

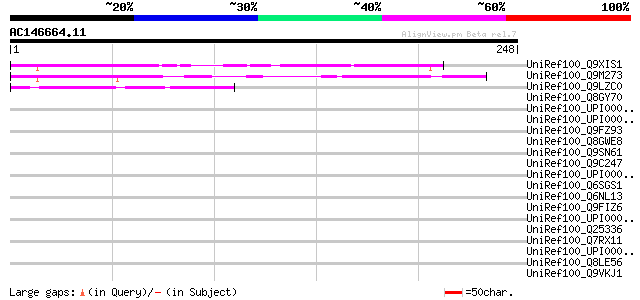
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146664.11 + phase: 0
(248 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9XIS1 F13O11.2 protein [Arabidopsis thaliana] 134 2e-30
UniRef100_Q9M273 Hypothetical protein F21F14.90 [Arabidopsis tha... 111 2e-23
UniRef100_Q9LZC0 Hypothetical protein F8F6_100 [Arabidopsis thal... 58 2e-07
UniRef100_Q8GY70 Hypothetical protein At3g10120/T22K18_5 [Arabid... 39 0.12
UniRef100_UPI0000431DBF UPI0000431DBF UniRef100 entry 36 0.79
UniRef100_UPI0000431DBE UPI0000431DBE UniRef100 entry 36 0.79
UniRef100_Q9FZ93 F3H9.15 protein [Arabidopsis thaliana] 36 0.79
UniRef100_Q8GWE8 Hypothetical protein At1g28190/F3H9_13 [Arabido... 36 0.79
UniRef100_Q9SN61 Hypothetical protein F25I24.120 [Arabidopsis th... 35 1.4
UniRef100_Q9C247 Related to a-agglutinin core protein AGA1 [Neur... 35 1.4
UniRef100_UPI0000364E62 UPI0000364E62 UniRef100 entry 35 1.8
UniRef100_Q6SGS1 Oxidoreductase, short-chain dehydrogenase/reduc... 35 1.8
UniRef100_Q6NL13 At5g37840 [Arabidopsis thaliana] 35 1.8
UniRef100_Q9FIZ6 Gb|AAD55501.1 [Arabidopsis thaliana] 35 1.8
UniRef100_UPI00002B84EA UPI00002B84EA UniRef100 entry 35 2.3
UniRef100_Q25336 Secreted acid phosphatase 2 (SAP2) precursor [L... 34 3.0
UniRef100_Q7RX11 Predicted protein [Neurospora crassa] 34 3.0
UniRef100_UPI0000362A11 UPI0000362A11 UniRef100 entry 33 5.1
UniRef100_Q8LE56 Hypothetical protein At4g37240/C7A10_120 [Arabi... 33 5.1
UniRef100_Q9VKJ1 CG4751-PA [Drosophila melanogaster] 33 5.1
>UniRef100_Q9XIS1 F13O11.2 protein [Arabidopsis thaliana]
Length = 203
Score = 134 bits (337), Expect = 2e-30
Identities = 92/217 (42%), Positives = 121/217 (55%), Gaps = 28/217 (12%)
Query: 1 MGNCLLGANSDP--DTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQSSKPLC 58
MGNCL G D D LIKVI DG ++EFY P+T F+++ F GHA+F D KPL
Sbjct: 1 MGNCLFGGLGDEEEDLLIKVIKSDGGVLEFYSPVTAGFVSHGFSGHALFSAVDLLWKPLA 60
Query: 59 QFDELVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPA 118
LV GQSYYL P ++S K + +C HVRS S + +
Sbjct: 61 HDHLLVPGQSYYLFP-NIVSDELK-TFVGSC---------------HVRSNSESLSAI-T 102
Query: 119 PYRMSLDYQYYQGVGLLKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCIT 178
PYRMSLDY + +LKR+ T+ FS ++ + K + RR+S K IWKV L I
Sbjct: 103 PYRMSLDYNH----RVLKRSYTDVFSRNSHIRTRQKEKKTRRRRTSSKG-AIWKVNLIIN 157
Query: 179 PEKLLEILSQEVSTKELIESVRIVAK---CGVTAASS 212
E+LL+ILS++ T ELIESVR VAK +T++SS
