C+S+K+++A++APNP KPYRV
Sbjct: 419 QHKWVSGAEQSQCASCRQPFGFTRKRHNCHNCGLVHCNACTSRKAVRAALAPNPAKPYRV 478
Query: 672 CDGCFNKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMESFK 731
CD CF KL +++ S++S++ E + ++ D R + S+++ +
Sbjct: 479 CDSCFLKLNNAVDS-----SAISKK-------KENVLRESNSDGRLTKAIIP-SNLDMIR 525
Query: 732 QVESRSSKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFN---PVFGSSKKFFSASVPGS 788
++S+++K+ KK + S +P N Q + +S + PV + + SV
Sbjct: 526 SLDSKAAKQGKKTDALSFLRTPQMNSLLQLRDIALSGGLDLNRPVPRAVRTTAVRSVN-- 583
Query: 789 RIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQ-VES 847
SRA SP SR+PSPPRSTTP PT GL+ K D+ KTN+ L+QEV +LR+Q V++
Sbjct: 584 --TSRAVSPFSRKPSPPRSTTPVPTTHGLSIGKGAADNLAKTNEMLNQEVERLRAQVVDN 641
Query: 848 LTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTA------QLKDMAERL 901
L + ++QE+EL++++K++++A+ + EE++K KAAKEVIKSLTA QLKDMAERL
Sbjct: 642 LRHRCEVQELELQKSAKKVQEAMTLVAEESSKSKAAKEVIKSLTAQVRPISQLKDMAERL 701
Query: 902 P--VGTAKSVKSPSIASFGSNELSFAAIDRLNIQATSPEADLTGSNTQLLSNGSSTVSNR 959
P G +S + EL A N ++ NT L+ G S +N
Sbjct: 702 PPDQGAYDGNESKQMHFPNGTELHAAIYSSTNGIHQLQNESISALNTPSLNTGRSLHANG 761
Query: 960 STGQNK-----------------------------------------QSQSDSTNRNGSR 978
+ Q+K +SD +N +R
Sbjct: 762 ISSQHKSLGSISEHSEVSTHSHRVSSPHDTELSNRRARISSDELFSASGKSDDSNNRDAR 821
Query: 979 TKD--------------SESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSE 1024
+ S ++ + EW+EQ EPGVYITLT+L G DLKRVRFSR+RF E
Sbjct: 822 SLQNGEDGYKPRGTVSLSSNQVQAEWIEQYEPGVYITLTTLRDGTRDLKRVRFSRRRFGE 881
Query: 1025 KQAENWWAENRVRVYEQYNV 1044
QAENWW ENR +VYE+YNV
Sbjct: 882 HQAENWWNENREKVYERYNV 901
>UniRef100_Q9SN72 Hypothetical protein F1P2.210 [Arabidopsis thaliana]
Length = 951
Score = 770 bits (1987), Expect = 0.0
Identities = 428/914 (46%), Positives = 571/914 (61%), Gaps = 65/914 (7%)
Query: 22 KEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDRSLDLICKDKDEAEVWFS 81
K+EK LKLS V+RII GQRT +F+RYP+P KEYQSFSLIY +RSLDL DKDEAE W +
Sbjct: 56 KKEKRLKLSSVTRIIPGQRTAVFRRYPQPTKEYQSFSLIYGERSLDL---DKDEAEFWLT 112
Query: 82 GLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSP----LHSPFGSNESSQKDSGD 137
L+AL+SR++ SRS + E + ++ S + S E ++K SG
Sbjct: 113 TLRALLSRNNSSALVLHSRSRSLAPENGEQSSSSQNSKSNIRSVSSDTSYEEHAKKASGS 172
Query: 138 HLRLHSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTM 197
H +P + G K VL LD H+ ++ + +
Sbjct: 173 HCN------TPQRLG--KVFSEVLSQTAVLKALSLDELVHKPHTSPPETIENRPTNHSSP 224
Query: 198 GMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDS 257
G+D + S+SSAVSSSSQGS +D +L DVF+WGE GDG++GGG H+ GS + DS
Sbjct: 225 GVDTSKYSISSAVSSSSQGSTFEDLKSLCDVFVWGESIGDGLLGGGMHKSGSSSSLMTDS 284
Query: 258 LFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDA 317
PK L+S V LD Q+I+CG +A LVTKQG+++SWGEESGGRLGHGV S V HPKLID
Sbjct: 285 FLPKVLKSHVALDAQSISCGTNYAVLVTKQGQMYSWGEESGGRLGHGVCSYVPHPKLIDE 344
Query: 318 LSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIH 377
+ + +EL CGE+HTCAVT SGDLY WG+G +N GLLG G+ SHW P R+ G +EGI+