Sbjct: 158 TEELLQILSEDGRTNELIESVRAVAKGETSSITSSSS 194
>UniRef100_Q9M273 Hypothetical protein F21F14.90 [Arabidopsis thaliana]
Length = 187
Score = 111 bits (277), Expect = 2e-23
Identities = 85/242 (35%), Positives = 111/242 (45%), Gaps = 70/242 (28%)
Query: 1 MGNCLLGANSDP-------DTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTND-- 51
MGNC+ N D+LIKV+T +G +ME +PPI FITNEF GH I +
Sbjct: 1 MGNCVFKGNGGSRKLYDKDDSLIKVVTPNGGVMELHPPIFAEFITNEFPGHVIHDSLSLR 60
Query: 52 QSSKPLCQFDELVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSV 111
SS PL +EL G YYLLP++ S ATA+ Q
Sbjct: 61 HSSPPLLHGEELFPGNIYYLLPLS----------SSAAATAQLDSSDQ------------ 98
Query: 112 PTTPLPAPYRMSLDYQYYQGVGLLKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIW 171
L PYRMS K+ I ++ S G+W
Sbjct: 99 ----LSTPYRMSF----------------------------GKTPIMAA--LSGGGCGVW 124
Query: 172 KVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLS 231
KV+L I+PE+L EIL+++V T+ L+ESVR VAKCG GCG + SDQ S++
Sbjct: 125 KVRLVISPEQLAEILAEDVETEALVESVRTVAKCG-----GYGCGGGVHSRANSDQLSVT 179
Query: 232 SS 233
SS
Sbjct: 180 SS 181
>UniRef100_Q9LZC0 Hypothetical protein F8F6_100 [Arabidopsis thaliana]
Length = 179
Score = 58.2 bits (139), Expect = 2e-07
Identities = 36/110 (32%), Positives = 59/110 (52%), Gaps = 10/110 (9%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQSSKPLCQF 60
MGNCL+ +IK++ DG ++E+ PI+V+ I +F GH+I N L
Sbjct: 1 MGNCLVMEKK----VIKIVRDDGKVLEYREPISVHHILTQFSGHSISHNNTH----LLPD 52
Query: 61 DELVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQS 110
+L++G+ YYLLP T+ K++ T A E G +++R+ S+S
Sbjct: 53 AKLLSGRLYYLLPTTM--TKKKVNKKVTFANPEVEGDERLLREEEDSSES 100
>UniRef100_Q8GY70 Hypothetical protein At3g10120/T22K18_5 [Arabidopsis thaliana]
Length = 173
Score = 38.9 bits (89), Expect = 0.12
Identities = 23/73 (31%), Positives = 39/73 (52%), Gaps = 4/73 (5%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQSSKPLCQF 60
MGNCL+ +IK++ DG ++E+ P+ V+ I +F H + ++ L
Sbjct: 1 MGNCLVMEKK----VIKIMRNDGKVVEYRGPMKVHHILTQFSPHYSLFDSLTNNCHLHPQ 56
Query: 61 DELVAGQSYYLLP 73
+L+ G+ YYLLP
Sbjct: 57 AKLLCGRLYYLLP 69
>UniRef100_UPI0000431DBF UPI0000431DBF UniRef100 entry
Length = 1762
Score = 36.2 bits (82), Expect = 0.79
Identities = 26/108 (24%), Positives = 50/108 (46%), Gaps = 4/108 (3%)
Query: 61 DELVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPAPY 120
D ++ Y + + N+ ++ P+ C + G Q +Q +++S S P +P
Sbjct: 506 DNVIFDNQCYATTPSSSNGNSDMEQPTHCNSRRCNGS-QAYQQQNMQSVSTPGSPTS--- 561
Query: 121 RMSLDYQYYQGVGLLKRTSTESFSCRTNRSSVNKSTISSSRRSSRKST 168
R+ L+Y+ + L K ES+S RT + + +S S++ ST
Sbjct: 562 RLLLEYEMHLRNTLAKGMDAESYSLRTFEALLTQSMEDLGSSSNKSST 609
>UniRef100_UPI0000431DBE UPI0000431DBE UniRef100 entry
Length = 1756
Score = 36.2 bits (82), Expect = 0.79
Identities = 26/108 (24%), Positives = 50/108 (46%), Gaps = 4/108 (3%)
Query: 61 DELVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPAPY 120
D ++ Y + + N+ ++ P+ C + G Q +Q +++S S P +P
Sbjct: 506 DNVIFDNQCYATTPSSSNGNSDMEQPTHCNSRRCNGS-QAYQQQNMQSVSTPGSPTS--- 561
Query: 121 RMSLDYQYYQGVGLLKRTSTESFSCRTNRSSVNKSTISSSRRSSRKST 168
R+ L+Y+ + L K ES+S RT + + +S S++ ST
Sbjct: 562 RLLLEYEMHLRNTLAKGMDAESYSLRTFEALLTQSMEDLGSSSNKSST 609
>UniRef100_Q9FZ93 F3H9.15 protein [Arabidopsis thaliana]
Length = 266
Score = 36.2 bits (82), Expect = 0.79
Identities = 22/85 (25%), Positives = 41/85 (47%), Gaps = 4/85 (4%)
Query: 16 IKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTN----DQSSKPLCQFDELVAGQSYYL 71
IK++ DG++ + P+ V+ +T +F H I ++ Q + L + + L G +Y+L
Sbjct: 39 IKLVRSDGSLQVYDRPVVVSELTKDFPKHKICRSDLLYIGQKTPVLSETETLKLGLNYFL 98
Query: 72 LPMTVLSPNNKIDYPSTCATAETGG 96
LP + +T T + GG
Sbjct: 99 LPSDFFKNDLSFLTIATLKTPQNGG 123
>UniRef100_Q8GWE8 Hypothetical protein At1g28190/F3H9_13 [Arabidopsis thaliana]
Length = 266
Score = 36.2 bits (82), Expect = 0.79
Identities = 22/85 (25%), Positives = 41/85 (47%), Gaps = 4/85 (4%)
Query: 16 IKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTN----DQSSKPLCQFDELVAGQSYYL 71
IK++ DG++ + P+ V+ +T +F H I ++ Q + L + + L G +Y+L
Sbjct: 39 IKLVRSDGSLQVYDRPVVVSELTKDFPKHKICRSDLLYIGQKTPVLSETETLKLGLNYFL 98
Query: 72 LPMTVLSPNNKIDYPSTCATAETGG 96
LP + +T T + GG
Sbjct: 99 LPSDFFKNDLSFLTIATLKTPQNGG 123
>UniRef100_Q9SN61 Hypothetical protein F25I24.120 [Arabidopsis thaliana]
Length = 136
Score = 35.4 bits (80), Expect = 1.4
Identities = 17/50 (34%), Positives = 31/50 (62%)
Query: 163 SSRKSTGIWKVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVTAASS 212
S ++ G+WK K+ I ++L EIL+ E +T LI+ +R A + +++S
Sbjct: 69 SPQRRNGVWKAKVVIGSKQLEEILAVEGNTHALIDQLRFAAAEALVSSTS 118
>UniRef100_Q9C247 Related to a-agglutinin core protein AGA1 [Neurospora crassa]
Length = 464
Score = 35.4 bits (80), Expect = 1.4
Identities = 42/168 (25%), Positives = 68/168 (40%), Gaps = 13/168 (7%)
Query: 75 TVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPAPYRMSLDYQYYQGVGL 134
T+ S + S ++A+ +Q S S + +P P + +