Sbjct: 345 FNGSTVELADCGEFHTCAVTASGDLYAWGDGDHNAGLLGLGSGASHWKPVRILGQMEGIY 404
Query: 378 VSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGV 437
V ISCGPWHTA VTS G+LFTFGDGTFGALGHGDR S ++PREVE+L G RT++A+CGV
Sbjct: 405 VKAISCGPWHTAFVTSEGKLFTFGDGTFGALGHGDRISTNIPREVEALNGCRTIKAACGV 464
Query: 438 WHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVA-LVEHNFCQV 496
WH+AAVV V SSGKLFTWGDGD GRLGHGD E +L+P+CV L +F QV
Sbjct: 465 WHSAAVVSVF-----GEATSSGKLFTWGDGDDGRLGHGDIECRLIPSCVTELDTTSFQQV 519
Query: 497 ACGHSLTVALTTSGHVYAMGSPVYGQLGNPQAD-GKLPTRVEGKLLKSFVEEIACGAYHV 555
ACG S+TVAL+ SG VYAMG+ +P D + P+ +EG L KSFV+E+ACG +H+
Sbjct: 520 ACGQSITVALSMSGQVYAMGT------ADPSHDIVRAPSCIEGGLGKSFVQEVACGFHHI 573
Query: 556 AVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKW 615
AVL + EVYTWGKG+NG+LGHGDT+ R PTLV ALK K V+ + CG+N+TA ICLHK
Sbjct: 574 AVLNSKAEVYTWGKGSNGQLGHGDTEYRCMPTLVKALKGKQVRKVVCGSNYTATICLHKP 633
Query: 616 VSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGC 675
++G D + CSGCR PFN+ RK HNCYNCG VFC+SC+SKKSL A+MAP N+PYRVCD C
Sbjct: 634 ITGTDSTKCSGCRHPFNYMRKLHNCYNCGSVFCNSCTSKKSLAAAMAPKTNRPYRVCDDC 693
Query: 676 FNKL---RKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMESFKQ 732
+ KL R++L T ++S + SL + D++ + QL R S + F+Q
Sbjct: 694 YIKLEGIRESLATPANS-------ARFSNASLPSSYEMDEIGITPQRQLLRVDSFDFFRQ 746
Query: 733 VESRSSKKNKKLEFNSSRVSPVPNGGSQRGALNIS--------KSFNPVFGSSKK----- 779
+ K + S S + + +G+ N+ SF+ V K+
Sbjct: 747 TKHADLKTIGETS-GGSCTSSIHSNMDIKGSFNLKGIRRLSRLTSFDSVQEEGKQRTKHC 805
Query: 780 ------------FFSASVPGSRIVSRATSPISRRPSPPRSTTPTPTL-GGLTTPKIVVDD 826
+ +P SR S P+S + SP S T +T +++ +
Sbjct: 806 ASKSDTSSLIRHSVTCGLPFSRRGSVELFPLSIKSSPVESVATTSDFTTDITDHELLQEV 865
Query: 827 AKKTNDSLSQEVIKLRSQVESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEV 886
KK+N LS E+ L++QVE LT K++ E EL +TSK+L+ A+ +A ++ K K+++E+
Sbjct: 866 PKKSNQCLSHEISVLKAQVEELTLKSKKLETELGKTSKKLEVAVLMARDDAEKIKSSEEI 925
Query: 887 IKSLTAQLKDMAER 900
++SLT QL + ++
Sbjct: 926 VRSLTLQLMNTTKK 939
>UniRef100_O23293 Disease resistance N like protein [Arabidopsis thaliana]
Length = 1996
Score = 750 bits (1936), Expect = 0.0
Identities = 447/1027 (43%), Positives = 609/1027 (58%), Gaps = 132/1027 (12%)
Query: 12 ERDIEQTFEW--KEEKHLKLSHVSR--IISGQRTP-IFQRYPRPEKEYQSFSLIYN--DR 64
+RDI+Q K + LK S R S + +P +F+RY RPEK+Y SFSLIY+ DR
Sbjct: 997 DRDIDQALVSLKKGTQLLKYSRKGRPKFRSFRLSPAVFRRYLRPEKDYLSFSLIYHNGDR 1056
Query: 65 SLDLICKDKDEAEVWFSGLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSP 124
SLDLICKDK E EVWF+GLK+LI ++ +++ ++E IP +S T R S
Sbjct: 1057 SLDLICKDKAETEVWFAGLKSLIRQNRNKQAKSE-----IPEIHDSDCFSTGRPSTASID 1111
Query: 125 FGSNESSQKDSGDHLRLHSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISS 184
F N + + + L + + SP K G S
Sbjct: 1112 FAPNNTRRGRTSIDLGIQN---SPTKFG-------------------------------S 1137
Query: 185 GGSDSMHGHMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGN 244
G+M D FR+S+SS S S+ SG DD ++LGDV++WGE DG+ G
Sbjct: 1138 SDVGYERGNMLRPSTDGFRISVSSTPSCSTGTSGPDDIESLGDVYVWGEVWSDGISPDG- 1196
Query: 245 HRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHG 304
V + VKID IACG RH ALVT+QGE+F+W EE+GGRLGHG
Sbjct: 1197 --VVNSTTVKID-----------------IACGVRHIALVTRQGEVFTWEEEAGGRLGHG 1237
Query: 305 VDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHW 364
+ DV PKL++ L+ TNI+ VACGEYHTCAV+ SGDL+TWG+G +N GLLGHG+ +SHW
Sbjct: 1238 IQVDVCRPKLVEFLALTNIDFVACGEYHTCAVSTSGDLFTWGDGIHNVGLLGHGSDLSHW 1297
Query: 365 VPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVES 424
+PKRV+GP+EG+ V ++CG WH+A+ T+ G+LFTFGDG FG LGHGDR+SVS P+EV+
Sbjct: 1298 IPKRVSGPVEGLQVLSVACGTWHSALATANGKLFTFGDGAFGVLGHGDRESVSYPKEVKM 1357
Query: 425 LKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPT 484
L GL+T++ +CGVWHT A+VEVM N + ++ SS KLFTWGDGDK RLGHG+KE L+PT
Sbjct: 1358 LSGLKTLKVACGVWHTVAIVEVM--NQTGTSTSSRKLFTWGDGDKNRLGHGNKETYLLPT 1415
Query: 485 CV-ALVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKS 543
CV +L+++NF Q+ACGH+ TVALTTSGHV+ MG +GQLG+ +DGKLP V+ +L+
Sbjct: 1416 CVSSLIDYNFNQIACGHTFTVALTTSGHVFTMGGTSHGQLGSSNSDGKLPCLVQDRLVGE 1475
Query: 544 FVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACG 603
FVEEI+CG +HVAVLT R+EV+TWGKG+NGRLGHGD DDR PTLV+AL+++HVKSI+C
Sbjct: 1476 FVEEISCGDHHVAVLTSRSEVFTWGKGSNGRLGHGDKDDRKTPTLVEALRERHVKSISC- 1534
Query: 604 TNFTAAICLHKWVSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAP 663
G DQS+CSGCR F F RKRHNCYNCGLV CH+CSSKK+LKA++AP
Sbjct: 1535 --------------GADQSVCSGCRQAFGFTRKRHNCYNCGLVHCHACSSKKALKAALAP 1580
Query: 664 NPNKPYRVCDGCFNKLRKTLETDSSSHSSVSRRGSIN-QGSLELIDKDDKLDTRSRNQLA 722
P KP+RVCD C+ KL+ +S +S+V+ R S SL+ + D+ D RS +++
Sbjct: 1581 TPGKPHRVCDACYTKLK---AGESGYNSNVANRNSTTPTRSLDGTGRPDR-DIRS-SRIL 1635
Query: 723 RFSSMESFKQVESRSSKKNKKLEFNSSRVSPVPNGGSQRGAL------NISKSFNPVFGS 776
E K E RSS+ E + R S VP R I +F PV S
Sbjct: 1636 LSPKTEPVKYSEVRSSRS----ESSIVRASQVPALQQLRDVAFPSSLSAIQNAFKPVASS 1691
Query: 777 SKKFFSASVPGSRIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQ 836
S + SRI SPPRS+ G + + ++D KK+N +++
Sbjct: 1692 STSTLPSGTRSSRI-----------SSPPRSS-------GFS--RGMIDTLKKSNGVINK 1731
Query: 837 EVIKLRSQVESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKD 896
E+ KL+SQ+++L K Q E++R K ++A +A + ++K KAA EV+KS+ L++
Sbjct: 1732 EMTKLQSQIKNLKEKCDNQGTEIQRLKKTAREASDLAVKHSSKHKAATEVMKSVAEHLRE 1791
Query: 897 MAERLPVGTAK-----SVKSPSIASFGSNELSFAAIDRLNIQATSPEADLTGSNTQLLSN 951
+ E+LP ++ S+ S + A ++E S + + ++ +NTQ +
Sbjct: 1792 LKEKLPPEVSRCEAFESMNSQAEAYLNASEASESCLPTTSL-GMGQRDPTPSTNTQ---D 1847
Query: 952 GSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVID 1011
+ S G N +SQ S S S E +EQ EPGVY+T G
Sbjct: 1848 QNIEEKQSSNGGNMRSQEPSGTTEAS---SSSKGGGKELIEQFEPGVYVTYVLHKNGGKI 1904
Query: 1012 LKRVRFS 1018
+RVRFS
Sbjct: 1905 FRRVRFS 1911
>UniRef100_Q84RS2 ZR1 protein [Medicago sativa]
Length = 1035
Score = 703 bits (1814), Expect = 0.0