Sbjct: 159 TIASSTQQPTTSSASSSAQATSSSSSAQQSTTSSSS---SSVPTPEPSTTTQAPPVTITT 215
Query: 135 LKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLEILSQEVSTKE 194
TST S T ++ KST SS+ ++S STG + E+ L ++S S
Sbjct: 216 TSTTSTTSTIPTTTSTTSIKSTSSSAPQASPSSTGTSVSE----SERTLVLISTSSSASS 271
Query: 195 LIESVRIVA------KCGVTAASSGGCGISSTTSIVSDQWSLSSSGRS 236
+ S V+ + GV++ +S G SST+S S SSS S
Sbjct: 272 VSSSASSVSSSEPSSEAGVSSTTSSSSGSSSTSSRSSSSLEPSSSSTS 319
>UniRef100_UPI0000364E62 UPI0000364E62 UniRef100 entry
Length = 1680
Score = 35.0 bits (79), Expect = 1.8
Identities = 44/171 (25%), Positives = 68/171 (39%), Gaps = 9/171 (5%)
Query: 76 VLSPNNKIDYPST---CATAETGGGGQIIRQGHVRSQSVPTTPLPAPYRMSLDYQYYQGV 132
VL P + T + + GGG R +SQ P++PLP+ + S+ + G+
Sbjct: 1351 VLQPTRQAQIVKTNQHMSVMQAGGGPVGSRMPGSKSQYAPSSPLPSRHMSSM-LKPGPGI 1409
Query: 133 GLLKRTSTESFSCRTNRSSVNKST----ISSSRRSSRKSTGIWKVKLCITPEKLLEILSQ 188
+ + NR + KS + S SS K G ++ P I S
Sbjct: 1410 DISLLSPPADEPVYGNRMPIAKSCENDGLQISHCSSSKVLGQDRLSAQSAPSLEGAICSG 1469
Query: 189 EVSTKELIESVRIVAKCGVTA-ASSGGCGISSTTSIVSDQWSLSSSGRSAP 238
+ E + A G T GG +SS+TS ++ SLS SG+ P
Sbjct: 1470 PGPQDKHAEPGKEAACGGATGWQGGGGMRVSSSTSSLASSSSLSDSGKLGP 1520
>UniRef100_Q6SGS1 Oxidoreductase, short-chain dehydrogenase/reductase family
[uncultured bacterium 463]
Length = 314
Score = 35.0 bits (79), Expect = 1.8
Identities = 19/58 (32%), Positives = 31/58 (52%), Gaps = 1/58 (1%)
Query: 191 STKELIESVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLSSSGRSAPSKVDALVLDI 248
ST E+IE V + KC + +S G G+ ++ S+ S ++ R A SK+D V +
Sbjct: 9 STSEVIEGVDLKGKCALVTGASSGLGVETSRSLASAGAAVIMVARDA-SKLDTAVAQV 65
>UniRef100_Q6NL13 At5g37840 [Arabidopsis thaliana]
Length = 214
Score = 35.0 bits (79), Expect = 1.8
Identities = 22/90 (24%), Positives = 40/90 (44%), Gaps = 9/90 (10%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQS-----SK 55
MGN ++ + +KV+ DG+I P+T + T E+ G + + +K
Sbjct: 1 MGNTIMVRRNK----VKVMKIDGDIFRLKTPVTASDATKEYPGFVLLDSETVKRLGVRAK 56
Query: 56 PLCQFDELVAGQSYYLLPMTVLSPNNKIDY 85
PL L +Y+L+ + + NK+ Y
Sbjct: 57 PLEPNQILKPNHTYFLVDLPPVDKRNKLPY 86
>UniRef100_Q9FIZ6 Gb|AAD55501.1 [Arabidopsis thaliana]
Length = 219
Score = 35.0 bits (79), Expect = 1.8
Identities = 22/90 (24%), Positives = 40/90 (44%), Gaps = 9/90 (10%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQS-----SK 55
MGN ++ + +KV+ DG+I P+T + T E+ G + + +K
Sbjct: 1 MGNTIMVRRNK----VKVMKIDGDIFRLKTPVTASDATKEYPGFVLLDSETVKRLGVRAK 56
Query: 56 PLCQFDELVAGQSYYLLPMTVLSPNNKIDY 85
PL L +Y+L+ + + NK+ Y
Sbjct: 57 PLEPNQILKPNHTYFLVDLPPVDKRNKLPY 86
>UniRef100_UPI00002B84EA UPI00002B84EA UniRef100 entry
Length = 277
Score = 34.7 bits (78), Expect = 2.3
Identities = 31/104 (29%), Positives = 44/104 (41%)
Query: 136 KRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLEILSQEVSTKEL 195
+R+S+ S S ++ SS + S+ SSS SSR S T S S+
Sbjct: 140 RRSSSSSSSSSSSSSSSSSSSSSSSSSSSRSSRSSRSSTSTSTSSSSRSSSSSSCSSSSS 199
Query: 196 IESVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLSSSGRSAPS 239
+ ++SS G G SSTT+ S SSS S+ S
Sbjct: 200 SSGGGGSSSSSSGSSSSSGSGSSSTTTKTRTSTSSSSSSSSSSS 243
Score = 33.5 bits (75), Expect = 5.1
Identities = 29/103 (28%), Positives = 46/103 (44%), Gaps = 17/103 (16%)
Query: 137 RTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLEILSQEVSTKELI 196
R S+ SCR++ SS + S+ SSS SS S+ S S+
Sbjct: 59 RRSSSRSSCRSSSSSSSSSSSSSSSSSSSSSSS-----------------SSSSSSSSSS 101
Query: 197 ESVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLSSSGRSAPS 239
S+ ++ +++SS SS++S S S+SSS R + S
Sbjct: 102 SSMSSSSRRSSSSSSSSSSSSSSSSSSSSSSSSMSSSSRRSSS 144
>UniRef100_Q25336 Secreted acid phosphatase 2 (SAP2) precursor [Leishmania mexicana]
Length = 888
Score = 34.3 bits (77), Expect = 3.0
Identities = 36/135 (26%), Positives = 57/135 (41%), Gaps = 9/135 (6%)
Query: 112 PTTPLPAPYRMS-LDYQYYQGVGLLKRTSTESFSCRTNRSSVNKSTISS--SRRSSRKST 168
P+ P Y +S +D+Q Y G + TS+ S T SS +T SS + SS + T
Sbjct: 425 PSKACPDSYILSAVDHQCYPGPDVTNPTSSSSSEGTTTSSSEGTATSSSDVTTTSSSEGT 484
Query: 169 GIWKVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVTAASSGGCGISSTTSIVSDQW 228
+ + S + +T ++ + G T++S S T+ SD
Sbjct: 485 ATSSSDATTSSSEGTATSSSDATTSSSSDATTTSSSEGTTSSS------SDATTSSSDAT 538
Query: 229 SLSSSGRSAPSKVDA 243
+ SSS +A S DA
Sbjct: 539 TTSSSEGTATSSSDA 553
>UniRef100_Q7RX11 Predicted protein [Neurospora crassa]
Length = 1165
Score = 34.3 bits (77), Expect = 3.0
Identities = 20/70 (28%), Positives = 37/70 (52%), Gaps = 7/70 (10%)
Query: 66 GQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPAPYRMSLD 125
GQ L T LS ++ +D+ ++ +T+ + I + H R Q PT+P + + +++D
Sbjct: 1038 GQLGPLSHATTLSLSDPLDHTNSASTSSS------ISKSHYRQQQKPTSPTKSDFSLAVD 1091
Query: 126 -YQYYQGVGL 134
Y Y G G+
Sbjct: 1092 NYNKYNGFGM 1101
>UniRef100_UPI0000362A11 UPI0000362A11 UniRef100 entry
Length = 302
Score = 33.5 bits (75), Expect = 5.1
Identities = 27/102 (26%), Positives = 47/102 (45%)
Query: 138 TSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLEILSQEVSTKELIE 197
+S+ S S ++ SS + S+ SSS SS S+ + +Q+ ++
Sbjct: 165 SSSSSSSSISSSSSSSSSSSSSSSSSSSSSSSSSTAAAAAAVAASVAAAAQQQQQQQQHS 224
Query: 198 SVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLSSSGRSAPS 239
S+ + ++SS ISS++S S S+SSS S+ S
Sbjct: 225 SISSSSSSSSISSSSSSSSISSSSSSSSSSSSISSSSSSSSS 266
>UniRef100_Q8LE56 Hypothetical protein At4g37240/C7A10_120 [Arabidopsis thaliana]
Length = 168
Score = 33.5 bits (75), Expect = 5.1
Identities = 29/107 (27%), Positives = 48/107 (44%), Gaps = 10/107 (9%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTN----DQSSKP 56
MG C ++ T K+I DG +MEF P+ V ++ ++ I ++ D +
Sbjct: 1 MGICSSSESTQVATA-KLILQDGRMMEFANPVKVGYVLLKYPMCFICNSDDMDFDDAVAA 59
Query: 57 LCQFDELVAGQSYYLLPMTVLSPNNKIDYPSTCATAET-----GGGG 98
+ +EL GQ Y+ LP+ L K + + A + GGGG
Sbjct: 60 ISADEELQLGQIYFALPLCWLRQPLKAEEMAALAVKASSALMRGGGG 106
>UniRef100_Q9VKJ1 CG4751-PA [Drosophila melanogaster]
Length = 1412
Score = 33.5 bits (75), Expect = 5.1
Identities = 54/225 (24%), Positives = 87/225 (38%), Gaps = 37/225 (16%)
Query: 45 AIFPT-NDQSSKPLCQFDELVAGQ---SYYLLPMTVLSPNNKIDYPST------CATAET 94
A PT ND + L Q +AGQ ++ + L + DY S+ A A+
Sbjct: 757 ACLPTANDLMAASLAQ----LAGQLPPNFLQGDLAALFQQQRKDYGSSSLNQLAAAAAKV 812
Query: 95 GGGGQIIRQGHVRSQSVPTTPLPAPYRMSLDYQYYQGVGLLKRTSTESFSCRTNRSS-VN 153
GG Q A Y S + GVG+ ++ + S ++S +
Sbjct: 813 GGSKQSNASASSGLNDPNVAAAVAAYSNSFNMPLPTGVGIGGSGNSNAHSSSKSKSERSS 872
Query: 154 KSTISSSRRSSRKSTGIWKVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVTAASSG 213
KS+ SSS +S ++ +K KL ++L E+ + + ELI S A A + G
Sbjct: 873 KSSSSSSSSNSSSTSNSYKTKLM---KELDELKNDPLKMSELIRSPEYAALLLQQAEALG 929
Query: 214 -------------------GCGISSTTSIVSDQWSLSSSGRSAPS 239
G G+ + ++ Q S SSS +S+ S
Sbjct: 930 ATTLGTLGFGSDYSYLTGAGLGVPAANALTGGQSSNSSSSKSSKS 974
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.130 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 407,937,886
Number of Sequences: 2790947
Number of extensions: 15953515
Number of successful extensions: 32607
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 32515
Number of HSP's gapped (non-prelim): 78
length of query: 248
length of database: 848,049,833
effective HSP length: 124
effective length of query: 124
effective length of database: 501,972,405
effective search space: 62244578220
effective search space used: 62244578220
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 73 (32.7 bits)
Medicago: description of AC146664.11