Identities = 433/1062 (40%), Positives = 597/1062 (55%), Gaps = 113/1062 (10%)
Query: 16 EQTFEW---KEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIY--NDRSLDLIC 70
E+ W K+EKHL+LS V++I+ G+ QR EKE S SLIY + LDLIC
Sbjct: 51 ERNLVWCSGKQEKHLRLSGVTKIVQGEGNIRSQRQNGTEKECHSSSLIYANGEHPLDLIC 110
Query: 71 KDKDEAEVWFSGLKALISRSHHRKWRTESRS-DGIPSEANSPRTYTRRSSPLHSPFGSNE 129
KDK +A WF GLKA+ISR K + RS G+ +SP RR N
Sbjct: 111 KDKAQAATWFVGLKAVISRCQQPKAFSSLRSCKGVQGCVSSPSGILRRKK--------NL 162
Query: 130 SSQKDSGDHLRLHSPYESPPKNGLDKAL-DVVLYAVPQKSFFPLDSASASVHSISSGGSD 188
D+ ++HS SP + ++ D + Y + F+ S+ +S+H I+
Sbjct: 163 GLLDDASQFTQVHSVCASPTLSLSERCFSDGLSY---KSDFYSSASSLSSIHGITDNSVP 219
Query: 189 S-------MHGHMKTMGMDA-FRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVV 240
S +H ++KT + ++ LSS S S + L DV IWG G G +V
Sbjct: 220 SSPYINPDIHSNIKTSRFEKEYKKELSSNRSIMPPASALVGNNVLKDVMIWGGGIGC-LV 278
Query: 241 GGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGR 300
G N R I SL PK LES +LDVQN+A GG HAA+VTKQGE++ WG+ GR
Sbjct: 279 GIVNERFVQN---SIYSLVPKLLESTAMLDVQNVALGGNHAAIVTKQGEVYCWGQGKCGR 335
Query: 301 LGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQ 360
LG +D D+ PK++D+LS+ ++ VACGEYHTCA+T SGD+YTWG + + L+ G
Sbjct: 336 LGQRIDMDISSPKIVDSLSDIRVKKVACGEYHTCALTDSGDVYTWGKDSCS-DLVDEGRI 394
Query: 361 VSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPR 420
S W+ + + P+EGI +S I+CG WHTA+V+S G+LFT+GDGTFG LGHG+ S S P+
Sbjct: 395 RSQWLTHKFSLPVEGISISSIACGEWHTAIVSSCGRLFTYGDGTFGVLGHGNYHSFSSPK 454
Query: 421 EVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAK 480
EVESLKGL +CG WHTAA++EV N S+GKLFTWGD D+GRLGH D K
Sbjct: 455 EVESLKGLCVRSVACGTWHTAAIIEVSA-ERFKYNTSTGKLFTWGDADEGRLGHADNVNK 513
Query: 481 LVPTCVA-LVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGK 539
LVPTCV+ LV+++F QV+CG +T+ALT G V+AMGS YGQLGNP + VEG
Sbjct: 514 LVPTCVSQLVDYDFVQVSCGRMMTLALTNMGKVFAMGSAKYGQLGNPHVKDRAVV-VEGM 572
Query: 540 LLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKS 599
L + +V+ I+CG+YHVAVLT VYTWGKG NG LG GDT++R P V+AL+D+ V +
Sbjct: 573 LKQEYVKMISCGSYHVAVLTSSGSVYTWGKGENGELGLGDTENRYTPCFVEALRDRQVDT 632
Query: 600 IACGTNFTAAICLHKWVSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKA 659
I CG +FT AICLHK +S DQS C+GCRLPF F RK+HNCYNCGL+FC SCSSKK L A
Sbjct: 633 ITCGPSFTVAICLHKPISISDQSSCNGCRLPFGFTRKKHNCYNCGLLFCRSCSSKKVLNA 692
Query: 660 SMAPNPNKPYRVCDGCFNKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRN 719
S+AP +K +RVC+ CFNK QGS E
Sbjct: 693 SLAPVKSKAFRVCESCFNK---------------------KQGSSER------------- 718
Query: 720 QLARFSSMESFKQVESRSSKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKK 779
SSM+S K SR+ + + + + V RG N++ P+ S+
Sbjct: 719 -----SSMDSSK---SRNCNNQQIQRHHQNMIGDVT---EDRGETNVTN--GPLLSLSQT 765
Query: 780 FFSASVPGSRIVSRATSPISRRPSPPRSTTPTPTLGGL------------TTPKIVVDDA 827
+ ++P R V + R ST + G T VV +
Sbjct: 766 CYRKNMPSGRKVWKIQHESQRDVEDSSSTLGSVIQCGQGQVPYSAQFRINCTENSVVHET 825
Query: 828 K--KTNDSLSQEVIKLRSQVESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKE 885
+ K++ L +EV +LR++ + L ++ +L+ E++ ++++++ ++A EE AKCKAAKE
Sbjct: 826 ETTKSDKLLIEEVQRLRAEAKRLEKQCELKNQEIQECQQKVEESWSVAKEEAAKCKAAKE 885
Query: 886 VIKSLTAQLKDMAERLPVGTAKSVKSPSIASFGSNELSFAAIDRLNIQATSPEADLTGSN 945
VIK+L +L ++ + G + + F N + ++ SP +N
Sbjct: 886 VIKALALRLHTISGKDNHGLEQKA---GLQEFLPN------LAPIHTDMNSPR----NAN 932
Query: 946 TQLLSNGSSTVSNRSTGQNKQSQSDSTNRNGSRTK---DSESRSETEWVEQDEPGVYITL 1002
T LSN S + S ++K +S ++ + TK E+ + EWVEQ E GVYITL
Sbjct: 933 TDSLSN--SPIIFSSALKSKFGRSILLKKDNNLTKAESQQENALKVEWVEQYENGVYITL 990
Query: 1003 TSLPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVYEQYNV 1044
T P G LKRVRFSRKRFS+K+AE WW EN+ +V+ +Y +
Sbjct: 991 TKSPSGEKGLKRVRFSRKRFSQKEAERWWEENQTKVHHKYEI 1032
>UniRef100_Q84RS0 ZR4 protein [Medicago sativa]
Length = 300
Score = 558 bits (1438), Expect = e-157
Identities = 293/300 (97%), Positives = 296/300 (98%)
Query: 712 KLDTRSRNQLARFSSMESFKQVESRSSKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFN 771
+LDTRSRNQLARFSSMESFKQVESRSSKKNKKLEFNSSRVSPVPNGGSQ GALNISKSFN
Sbjct: 1 RLDTRSRNQLARFSSMESFKQVESRSSKKNKKLEFNSSRVSPVPNGGSQWGALNISKSFN 60
Query: 772 PVFGSSKKFFSASVPGSRIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTN 831
PVFGSSKKFFSASVPGSRIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTN
Sbjct: 61 PVFGSSKKFFSASVPGSRIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTN 120
Query: 832 DSLSQEVIKLRSQVESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLT 891
DSLSQEVIKLRSQVESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLT
Sbjct: 121 DSLSQEVIKLRSQVESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLT 180
Query: 892 AQLKDMAERLPVGTAKSVKSPSIASFGSNELSFAAIDRLNIQATSPEADLTGSNTQLLSN 951
AQLKDMAERLPVG AKSVKSPSIASFGSNELSFAAIDRLNIQATSPEADLTGSNTQLLSN
Sbjct: 181 AQLKDMAERLPVGAAKSVKSPSIASFGSNELSFAAIDRLNIQATSPEADLTGSNTQLLSN 240
Query: 952 GSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVID 1011
GSSTVS+RSTGQNKQ+QSDSTNRNGSRTKDSESRSETEWVEQDE GVYITLTSLPGG ID
Sbjct: 241 GSSTVSSRSTGQNKQNQSDSTNRNGSRTKDSESRSETEWVEQDEAGVYITLTSLPGGAID 300
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.130 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,810,505,602
Number of Sequences: 2790947
Number of extensions: 80223009
Number of successful extensions: 257048
Number of sequences better than 10.0: 2109
Number of HSP's better than 10.0 without gapping: 1069
Number of HSP's successfully gapped in prelim test: 1061
Number of HSP's that attempted gapping in prelim test: 237106
Number of HSP's gapped (non-prelim): 8339
length of query: 1061
length of database: 848,049,833
effective HSP length: 138
effective length of query: 923
effective length of database: 462,899,147
effective search space: 427255912681
effective search space used: 427255912681
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 80 (35.4 bits)
Medicago: description of AC146692.2


